Abstract
Risk for alcohol dependence in humans has substantial genetic contributions. Successful rodent models generally attempt to address only selected features of the human diagnosis. Most such models target the phenotype of oral administration of alcohol solutions, usually consumption of or preference for an alcohol solution versus water. Data from rats and mice for more than 50 years have shown genetic influences on preference drinking and related phenotypes. This paper summarizes some key findings from that extensive literature. Much has been learned, including the genomic location and possible identity of several genes influencing preference drinking. We report new information from congenic lines confirming QTLs for drinking on mouse chromosomes 2 and 9. There are many strengths of the various phenotypic assays used to study drinking, but there are also some weaknesses. One major weakness, the lack of drinking excessively enough to become intoxicated, has recently been addressed with a new genetic animal model, mouse lines selectively bred for their high and intoxicating blood alcohol levels after a limited period of drinking in the circadian dark. We report here results from a second replicate of that selection and compare them with the first replicate.
Keywords: Selected mouse lines, High Drinking in the Dark (HDID) mice, Ethanol preference, QTL, Review, Alcohol drinking
Introduction
An individual's risk for earning a diagnosis of dependence on alcohol (ethanol) has been shown over many years to reflect both genetic and environmental influences to an approximately equal degree (Enoch et al. 2003). Genetic risk is also clearly conditional upon multiple environmental circumstances, making it difficult to estimate any particular individual's specific risk (Johnson et al. 1998). Studies of gene-environment interaction are in theory much easier to perform with suitable animal models, where matings can be arranged and environmental control is relatively easy. So too, are studies whose goal is to identify any of the multiple genes affecting risk. The advantages of rodents for modeling alcohol dependence phenotypes were recognized in the late 1940s, when Jorge Mardones began to selectively breed rat lines with high (UChB) or low (UChA) relative preference for 10% alcohol solutions when water was freely available (Mardones and Segovia-Riquelme 1983). Over the course of several days, animals develop a stable and characteristic intake of ethanol, which can be indexed as either the relative preference for ethanol (the preference ratio, or PR, equals the percentage of total daily fluid consumed from the ethanol bottle) or as the dose of ethanol ingested, usually expressed as g ethanol/kg body weight per 24-h or during a limited time period per day. The latter index is preferable because it is relatively independent of the percentage of alcohol offered. The success of this first selection proved, ipso facto, that genes contribute to alcohol preference drinking, and the project spawned a large number of other selected lines of rats and mice bred for the same phenotypic difference in preference drinking.
The unique characteristics of inbred mouse strains for dissecting environmental from genetic influences were first exploited by Gerald McClearn and David Rodgers in 1959 when they showed that 5 inbred mouse strains differed in alcohol preference (McClearn and Rodgers 1959). This paper will reveal that Gerald McClearn posed many of the most important questions regarding the control of drinking in mice many years ago. As the Human Genome Project spurred the development of tools allowing the rapid mapping of genes that influence quantitative traits (QTLs), and molecular biological techniques allowed specific genes to be manipulated for over or under expression starting in the late 1980s, mice began their inexorable ascendance as the species of choice for attacking many questions in neuro-science, including the neuroscience of the addictions.
In this paper, we first sketch selected highlights gleaned from the 60 years of biomedical research into the genetics of alcohol drinking in rodents. We next review evidence on the genomic location of genes affecting ethanol preference drinking from QTL mapping studies in mice and rats, and present new information for two murine QTLs. Finally, we provide a progress report on a project creating a new mouse model for binge-like drinking, the High Drinking in the Dark (HDID) selected lines.
Alcohol preference drinking
Alcohol drinking is a prototypic complex genetic trait, one that is multigenic, oligogenic, and probably polygenic in nature. Several issues complicate our attempts to understand rodents’ relative avidity for drinking. One main issue is taste. Rodents cannot vomit, and have therefore, presumably through evolutionary pressures, developed exquisite sensitivity to novel flavors, and generally avoid them or approach them with caution. However, they greatly prefer sweet solutions, and the preference for sweet solutions is partially genetically influenced (Boughter Jr and Bachmanov 2007). The genetic contributions to preference for alcohol and sweet solutions are overlapping (Rodgers and McClearn 1964). Novelty per se is another factor that must be considered, as animals differ genetically in their response to novel situations (McClearn 1959). Learning may play a role in changes in consumption over time (Blizard et al. 2008), and ingestion of too much alcohol can lead animals to avoid drinking it when it is later offered again (Belknap et al. 1978). Also, alcohol may be ingested simply because it provides calories (Rodgers et al. 1963). In addition, the role of ethanol metabolism must be considered. Thus, although hamsters drink great quantities of alcohol in preference tests, their rate of metabolism is also extraordinarily high, so the blood alcohol levels they achieve are quite modest (Blum et al. 1982). Early studies addressing these issues were nearly all conducted in two inbred mouse strains, the highly preferring C57BL/6 strain and the avoiding DBA/2 strain, and have been reviewed many times [for an excellent discussion of the early work, see (McClearn 1968)].
The issues delineated above could be described as reflecting our uncertainty about what the preference drinking assay is modeling. Certainly it is not “alcoholism” in its entirety, for even long-term preference drinking does not generally lead to clear signs of withdrawal when ethanol is removed, and the drinking lacks the compulsive features consistent with human substance abuse. A more realistic assessment is that it may model selective features of the human disorder (McClearn 1979). But which? The human diagnosis is largely behavioral: 5 of the 7 symptoms that can contribute to a diagnosis of alcohol dependence are behavioral rather than strictly medical. Principal among them are such symptoms as the feeling of loss of control over drinking, the supplanting of normal activities or the destruction of family and career by drinking-related behaviors. None of these is convincingly modeled in rodents. Oddly, the diagnostic criteria for alcohol dependence in the USA do not include either the quantity or frequency of drinking (Hasin et al. 2006), but there is much current interest in incorporating these indices into the forthcoming new diagnostic scheme, DSM-V (Hasin and Beseler 2009). A crucial feature of preference drinking is that even high-drinking genotypes seem generally to limit their intake to blood alcohol levels below those leading to overt intoxication, quite unlike human alcoholics (Rhodes et al. 2005). Nevertheless, the work on preference drinking has provided many important insights.
Selective breeding
Bidirectional selection for high preferring vs low preferring rat and mouse lines has been conducted many times (see Table 1). Interestingly, nearly all of these studies have used almost exactly the same protocol for assigning individual phenotypic values to animals. In the rat selections, a period of a few days of forced exposure to alcohol is followed by up to 3 weeks of two-bottle preference for 10% ethanol vs water. The mouse selection studies have been conducted similarly, minus the forced exposure period. All such selections have succeeded, yielding divergent phenotypes for high and low preference drinking, and many have been replicated, following the recommendations of the authors of a selection project for activity in mice (DeFries et al. 1966). Replication has allowed us to have greater confidence that the results of these efforts are principally due to changes in the gene frequencies at trait-relevant loci rather than genetic drift due to the necessarily small population sizes. The main advantage of selective breeding is that any other traits that differ in the divergently selected lines are presumably due to pleiotropic influences of the genes affecting the selection phenotype. These are called genetically correlated responses to selection, and should offer clues to the neurobiology of the trait of interest. Discussions of the technical requirements for interpreting selection studies for behavioral traits have been published (Crabbe et al. 1990; Crabbe 1999; Henderson 1989).
Table 1.
Selectively bred and other genetic rat and mouse models of high or low alcohol drinking (includes high-drinking inbred and hybrid strains)
| Lines | Acronym | Phenotype | Reference | |
|---|---|---|---|---|
| Rats | University of Chile alcohol drinker and non-drinker (derived from Wistar rats; Chile) | UChB/UChA | High/low consumption 10% ethanol vs water | Quintanilla et al. (2006) |
| ALKO Alcohol and non-alcohol (derived from a foundation stock including Wistar and Sprague–Dawley rats; Lewis and Brown Norwegian genotype introduced later; Finland) | AA/ANA | High/low consumption and preference 10% ethanol vs water | Sommer et al. (2006) | |
| Alcohol preferring and nonpreferring (derived from Wistar rats; United States); inbred P, inbred NPa | P/NP iP/iNPa |
High/low consumption and preference 10% ethanol vs water | Bell et al. (2006) | |
| Sardinian alcohol preferring and nonpreferring (derived from Wistar rats; Italy) | sP/sNP | High/low consumption and preference 10% ethanol vs water | (Colombo et al. (2006) | |
| Marchigian Sardinian alcohol preferring (derived from 13th generation sP rats; Italy) | msP | High consumption and preference 10% ethanol vs water | Ciccocioppo et al. (2006) | |
| High/low alcohol drinking (derived from the N/Nih 8-way inbred strain cross; United States) | HAD-1/LAD-1 HAD-2/LAD-2 |
High/low consumption and preference for 10% ethanol vs water | Murphy et al. (2002) | |
| Fawn Hoodedb | FH/Wjdb | High consumption 10% ethanol vs water | Overstreet et al. (2007) | |
| High ethanol preferring (derived from a foundation stock created by crossing alcohol Preferring P rats and Harlan Sprague–Dawley; United States) | HEP | High consumption and preference for 9–20% ethanol vs water | Myers et al. (1998) | |
| High/low addiction research foundation (derived from the N/Nih 8-way inbred strain cross; United States) | HARF/LARF | High/low consumption 12% ethanol during a 20 min period of limited access | Le et al. (2001) | |
| Mice | Short-term high/low alcohol preference (derived from the F2 cross of C57BL/6J and DBA/2J inbred strains; United States) | High/Low | High/low preference 10% ethanol vs water | Belknap et al. (1997) |
| High/low alcohol consumption (derived from the HS/Ibg 8-way inbred strain cross; United States) | HAP-1/LAP-1 HAP-2/LAP-2 HAP-3/LAP-3 cHAPc (HAP-1 is extinct) |
High/low consumption 10% ethanol vs water | Green and Grahame (2008) | |
| Short-term high/low alcohol consumption (derived from the F2 cross of C57BL/6J and DBA/2J inbred strains; United States) | STDRHI/STDRLO | High/low consumption 10% ethanol vs water | Phillips et al. (2005) | |
| C57BL/6Jd | B6d | High consumption and preference 10% ethanol vs water | Wahlsten et al. (2006) | |
| (C57BL/6J X FVB/NJ)-F1e | B6FVB <mi></mi> | High consumption and preference 10% ethanol vs water | Blednov et al. (2005) | |
| High Drinking in the Darkf (derived from the HS/Npt 8-way inbred strain cross; United States) | HDID-1f HDID-2 |
High blood alcohol levels after drinking 20% ethanol in a single-bottle, limited access exposure | Crabbe et al. (2009) | |
| Short-term high/low alcohol consumption (derived from the HS4 4-way inbred strain cross; United States) | High/Low | High/Low consumption 10% ethanol vs water | Hitzemann et al. (2009) |
iP and iNP lines were inbred from P and NP, respectively, after many generations of selective breeding
FH/Wjd were inbred from one of the FH outbred stocks; several substrains of FH exist, but FH/Wjd show higher intake than FH/Har, FHH/Eur, and FHL/Eur
cHAP mice were produced by intercrossing HAP-1 and HAP-2 lines after many generations of selective breeding, and have since been under continued selection
This inbred, and most others from the C57/C58 lineage (Petkov et al. 2004), consistently tops the lists in comparisons of inbred strains for ethanol preference and consumption
This is the hybrid cross of the high ethanol intake strain, C57BL/6J, and a lower intake strain, FVB/NJ; the hybrid strain drinks more ethanol in the two bottle preference test than the C57BL/6J inbred, an example of heterosis
Selection is only in the direction of high drinking: the HS/Npt heterogeneous stock from which the lines were derived serves as the control population
Most of the work with these lines, covering many years, has been collated in a set of recent reviews (See Table 1) and one of us recently summarized that collection of reviews (Crabbe 2008). Some findings have emerged robustly across most or all pairs of selected lines. High drinking genotypes clearly have reduced serotonin function in limbic brain areas, accompanied by anatomical differences in this neurotransmitter system, but evidence for differences in several other transmitters have remained equivocal. Behaviorally, high drinkers consistently are shown to develop a greater degree of tolerance to alcohol's intoxicating effects and that tolerance is longer lasting. As serotonin is known to mediate some aspects of alcohol tolerance (Kalant 1998), these two results are consistent.
All the high drinking lines continue to drink until they reach a blood alcohol level of approximately 60–70 mg%. While this level is slightly above the 50 mg% criterion for driving under the influence in the USA, it is not sufficient to induce overt intoxication in rodents. Under some conditions (e.g., when alcohol is administered for months, or offered sporadically), some animals can be induced to drink excessive amounts. The effective procedures, however, are arduous (Rodd et al. 2009). The most effective method, and in our view the method with the most face validity, is to combine periods of free access to alcohol with periods where alcohol is forced upon the animals, usually through forced inhalation of alcohol vapor. Alternating these conditions can lead to a substantial increase in preference drinking, which has been proposed to be due to self-medication of an alcohol withdrawal state (Becker and Lopez 2004; Spanagel 2009; O'Dell et al. 2004).
Selective breeding has been an effective tool for detecting genetic correlations between different phenotypes. For example, Metten et al. (1998) reviewed evidence from several studies showing that alcohol preference drinking and withdrawal severity in physically dependent mice are inversely genetically correlated: thus, high preferring genotypes are genetically predisposed to show low withdrawal severity and vice versa. Precisely which genes are responsible for the negative genetic correlation are unknown. Using short-term selective breeding for each of the two traits starting from a heterogeneous stock (derived from a cross of four inbred strains, including C57BL/6J and DBA/2J), Hitzemann et al. (2009) found seven QTLs that affected both traits in the expected inverse direction, and offered a number of plausible candidate genes worthy of further investigation.
Inbred strains
The first demonstration of inbred strain differences in preference drinking in 1959 compared male mice from 5 strains. The data revealed several more subtle differences as well. Within each strain, there were apparent individual differences among animals’ preference ratios. For example, of the four mice in the C3H/NT strain, two were relatively high drinkers and two relatively low, so this strain's “average” drinking had relatively high variability. For preferring strains, preference tended to increase over a period of 2–3 weeks until it settled at a plateau. Whenever the side on which the alcohol bottle was placed on the cage top was switched, there was a temporary reduction in intake of alcohol during the following 1–2 days, but animals quickly reestablished their strain-specific preference ratio (McClearn and Rodgers 1959). Many subsequent studies, published between 1966 and 2004, have employed relatively large panels of inbred strains. A recent review of several of those studies was able to compare relative strain preference ratios for 10% ethanol across decades, laboratories, and small variations in the procedures employed. Despite the inevitable genetic drift due to differential new mutations within strains over more than 150 generations, the correlations of strain mean preference ratios exceeded r = 0.90 for any pairwise correlation between data sets. Not only is this a highly heritable trait, it is highly genetically stable over time and laboratories (Wahlsten et al. 2006). As this animal model represents an aspect of the human disease, these results indicate that the genetic contribution to quantity of alcohol intake is profoundly important and supports further consideration of including this index in the scheme used to diagnose human alcohol use disorders.
The common inbred mouse strains have been assigned to seven lineages based on detailed pedigree records and, more recently, extensive single nucleotide polymorphism comparisons (Petkov et al. 2004). Strains from the C57/C58 lineage are generally among the highest drinkers, while those related to the DBA lineage are low preferring (Wahlsten et al. 2006). Recently, it was fortuitously discovered by Yuri Blednov that an F1 hybrid between the C57BL/6J and FVB/NJ inbred strains drank even more ethanol than C57BL/6J mice in standard 2-bottle preference tests (Blednov et al. 2005). Other strains in the FVB (“Swiss”) lineage show wide variability in drinking—for example, RIIIs/J are above average drinkers, while BUB/BnJ are below average (Yoneyama et al. 2008). This result suggests a role for epistatic interactions between alleles common in the C57/C58 lineage and those common in the Swiss lineage. We are currently mapping QTLs responsible for the excessive drinking of mice that are heterozygous for the C57BL/6J and FVB/NJ alleles when compared to those that are homozygous for one or the other allele. To do this, the genome was searched for individual QTLs showing overdominance in a C57BL/6J X FVB/NJ F2 population. Our initial results have identified three QTL regions showing overdominance in the expected direction (Phillips et al. 2010).
The utility of inbred strains for genetic correlational inferences (see “The Complexity of Preference Drinking: Multivariate Approaches” section) has taken great strides forward with the formation of a Mouse Phenome Database at The Jackson Laboratory (Grubb et al. 2004). This informatics project had 2081 phenotypes stored for 4 or more common inbred strains (average = 19 strains/phenotype) on June 3, 2009. In the alcohol field, strain mean correlations between preference drinking and two other phenotypes have prompted further studies. Inbred strains that have high alcohol preference also are genetically predisposed to have low withdrawal severity after being rendered physically dependent on alcohol (Metten et al. 1998), which supports the numerous studies reviewed above with lines of mice or rats selected for high vs low withdrawal severity or high vs low preference and tested on the alternate trait.
The complexity of preference drinking: multivariate approaches
The wealth of data on inbred strains in particular has enabled the use of multivariate approaches to understanding drinking (see Phillips and Belknap 2002). Trait (phenotype) data collected on a dozen or more inbred strains are valuable for exploring common genetic influences among multiple traits by correlational and multivariate analyses. Parallel patterns of strain differences among traits indicate common genetic influences which can implicate common genetically-mediated mechanisms for further investigation. In a recent meta-analysis (Belknap et al. 2008), we analyzed genetic commonalities across four different drugs subject to abuse (ethanol, morphine, pentobarbital, and diazepam) tested on 14–15 standard inbred mouse strains for a series of traits appropriate for all four drugs across multiple doses. A correlation matrix based on strain means was first constructed (bivariate analysis), followed by multivariate analyses based on clusters of correlated traits as the unit of analysis. Multidimensional scaling and cluster analysis showed that for two-bottle choice preference drinking, morphine and ethanol were similar in their pattern of strain differences, but different from diazepam and pentobarbital. This finding was not anticipated based on the pharmacological classes to which these drugs belong (pharmacology alone would have predicted similarity of genetic control across the three drugs sharing strong effects on GABA-A receptors, i.e., ethanol, diazepam and pentobarbital). Data from the literature for a series of sweet and bitter tastants without known pharmacological effects were also examined. Preference for the tastants was similar to drug preferences only in the case of ethanol, where the sweet tastants (sucrose, saccharin) were significantly genetically similar to ethanol preference drinking across strains. This trend was strongest for the 3% ethanol concentration, and less evident for 6 and 10% ethanol. This is consistent with the hypothesis that sweet taste preference drinking partially contributes to ethanol preference drinking in mice, especially at the lower ethanol concentrations (Belknap et al. 1993; Boughter and Bachmanov 2007).
Gene targeting
In 1996, the first gene-targeted mouse to be tested for alcohol preference drinking was found to drink more than it's wild-type counterpart, suggesting that the serotonin 1B receptor gene might play a role in preference drinking (Crabbe et al. 1996). However, several subsequent studies showed that this single gene effect was strongly dependent on the genetic background of the mice, and we have not to date been able to ascertain the physiological basis of the role of this gene in drinking (Phillips et al. 1999; Phillips and Belknap 2002; Crabbe et al. 1999). Since then, many additional over expression or null mutants have been created and tested for alcohol preference drinking. In 2006, we reviewed results for 76 targeted genes (Crabbe et al. 2006). Since that review, mutants for at least 10 additional genes have been tested. The review covered standard gene knockouts, over expression transgenics, and targeted point mutations, most expressed throughout development but some altered conditionally or in a brain region specific manner. These various manipulations of 23 genes increased preference drinking significantly; for 30 genes, preference was significantly reduced; and for 33 genes, no substantial difference from controls was seen. Analysis of this distribution yielded χ2 = 1.84, p = 0.4 (NS). Our interpretation of this pattern is that while a specific gene or gene pathway may well affect preference drinking when looked at in isolation, there are so many potentially influential genes that it is not easy to see how this oligogenic trait will yield its secrets to a one gene at a time analysis.
Stepping back from the big picture, how is one to determine where the signal resides, and what is noise in the system? One good approach is to explore results suggested in null mutant studies with further investigations using drugs and other research tools. For example, several different lines of mice engineered to lack the CB1 receptor gene showed reduced alcohol preference, and null mutants for the FAAH gene that had increased endogenous cannabinoids showed elevated preference. The potential role of the cannabinoid system was also supported by studies with CB1 receptor antagonist drugs, and studies with AA and ANA selected rat lines. The cannabinoid antagonist rimonibant is currently under investigation as a potential therapeutic agent for alcohol dependence (Crabbe et al. 2006).
The search for QTLs for alcohol consumption
A mapped QTL defines the location of a gene (or more than one gene) that influences a complex trait (i.e., one that is influenced by multiple genes, by environmental factors and can be influenced by all possible interactions of these variables). The principles and complexities of QTL mapping, and the importance of a systems genetics approach, have been recently presented in an excellent review (Mackay et al. 2009). Gerald McClearn was among the first to consider gene–gene interaction effects on alcohol preference drinking in mice as one of the complicating factors in identifying genetic influences (Fernandez et al. 2000). QTL mapping results for murine ethanol consumption were examined for replicability using meta-analysis in 2001 (Belknap and Atkins 2001). The analysis included only QTL studies that used populations derived from the C57BL/6J and DBA/2J inbred strains. These strains are typically at the top and bottom, respectively, of lists comparing inbred strains for voluntary alcohol consumption/preference (Belknap et al. 1993; Wahlsten et al. 2006; Yoneyama et al. 2008). In all 9 studies, data were available that compared the consumption of 10% ethanol when it was offered versus water. This analysis provided the strongest support for QTLs on mouse chromosomes 2, 3, 4 and 9 (Belknap and Atkins 2001). Since then, additional evidence has been provided supporting the existence of QTLs in some of the same regions, particularly chromosomes 2 (Fehr et al. 2005) and 9 (Hitzemann et al. 2004).
Other mouse genotype combinations have also been used to map QTLs for ethanol consumption and have provided strong or suggestive evidence for the same regions and for additional regions (Bachmanov et al. 2002; Bice et al. 2006; Gill and Deitrich 1998; Gill and Boyle 2005). There has also been QTL mapping accomplished for ethanol consumption using rats (Bice et al. 1998; Carr et al. 1998, 2003; Foroud et al. 2003; Terenina-Rigaldie et al. 2003b). In one study, following a QTL search in an F2 cross of low (Wistar-Kyoto; WKY) and high (High-Ethanol Preferring; HEP) alcohol consuming lines, a region of rat chromosome 4 was genotyped and rats with alleles from the HEP or WKY lines were each interbred. Analysis of offspring ethanol drinking data confirmed the presence of a QTL that influences ethanol consumption on rat chromosome 4. The same or a linked gene(s) in the same region also influenced saccharin and quinine intake, such that rats with the high ethanol preference genotype consumed more of both tastants (Terenina-Rigaldie et al. 2003a). A chromosome 4 QTL was also found in a study using the intercross of the selectively bred preferring (P) and non-preferring (NP) rat lines (Carr et al. 1998) and following further exploration, genes in the region including neuropeptide Y (Npy), α-synuclein (Snca), and corticotrophin-releasing hormone receptor 2 (Crhr2) have been suggested as candidates for the linkage signal (Spence et al. 2009).
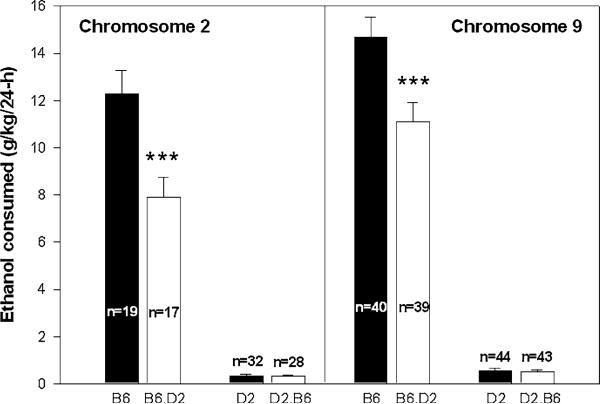
One strategy for QTL confirmation uses congenic mice that are entirely C57BL/6J in genotype with the exception of a region on a single chromosome from the DBA/2J genotype or vice versa. This approach was used by Blizard and McClearn (Blizard and McClearn 2000) to substantiate the genetic association of ethanol and sucrose intake. We have tested two chromosome 2 congenics vs their matched C57BL/6J and DBA/2J background mice. The background strain mice were originally obtained from The Jackson Laboratory, and then used for the backcrossing required to create congenics (Bennett 2000), and bred alongside the congenics during the same time span. Adult mice were tested in a 24-h choice procedure for their consumption and preference for a 10% (v/v) ethanol solution when offered vs water, using methods for measuring consumption that are standard in our laboratory (Phillips et al. 2005). The 10% ethanol solution was offered for 4–10 days. As shown in the left panel of Fig. 1, chromosome 2 congenic data supported QTL capture on the C57BL/6J background (introgression of the DBA/2J region reduced ethanol consumption), but not on the DBA/2J background (introgression of the C57BL/6J region did not significantly increase ethanol consumption). Similar results were obtained in chromosome 9 congenic mice (Fig. 1, right panel). In both cases, the data suggest that a single QTL region was unable to alter the ethanol drinking phenotype on a genetic background that shows extreme avoidance of ethanol.
Fig. 1.
Introgression of a genetic DBA/2J (D2) strain segment onto a pure C57BL/6J strain background reduces ethanol consumption. Shown is mean SEM consumption of 10% ethanol in Chromosome 2 (left) or Chromosome 9 (right) congenic and background strain mice. Introgression of a B6 segment on the alcohol avoiding D2 background did not significantly alter ethanol consumption. Congenic interval: B6.D2 Chromosome 2: 10–86 cM; D2.B6 Chromosome 2: 37–71 cM; both B6.D2 and D2.B6 Chromosome 9: 9–58 cM. *** p < 0.01 for the comparison of B6 to B6.D2 congenic mice
Others using the congenic approach have consistently created congenics using C57BL/6J or another C57 substrain as the background strain. For example, Whatley et al. used a panel of 21 C57BL/6J background congenics with DBA/2J substituted regions and confirmed QTLs for 10% ethanol consumption on chromosomes 1 and 2 (Whatley et al. 1999). Gill and Boyle used a panel of 20 C57BL/6J congenic strains with substituted genetic regions of various lengths from the A/J strain to confirm provisional QTLs for 10% ethanol consumption on chromosomes 2 and 15 (Gill and Boyle 2005). Lesscher et al. also confirmed a chromosome 2 QTL using an A/J substituted region for a limited access (rather than 24-h access) ethanol drinking trait (Lesscher et al. 2009). Finally, C57BL/6ByJ quasi-congenics [these mice carry genetic passenger regions in addition to the introgressed segment; see (Bennett 2000)] with substituted regions from BALB/cJ and CXBI/ByJ strains were used to provide evidence for QTLs on chromosome 6 and 12 for the consumption of a 12% ethanol solution offered vs water (Vadasz et al. 2007).
While the original QTL mapping work was nearly all based on linkage and association analysis of numerous polymorphisms across the genome, the use of DNA chips and microarray analyses to identify QTLs was soon adopted. Differences in gene expression have therefore also helped to identify the specific genes in these and other regions in both mice and rats that may be responsible for differences in ethanol consumption (Carr et al. 2007; Hitzemann et al. 2004; Mulligan et al. 2006; Tabakoff et al. 2008; Treadwell et al. 2004; Weng et al. 2009a, b; Worst et al. 2005). Gene sequence data have also been examined (Fehr et al. 2005; Boyle and Gill 2008). To date, none of the evidence can be considered definitive with regard to concluding that a gene accounting for differences in ethanol consumption has been identified. However, compelling candidates include syntaxin binding protein 1 (Stxbp1) on chromosome 2 and sodium channel, type IV, (Scn4b) on chromosome 9, among many others, including some mentioned above. A significant development corresponding with the ability to examine the expression of thousands of genes at a single time is the move toward more comprehensive analyses of gene networks (Mulligan et al. 2006; Green et al. 2007; Kerns and Miles 2008; Song et al. 2009). Genetic perturbation of interconnected pathways regulated by multiple genes is more likely than are large major gene effects to explain genetic components for complex alcohol use disorders and even seemingly simpler traits used to model aspects of excessive drinking.
Beyond alcohol preference drinking
Strengths and weaknesses of the two-bottle test
We have learned a great deal about ethanol preference drinking from the genetic animal model literature. Yet, as the field of the biology of alcoholism has co-evolved with that specialized in alcohol drinking studies, we must ask, what are the principal strengths and weaknesses of the phenotype, preference drinking, vis à vis compulsive human drinking? One approach to asking this question was taken by a review of the multitude of data from inbred strains and from lines selected for preference. Nearly all the selected lines were selected for essentially the same trait, two-bottle preference drinking. These authors asked whether the patterns of correlated responses for other measures of ethanol reinforcement seen across the divergently-selected drinking lines were similar. For example, did high preference-drinking genotypes also show a greater sensitivity than low drinking genotypes to alcohol's stimulus properties in conditioning paradigms, or would they work harder in an operant task to obtain access to alcohol (Green and Grahame 2008)? It is reassuring to find that the animals with high preference genotypes indeed would work harder in oral operant self-administration studies. Another commonly employed behavioral assay depends on Pavlovian conditioning. When an alcohol injection is paired with drinking a novel fluid, animals will sometimes gradually develop an aversion for the novel fluid. This conditioned taste aversion is taken as an index of ethanol's hedonic stimulus properties, and sensitivity to this effect of alcohol has been shown by some to correspond with avoidance of alcohol intake in low alcohol consuming lines. Thus, aversive effects of relatively high doses of alcohol may tend to limit consumption to a greater extent in non-preferring genotypes. Finally, if alcohol injections are paired with a novel location and the animal is subsequently given a choice of locations, it may show a conditioned place preference, and high preference drinking genotypes were more likely to do so than non-preferring genotypes. Overall, these results suggest that preference drinking is a reasonable model of ethanol's reinforcing effects (Green and Grahame 2008).
Among the other strengths of the two-bottle preference drinking test is the fact that it is very easy to implement. We routinely have high school students performing this test within 2 weeks of their entering our laboratories for summer research experiences. It is also possible to achieve very high throughput when collecting data, which is an enormous advantage for complex genetics studies. As we have mentioned above, we have nearly 60 years of rat data, and 50 of mouse data. The existence of numerous selectively bred lines has allowed us to achieve convergent validity for many correlates of preference drinking. The trait has a relatively high heritability compared to other behaviors, which also has facilitated genetic studies, and some QTLs have been found reliably across time and laboratories.
However, like all methods, there are some inherent weaknesses, or at least limitations to the method's application. There is limited to no predictive validity for drugs with clinical efficacy at treating alcoholism (Egli 2005). This is principally because many compounds are “false-positives” in the two-bottle test, reducing preference drinking but subsequently proving to have limited to no efficacy in tests with human alcohol-abusing populations. Even after all these years, we still do not know precisely what the motivation for drinking is. Why do some animals prefer and others avoid? Although the trait is a reasonable surrogate for alcohol's reinforcing properties, this is not the same as knowing that this is why animals elect to drink. Our inability to determine motivation definitively is compounded by the strong innate tendencies regarding novel flavors, and novel situations in general. As reviewed above, the genetics of alcohol preference is clearly intertwined with genetic approach and avoidance tendencies for other tastants. However, the principal limitation of two-bottle preference drinking in our opinion is that under most circumstances, even high preferring genotypes will not drink enough to become overtly intoxicated. For mice, it requires a blood ethanol concentration (BEC) of approximately 100 mg% before behavioral intoxication can be demonstrated unequivocally (Crabbe et al. 2005, 2008). There are numerous methods that can be employed to elevate voluntary drinking to a degree that this threshold is surpassed. However, they require very long-term access to alcohol solutions (e.g., months), generally employ periods of deprivation alternated with access, and/or require extensive training. Alternatively, if alcohol is sweetened, animals will readily drink to intoxication, but the specific role alcohol plays in directing their drinking is questionable. Many of these alternative methods are described in a paper that first reported the development of a new model, “Drinking in the Dark” or DID (Rhodes et al. 2005).
Drinking in the Dark—a new genetic animal model
If mice are offered a single bottle of a high concentration of alcohol (20%) for 2–4 h starting soon after the onset of their circadian dark cycle, some will drink a great deal of alcohol and achieve measurable BECs (Rhodes et al. 2005). Strains differ markedly in DID, with C57BL/6J drinking the most and DBA/2J among the least; the trait is heritable, as inbred strains differ markedly in their BECs after a DID test (Rhodes et al. 2007). The DID phenotype, and the selected lines developed based upon high BECs during this test (see next section), are not intended as an animal model of alcoholism. Rather, we have targeted a single deficiency of the older models, i.e., the failure to show a pattern of binge-like drinking that leads to intoxication. A principal limitation of the phenotype is that during the DID test, no choice is offered—the animals, for a 2 or 4 h period, have only access to ethanol. While they do not have to drink in order to maintain a normal physiological state [i.e., they are not fluid-deprived (Toth and Gardiner 2000)], the face validity of the model would be improved if a choice were offered. However, in the F2 cross of C57BL/6J and DBA/2J, ethanol intake was reduced when water was offered as an alternative drinking solution (Phillips et al. 2010). Todd Thiele's group has performed studies in C57BL/6J mice where periods of food deprivation were shown not to affect intake or BEC in DID drinking (Lyons et al. 2008). Peripheral administration of the anorectic agent leptin had no effect on DID intake or BECs, though separate tests showed its effectiveness in reducing food intake. Ghrelin similarly enhanced eating, but showed only a modest tendency to increase alcohol intake/BEC. The alcohol drinking experiments were conducted with food removed. These studies suggest that the DID phenotype is not primarily related to feeding behavior, or at least to calorie-seeking (Lyons et al. 2008). In another application of the method, Boehm et al. have used the DID procedure to offer high ethanol concentrations to pregnant C57BL/6J mice in an attempt to model binge-like consumption (Boehm et al. 2008). These animals reached BECs of 118–182 mg%, sufficient to produce behavioral changes in adolescent offspring. The authors suggest that the procedure may be useful for animal model studies of Fetal Alcohol Spectrum Disorders.
Selection for high blood ethanol concentrations
To provide a new model of binge-like drinking, we bred a mouse line for high blood ethanol levels due to High DID (HDID), starting with a large population of a genetically heterogeneous stock, HS/Npt, derived from intercrossing 8 standard inbred strains. Each generation, mice were given a 2 day DID test, with access to 20% ethanol (v/v) in a single bottle for two hr on the first day, and four hr on the second day, starting 3 h after the onset of their circadian dark. Each generation, selection of breeders was based on their BEC at the end of the second day of drinking, and mice with the highest BECs were mated to produce the next generation. We recently reported the results from the first replicate line (HDID-1) for this selection project (Crabbe et al. 2009). When compared with mice offered water, the HDID-1 mice were shown to be intoxicated in the balance beam test. The heritability estimated from the realized response to selection after 11 selected generations was h2 = 0.09, and the relatively slow rate of response suggests that the trait is multigenic and perhaps highly polygenic (influenced by genes that each have a small effect on the phenotype). Mice achieved higher BECs than the HS/Npt population from which they were selected—this stock serves as the control line, as this selection was not bidirectional.
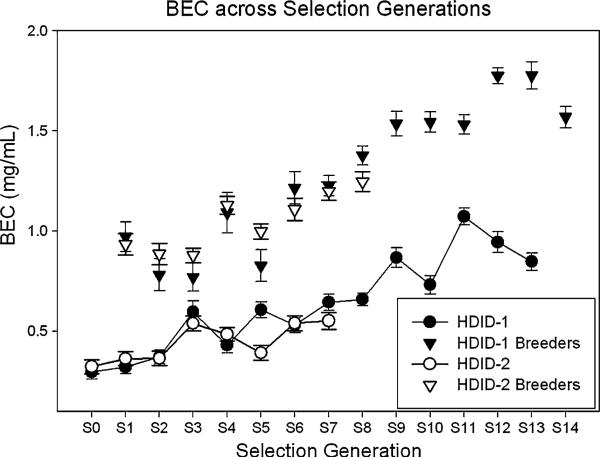
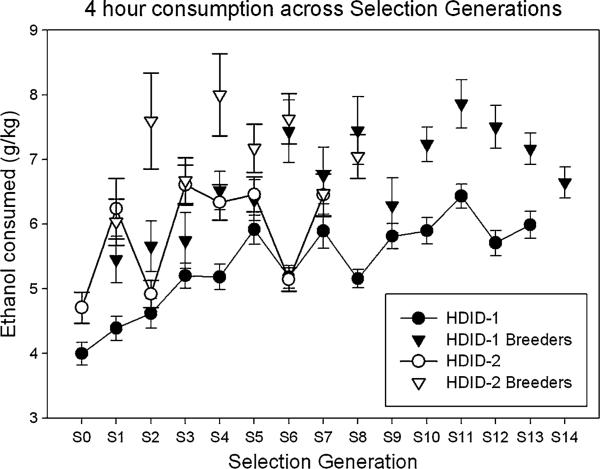
To provide a replicate that could be used to verify any correlated responses to selection, we initiated a second line. The HDID-2 line was selected in the same way, for the same trait. The only exception was that we started the HDID-1 selection using a rotational breeding scheme (Halcomb et al. 1975), and then to accelerate the rate of response, we shifted to individual selection after 5 generations. The HDID-2 line was subjected to individual selection throughout. Figure 2 shows the response to selection in both replicates. The HDID-2 selection response appears to resemble the response in HDID-1 very closely. For example, the S7 HDID-2 mean BEC was 0.55 ± 0.04 mg/ml, and the S7 HDID-1 mean was 0.64 ± 0.04 mg/ml. In both lines, the animals achieved higher BECs by ingesting more alcohol. Although selection was based only on BEC and not on intake, the intakes have increased as a genetically correlated response to selection (Fig. 3). The S7 HDID-2 mean ethanol intake was 6.45 ± 0.33 g/kg, while that for the S7 HDID-1 mean was 5.89 ± 0.26 g/kg. That mice achieving higher BECs drank more makes logical sense. That there was a smaller increase in intake than there was in BEC over generations is consistent with there being partial, but not total, overlap between the pools of genes affecting the two traits. Within genetically homogeneous C57BL/6J mice, the correlation (which is traditionally taken as environmental in origin) between intake and BEC was r = 0.71 (Rhodes et al. 2005). Within HDID-1 mice of the S11 generation, where the correlation represents a mix of genetic and environmental influences depleted to the extent that selection has fixed some relevant genes, we found r = 0.45 (Crabbe et al. 2009). In the foundation population (S0) of HS/Npt animals, the correlation was r = 0.44 (unpublished data). We believe that across populations and estimates, intake typically explains about 25% of BEC variation, whether considered phenotypically or ‘genetically’ [i.e., from inbred strain means (Crabbe et al. 2009)].
Fig. 2.
Realized response to selection for high blood ethanol concentration (BEC) in High Drinking in the Dark (HDID-1 and HDID-2) mice. Response is shown across 13 selected generations for HDID-1 mice and 7 generations for HDID-2 mice. Mean ± SEM BEC is shown. Circles represent the total population tested each generation. Inverted triangles give values of the animals chosen as parents from the preceding generation: their offspring are represented in circles directly below. For a detailed discussion of the first 11 generations of the HDID-1 selection, as well as the specific methods used, see (Crabbe et al. 2009)
Fig. 3.
Increased consumption in the HDID-1 and HDID-2 selected lines across generations. Mean ± SEM g kg ethanol ingested during the second day of DID testing is shown. Intake is a correlated response to selection on the BEC levels depicted in Fig. 2. In HDID-1 mice, as BEC tripled across the first 11 generations, intake increased by 50%
After 9 generations of selection for high DID, the HDID-1 mice were tested in a standard DID test, except they were offered two bottles, one containing water and one containing alcohol. They drank more alcohol than controls (and, in fact, nearly the amount they usually drank in a single-bottle DID test), but they also drank some water, and their BECs were significantly lower (Crabbe et al. 2009). We have not yet tested the HDID-2 line for two-bottle DID, or for intoxication after drinking. Because selection is unidirectional, we are dependent upon finding differences between the HDID lines and the non-selected control for concluding that there are correlated response differences for a given trait. Control line values can differ considerably depending on the phenotype, and genetic confounds due to accidental fixation of genes in the control lines can lead to false positive or false negative conclusions. One advantage of finding that the HDID-2 line is showing increasing BECs at nearly exactly the same rate as the HDID-1 line is that we can perform correlated response tests using all three lines simultaneously, while predicting the rank order of differences among lines (HDID-1 ≥ HDID-2 > Control, for example).
Pharmacological modulation of drinking in the dark
The DID paradigm has been used in several laboratories to explore pharmacological control of this novel drinking paradigm. To date, most such studies have employed peripheral administration of compounds to C57BL/6J mice, typically before the second day of DID testing. Studies have shown that the mu opioid antagonist naltrexone and the dopamine transporter antagonist GBR 12909 both reduced DID intake of ethanol at doses without effect on intake of plain water (Kamdar et al. 2007). GBR, but not naltrexone, also reduced sweetened water intake. A subsequent study showed that acamprosate, which blocks NMDA and/or metabotropic glutamate receptors, and the specific mGluR5 receptor antagonist MPEP both reduced DID ethanol drinking. Neither compound affected water or sugar water drinking at the doses tested (Gupta et al. 2008). The GABA-B agonist baclofen dose-dependently increased binge-like ethanol intake, without affecting water drinking, while both the GABA-A agonists muscimol and THIP reduced ethanol intake, but also water (Moore et al. 2007). Nicotinic acetylcholine receptor function may also be involved as shown in a recent study using mecamylamine, hexamethonium, dihydro-beta-erythroidine (DHβE), methyllycaconitine (MLA), nicotine and cytisine in a modified DID assay (Hendrickson et al. 2009). The non-specific antagonist mecamylamine reduced ethanol intake and lowered BECs, while the peripheral antagonist hexamethonium did not. Neither the specific competitive nicotinic antagonist DHβE nor the α7 selective antagonist MLA reduced alcohol intake. Nicotine, and cytisine, the β4* receptor full antagonist and β2 partial agonist, also reduced ethanol drinking. Some of these treatments had modest effects on sucrose drinking as well. These authors also performed a rough analysis of drinking patterns, averaging intake into 15 min bins. Nicotine pretreatment predominantly reduced ethanol drinking during the first hour, while mecamylamine affected drinking during the second hour. cFos/tyrosine hydroxylase double labeling experiments suggested that mecamylamine pretreatment before ethanol DID drinking reduced the number of double-labeled cells in the ventral tegmental area while nicotine pretreatment did not (Hendrickson et al. 2009). In addition, one study has shown that the CRF1 antagonist CP-154,526 reduced ethanol intake and BEC during 4 h DID sessions that resulted in relatively high intake and BECs, but was without effect in shorter sessions leading to lower intakes/BECs (Sparta et al. 2008).
We are aware of only three studies to date that have employed a modification of the DID model to deliver a compound directly to a specific brain nucleus in C57BL/6J mice. Infusions of the neuropeptide urocortin 1 (Ucn1) into the lateral septum altered the acquisition and expression of ethanol intake with the DID procedure (Ryabinin et al. 2008). The GABA-B receptor agonist baclofen reduced DID when infused into the anterior, but not the posterior, ventral tegmental area. Infusion did not affect drinking of either water or sugar water (Moore and Boehm 2009). More complex results were seen using the cannabinoid receptor agonist WIN 55-212,2. This compound increased DID drinking at the lowest dose, but suppressed it at two higher doses, but only if delivered to the posterior ventral tegmental area; all doses were ineffective in the anterior ventral tegmental area. These findings were complicated by effects on locomotor activity in some conditions, which may have competed with the consummatory drinking (Linsenbardt and Boehm 2009). In summary, the DID model has been used by several investigators, and a wide range of neural systems have been provisionally implicated by the pharmacological studies performed thus far.
Future directions
The tendency of some rodents to prefer and others to avoid alcohol solutions has provided the cornerstone for behavioral neuroscience research into alcohol use disorders since its discovery more than 60 years ago. The early demonstration that those differences were in great part genetically based has led to the prominent role of behavioral genetics in preclinical research on alcohol. As genetics, genomics, and bioinformatics tools proliferated and grew markedly in sophistication, we have made substantial progress toward identifying some of the genes responsible for differential avidity for alcohol solutions. This has led to pursuit of some novel new drugs with potential therapeutic value. It has also become abundantly clear from studies of genes and their expression patterns how complex the genetic contributions to drinking are. Curiously, the phenotypes studied in rodents remain much the same as they were at the outset. In part, this is because alcoholism is a developmental, chronically relapsing disorder, and it is both difficult and expensive to model in the laboratory across the life span, even in rodents that typically live only 2–3 years. Also, we believe that this is due to a disconnection between the phenotypic targets chosen for genetic studies by clinical and preclinical researchers. The former tend to concentrate on diagnostic categories, while the latter can provide believable models for only selected features of clinically-relevant symptom clusters. Some of the crucial features of alcohol use disorders (e.g., the tendency for preoccupation with the drug to interfere with work or peer relationships) are unlikely to be modeled realistically in laboratory rodents.
Several investigators recently made an effort to review the state of the art in human and animal research across many domains important for alcohol use disorder epidemiology, pathophysiology and treatment (Crabbe 2010). One domain considered was the assessment of ethanol consumption (Leeman et al. 2010). The work group concluded that three categories could encompass most of the relevant phenotypes: abstinence (or the decision to drink or not to drink); the amount consumed; and heavy drinking. Laboratory studies of the decision making process are sparse in both humans and laboratory animals. Human studies were encouraged that consider excessive drinking according to standard guidelines (reaching > 80 mg% within 2 h), and the DID model is a promising start in rodents. More attention was encouraged in all species to studies of the patterning of drinking both within a drinking session, with respect to circadian time, and across drinking sessions. Finally, the need for measuring BECs in all studies was emphasized (Leeman et al. 2010). Although alcohol consumption is only one area of importance for understanding and treating alcohol use disorders, it is one where the field of alcohol research offers a strong opportunity for clinical and preclinical approaches to find common ground in genetics. The genetics traditions at both levels of investigation are strongly historically based, and the target population that would benefit from biomedical progress is very large.
Acknowledgments
This paper is based on a presentation at a festschrift in honor of Gerald E. McClearn at The Pennsylvania State University, May, 2009. Preparation of this manuscript was supported by three grants from the US Department of Veterans Affairs; by NIAAA Grants AA10760, AA06243, and AA016655. The HDID selection is supported by Integrative Neuroscience Initiative on Alcoholism-West consortium Grant AA013519. We thank Lauren Brown and Pamela Metten for assistance with the breeding of HDID mice. We thank Sue Burkhart-Kasch and Jeanna Wheeler for assistance with congenic study data collection.
Contributor Information
John C. Crabbe, Portland Alcohol Research Center, Department of Behavioral Neuroscience, Oregon Health & Science University, Portland, OR 97239, USA VA Medical Center (R&D 12), Portland, OR 97239, USA.
Tamara J. Phillips, Portland Alcohol Research Center, Department of Behavioral Neuroscience, Oregon Health & Science University, Portland, OR 97239, USA VA Medical Center (R&D 12), Portland, OR 97239, USA.
John K. Belknap, Portland Alcohol Research Center, Department of Behavioral Neuroscience, Oregon Health & Science University, Portland, OR 97239, USA VA Medical Center (R&D 12), Portland, OR 97239, USA.
References
- Bachmanov AA, Reed DR, Li X, Li S, Beauchamp GK, Tordoff MG. Voluntary ethanol consumption by mice: genome-wide analysis of quantitative trait loci and their interactions in a C57BL/6ByJ × 129P3/J F2 intercross. Genome Res. 2002;12:1257–1268. doi: 10.1101/gr.129702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HC, Lopez MF. Increased ethanol drinking after repeated chronic ethanol exposure and withdrawal experience in C57BL/6 mice. Alcohol Clin Exp Res. 2004;28:1829–1838. doi: 10.1097/01.alc.0000149977.95306.3a. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Atkins AL. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mamm Genome. 2001;12:893–899. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Coleman RR, Foster K. Alcohol consumption and sensory threshold differences between C57BL/6J and DBA/2J mice. Physiol Psychol. 1978;6:71–74. [Google Scholar]
- Belknap JK, Crabbe JC, Young ER. Voluntary consumption of ethanol in 15 inbred mouse strains. Psychopharmacology. 1993;112:503–510. doi: 10.1007/BF02244901. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Richards SP, O'Toole LA, Helms ML, Phillips TJ. Short-term selective breeding as a tool for QTL mapping: ethanol preference drinking in mice. Behav Genet. 1997;27:55–66. doi: 10.1023/a:1025615409383. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Metten P, Beckley EH, Crabbe JC. Multivariate analyses reveal common and drug-specific genetic influences on responses to four drugs of abuse. Trends Pharmacol Sci. 2008;29:537–543. doi: 10.1016/j.tips.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RL, Rodd ZA, Lumeng L, Murphy JM, McBride WJ. The alcohol-preferring P rat and animal models of excessive alcohol drinking. Addict Biol. 2006;11:270–288. doi: 10.1111/j.1369-1600.2005.00029.x. [DOI] [PubMed] [Google Scholar]
- Bennett B. Congenic strains developed for alcohol- and drug-related phenotypes. Pharmacol Biochem Behav. 2000;67:671–681. doi: 10.1016/s0091-3057(00)00412-3. [DOI] [PubMed] [Google Scholar]
- Bice P, Foroud T, Bo R, Castelluccio P, Lumeng L, Li T-K, Carr LG. Genomic screen for QTLs underlying alcohol consumption in the P and NP rat lines. Mamm Genome. 1998;9:949–955. doi: 10.1007/s003359900905. [DOI] [PubMed] [Google Scholar]
- Bice PJ, Foroud T, Carr LG, Zhang L, Liu L, Grahame NJ, Lumeng L, Li T-K, Belknap JK. Identification of QTLs influencing alcohol preference in the High Alcohol Preferring (HAP) and Low Alcohol Preferring (LAP) mouse lines. Behav Genet. 2006;36:248–260. doi: 10.1007/s10519-005-9019-6. [DOI] [PubMed] [Google Scholar]
- Blednov YA, Metten P, Finn DA, Rhodes JS, Bergeson SE, Harris RA, Crabbe JC. Hybrid C57BL/6J × FVB/NJ mice drink more alcohol than do C57BL/6J mice. Alcohol Clin Exp Res. 2005;29:1949–1958. doi: 10.1097/01.alc.0000187605.91468.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blizard DA, McClearn GE. Association between ethanol and sucrose intake in the laboratory mouse: exploration via congenic strains and conditioned taste aversion. Alcohol Clin Exp Res. 2000;24:253–258. [PubMed] [Google Scholar]
- Blizard DA, Vandenbergh DJ, Lionikas A, McClearn GE. Learning in the 2-bottle alcohol preference test. Alcohol Clin Exp Res. 2008;32:2041–2046. doi: 10.1111/j.1530-0277.2008.00791.x. [DOI] [PubMed] [Google Scholar]
- Blum K, Briggs AH, Elston SFA, DeLallo L, Sheridan PJ. Reduced leucine-enkephalin-like immunoreactive substance in hamster basal ganglia after long-term ethanol exposure. Science. 1982;216:1425–1427. doi: 10.1126/science.7089531. [DOI] [PubMed] [Google Scholar]
- Boehm SL, Moore EM, Walsh CD, Gross CD, Cavelli AM, Gigante E, Linsenbardt DN. Using drinking in the dark to model prenatal binge-like exposure to ethanol in C57BL/6J mice. Dev Psychobiol. 2008;50:566–578. doi: 10.1002/dev.20320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughter JD, Jr, Bachmanov AA. Behavioral genetics and taste. BMC Neurosci. 2007;8(Suppl 3):S3. doi: 10.1186/1471-2202-8-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AE, Gill KJ. Confirmation of provisional quantitative trait loci for voluntary alcohol consumption: genetic analysis in chromosome substitution strains and F2 crosses derived from A/J and C57BL/6J progenitors. Pharmacogenet Genomics. 2008;18:1071–1082. doi: 10.1097/FPC.0b013e32831367f0. [DOI] [PubMed] [Google Scholar]
- Carr LG, Foroud T, Bice P, Gobbett T, Ivashina J, Edenberg HJ, Lumeng L, Li T-K. A quantitative trait locus for alcohol consumption in selectively bred rat lines. Alcohol Clin Exp Res. 1998;22:884–887. [PubMed] [Google Scholar]
- Carr LG, Habegger K, Spence J, Ritchotte A, Liu L, Lumeng L, Li T-K, Foroud T. Analyses of quantitative trait loci contributing to alcohol preference in HAD1/LAD1 and HAD2/ LAD2 rats. Alcohol Clin Exp Res. 2003;27:1710–1717. doi: 10.1097/01.ALC.0000097161.51093.71. [DOI] [PubMed] [Google Scholar]
- Carr LG, Kimpel MW, Liang T, McClintick JN, McCall K, Morse M, Edenberg HJ. Identification of candidate genes for alcohol preference by expression profiling of congenic rat strains. Alcohol Clin Exp Res. 2007;31:1089–1098. doi: 10.1111/j.1530-0277.2007.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccocioppo R, Economidou D, Cippitelli A, Cucculelli M, Ubaldi M, Soverchia L, Lourdusamy A, Massi M. Genetically selected Marchigian Sardinian alcohol-preferring (msP) rats: an animal model to study the neurobiology of alcoholism. Addict Biol. 2006;11:339–355. doi: 10.1111/j.1369-1600.2006.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo G, Lobina C, Carai MA, Gessa GL. Phenotypic characterization of genetically selected Sardinian alcohol-preferring (sP) and -non-preferring (sNP) rats. Addict Biol. 2006;11:324–338. doi: 10.1111/j.1369-1600.2006.00031.x. [DOI] [PubMed] [Google Scholar]
- Crabbe JC. Psychopharmacogenetics of alcohol. In: Mormede P, Jones BC, editors. Neurobehavioral genetics: methods and applications. CRC Press; Boca Raton, FL: 1999. pp. 329–339. [Google Scholar]
- Crabbe JC. Neurogenetic studies of alcohol addiction. Philos Trans R Soc Lond B Biol Sci. 2008;363:3201–3211. doi: 10.1098/rstb.2008.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC. Consilience of rodent and human phenotypes relevant for alcohol dependence. Addict Biol. 2010;15:103–108. doi: 10.1111/j.1369-1600.2009.00188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Kosobud A, Belknap JK. Estimation of genetic correlation: interpretation of experiments using selectively bred and inbred animals. Alcohol Clin Exp Res. 1990;14(2):141–151. doi: 10.1111/j.1530-0277.1990.tb00461.x. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Feller DJ, Hen R, Wenger CD, Lessov CN, Schafer GL. Elevated alcohol consumption in null mutant mice lacking 5-HT1B serotonin receptors. Nat Genet. 1996;14:98–101. doi: 10.1038/ng0996-98. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Wahlsten D, Dudek BC. Genetics of mouse behavior: interactions with laboratory environment. Science. 1999;284:1670–1672. doi: 10.1126/science.284.5420.1670. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Metten P, Cameron AJ, Wahlsten D. An analysis of the genetics of alcohol intoxication in inbred mice. Neurosci Biobehav Rev. 2005;28:785–802. doi: 10.1016/j.neubiorev.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Cameron AJ, Munn E, Bunning M, Wahlsten D. Overview of mouse assays of ethanol intoxication. Curr Protoc Neurosci. 2008:9.26.1–9.26.18. doi: 10.1002/0471142301.ns0926s42. Unit. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Metten P, Rhodes JS, Yu C-H, Brown LL, Phillips TJ, Finn DA. A line of mice selected for high blood ethanol concentrations shows drinking in the dark to intoxication. Biol Psychiat. 2009;65:662–670. doi: 10.1016/j.biopsych.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFries JC, Hegmann JP, Weir MW. Open-field behavior in mice: evidence for a major gene effect mediated by the visual system. Science. 1966;154:1577–1579. doi: 10.1126/science.154.3756.1577. [DOI] [PubMed] [Google Scholar]
- Egli M. Can experimental paradigms and animal models be used to discover clinically effective medications for alcoholism? Addict Biol. 2005;10:309–319. doi: 10.1080/13556210500314550. [DOI] [PubMed] [Google Scholar]
- Enoch MA, Schuckit MA, Johnson BA, Goldman D. Genetics of alcoholism using intermediate phenotypes. Alcohol Clin Exp Res. 2003;27:169–176. doi: 10.1097/01.ALC.0000052702.77807.8C. [DOI] [PubMed] [Google Scholar]
- Fehr C, Shirley RL, Crabbe JC, Belknap JK, Buck KJ, Phillips TJ. The syntaxin binding protein 1 gene (Stxbp1) is a candidate for an ethanol preference drinking locus on mouse chromosome 2. Alcohol Clin Exp Res. 2005;29:708–720. doi: 10.1097/01.alc.0000164366.18376.ef. [DOI] [PubMed] [Google Scholar]
- Fernandez JR, Tarantino LM, Hofer SM, Vogler GP, McClearn GE. Epistatic quantitative trait loci for alcohol preference in mice. Behav Genet. 2000;30:431–437. doi: 10.1023/a:1010232900342. [DOI] [PubMed] [Google Scholar]
- Foroud T, Ritchotte A, Spence J, Liu L, Lumeng L, Li T-K, Carr LG. Confirmation of alcohol preference quantitative trait loci in the replicate high alcohol drinking and low alcohol drinking rat lines. Psychiatr Genet. 2003;13:155–161. doi: 10.1097/00041444-200309000-00004. [DOI] [PubMed] [Google Scholar]
- Gill K, Boyle AE. Genetic analysis of alcohol intake in recombinant inbred and congenic strains derived from A/J and C57BL/6J progenitors. Mamm Genome. 2005;16:319–331. doi: 10.1007/s00335-004-2239-x. [DOI] [PubMed] [Google Scholar]
- Gill K, Deitrich RA. Acute tolerance to the ataxic effects of ethanol in short-sleep (SS) and long-sleep (LS) mice. Psycho-pharmacology. 1998;136:91–98. doi: 10.1007/s002130050543. [DOI] [PubMed] [Google Scholar]
- Green AS, Grahame NJ. Ethanol drinking in rodents: is free-choice drinking related to the reinforcing effects of ethanol? Alcohol. 2008;42:1–11. doi: 10.1016/j.alcohol.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green ML, Singh AV, Zhang Y, Nemeth KA, Sulik KK, Knudsen TB. Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Dev Dyn. 2007;236:613–631. doi: 10.1002/dvdy.21048. [DOI] [PubMed] [Google Scholar]
- Grubb SC, Churchill GA, Bogue MA. A collaborative database of inbred mouse strain characteristics. Bioinformatics. 2004;20:2857–2859. doi: 10.1093/bioinformatics/bth299. [DOI] [PubMed] [Google Scholar]
- Gupta T, Syed YM, Revis AA, Miller SA, Martinez M, Cohn KA, Demeyer MR, Patel KY, Brzezinska WJ, Rhodes JS. Acute effects of acamprosate and MPEP on ethanol Drinking-in-the-Dark in male C57BL/6J mice. Alcohol Clin Exp Res. 2008;32:1992–1998. doi: 10.1111/j.1530-0277.2008.00787.x. [DOI] [PubMed] [Google Scholar]
- Halcomb RA, Hegmann JP, DeFries JC. Open-field behavior in mice: a diallel analysis of selected lines. Behav Genet. 1975;5(3):217–231. doi: 10.1007/BF01066174. [DOI] [PubMed] [Google Scholar]
- Hasin DS, Beseler CL. Dimensionality of lifetime alcohol abuse, dependence and binge drinking. Drug Alcohol Depend. 2009;101:53–61. doi: 10.1016/j.drugalcdep.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin D, Hatzenbuehler ML, Keyes K, Ogburn E. Substance use disorders: Diagnostic and Statistical Manual of Mental Disorders, fourth edition (DSM-IV) and International Classification of Diseases, tenth edition (ICD-10). Addiction. 2006;101(Suppl 1):59–75. doi: 10.1111/j.1360-0443.2006.01584.x. [DOI] [PubMed] [Google Scholar]
- Henderson ND. Interpreting studies that compare high- and low-sele cted lines on new characters. Behav Genet. 1989;19:473–502. doi: 10.1007/BF01066250. [DOI] [PubMed] [Google Scholar]
- Hendrickson LM, Zhao-Shea R, Tapper AR. Modulation of ethanol drinking-in-the-dark by mecamylamine and nicotinic acetylcholine receptor agonists in C57BL/6J mice. Psychopharmacology (Berl) 2009;204:563–572. doi: 10.1007/s00213-009-1488-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R, Reed C, Malmanger B, Lawler M, Hitzemann B, Cunningham B, McWeeney S, Belknap J, Harrington C, Buck K, Phillips T, Crabbe J. On the integration of alcohol-related quantitative trait loci and gene expression analyses. Alcohol Clin Exp Res. 2004;28:1437–1448. doi: 10.1097/01.alc.0000139827.86749.da. [DOI] [PubMed] [Google Scholar]
- Hitzemann R, Edmunds S, Wu W, Malmanger B, Walter N, Belknap J, Darakjian P, McWeeney S. Detection of reciprocal quantitative trait loci for acute ethanol withdrawal and ethanol consumption in heterogeneous stock mice. Psychopharmacology (Berl) 2009;203:713–722. doi: 10.1007/s00213-008-1418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EO, van den Bree M, Gupman AE, Pickens RW. Extension of a typology of alcohol dependence based on relative genetic and environmental loading. Alcohol Clin Exp Res. 1998;22:1421–1429. doi: 10.1111/j.1530-0277.1998.tb03930.x. [DOI] [PubMed] [Google Scholar]
- Kalant H. Research on tolerance: What can we learn from history? Alcohol Clin Exp Res. 1998;22:67–76. doi: 10.1111/j.1530-0277.1998.tb03618.x. [DOI] [PubMed] [Google Scholar]
- Kamdar NK, Miller SA, Syed YM, Bhayana R, Gupta T, Rhodes JS. Acute effects of naltrexone and GBR 12909 on ethanol drinking-in-the-dark in C57BL/6J mice. Psychopharmacology. 2007;192:207–217. doi: 10.1007/s00213-007-0711-5. [DOI] [PubMed] [Google Scholar]
- Kerns RT, Miles MF. Microarray analysis of ethanol-induced changes in gene expression. Meth Mol Biol. 2008;447:395–410. doi: 10.1007/978-1-59745-242-7_26. [DOI] [PubMed] [Google Scholar]
- Le AD, Israel Y, Juzytsch W, Quan B, Harding S. Genetic selection for high and low alcohol consumption in a limited-access paradigm. Alcohol Clin Exp Res. 2001;25:1613–1620. doi: 10.1111/j.1530-0277.2001.tb02168.x. [DOI] [PubMed] [Google Scholar]
- Leeman RF, Heilig M, Cunningham CL, Stephens DN, Duka T, O'Malley SS. Ethanol consumption: how should we measure it? Achieving consilience between human and animal phenotypes. Addict Biol. 2010;15:109–124. doi: 10.1111/j.1369-1600.2009.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesscher HM, Kas MJ, van der ES, van Lith HA, Vanderschuren LJ. A grandparent-influenced locus for alcohol preference on mouse chromosome 2. Pharmacogenet Genomics. 2009;19:719–729. doi: 10.1097/FPC.0b013e3283311320. [DOI] [PubMed] [Google Scholar]
- Linsenbardt DN, Boehm SL. Agonism of the endocannabinoid system modulates binge-like alcohol intake in male C57BL/6J mice: involvement of the posterior ventral tegmental area. Neuroscience. 2009;164:424–434. doi: 10.1016/j.neuroscience.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons AM, Lowery EG, Sparta DR, Thiele TE. Effects of food availability and administration of orexigenic and anorectic agents on elevated ethanol drinking associated with drinking in the dark procedures. Alcohol Clin Exp Res. 2008;32:1962–1968. doi: 10.1111/j.1530-0277.2008.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nat Rev Genet. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- Mardones J, Segovia-Riquelme N. Thirty-two years of selection of rats by ethanol preference: UChA and UChB strains. Neurobehav Toxicol Teratol. 1983;5:171–178. [PubMed] [Google Scholar]
- McClearn GE. The genetics of mouse behavior in novel situations. J Comp Physiol Psychol. 1959;52:62–67. doi: 10.1037/h0044664. [DOI] [PubMed] [Google Scholar]
- McClearn GE. Genetics and motivation of the mouse. In: Arnold WJ, editor. Nebraska Symposium on Motivation. University of Nebraska Press; Lincoln, Nebraska: 1968. pp. 47–83. [Google Scholar]
- McClearn GE. Genetics and alcoholism simulacra. Alcohol Clin Exp Res. 1979;3:255–258. doi: 10.1111/j.1530-0277.1979.tb05310.x. [DOI] [PubMed] [Google Scholar]
- McClearn GE, Rodgers DA. Differences in alcohol preference among inbred strains of mice. Q J Stud Alcohol. 1959;20:691–695. [Google Scholar]
- Metten P, Phillips TJ, Crabbe JC, Tarantino LM, McClearn GE, Plomin R, Erwin VG, Belknap JK. High genetic susceptibility to ethanol withdrawal predicts low ethanol consumption. Mamm Genome. 1998;9:983–990. doi: 10.1007/s003359900911. [DOI] [PubMed] [Google Scholar]
- Moore EM, Boehm SL. Site-specific microinjection of baclofen into the anterior ventral tegmental area reduces binge-like ethanol intake in male C57BL/6J mice. Behav Neurosci. 2009;123:555–563. doi: 10.1037/a0015345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore EM, Serio KM, Goldfarb KJ, Stepanovska S, Linsenbardt DN, Boehm SL. GABAergic modulation of binge-like ethanol intake in C57BL/6J mice. Pharmacol Biochem Behav. 2007;88:105–113. doi: 10.1016/j.pbb.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan MK, Ponomarev I, Hitzemann RJ, Belknap JK, Tabakoff B, Harris RA, Crabbe JC, Blednov Y, Grahame NJ, Phillips TJ, Finn DA, Hoffman PL, Iyer VR, Koob GF, Bergeson SE. Toward understanding the genetics of alcohol drinking through transcriptome meta-analysis. Proc Natl Acad Sci USA. 2006;103:6368–6373. doi: 10.1073/pnas.0510188103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JM, Stewart RB, Bell RL, Badia-Elder NE, Carr LG, McBride WJ, Lumeng L, Li T-K. Phenotypic and genotypic characterization of the Indiana University rat lines selectively bred for high and low alcohol preference. Behav Genet. 2002;32:363–388. doi: 10.1023/a:1020266306135. [DOI] [PubMed] [Google Scholar]
- Myers RD, Robinson DE, West MW, Biggs TA, McMillen BA. Genetics of alcoholism: rapid development of a new high-ethanol-preferring (HEP) strain of female and male rats. Alcohol. 1998;16:343–357. doi: 10.1016/s0741-8329(98)00031-7. [DOI] [PubMed] [Google Scholar]
- O'Dell LE, Roberts AJ, Smith RT, Koob GF. Enhanced alcohol self-administration after intermittent versus continuous alcohol vapor exposure. Alcohol Clin Exp Res. 2004;28:1676–1682. doi: 10.1097/01.alc.0000145781.11923.4e. [DOI] [PubMed] [Google Scholar]
- Overstreet DH, Rezvani AH, Djouma E, Parsian A, Lawrence AJ. Depressive-like behavior and high alcohol drinking co-occur in the FH/WJD rat but appear to be under independent genetic control. Neurosci Biobehav Rev. 2007;31:103–114. doi: 10.1016/j.neubiorev.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14:1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Belknap JK. Complex-trait genetics: emergence of multivariate strategies. Nat Rev Neurosci. 2002;3:478–485. doi: 10.1038/nrn847. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Hen R, Crabbe JC. Complications associated with genetic background effects in research using knockout mice. Psychopharmacology. 1999;147:5–7. doi: 10.1007/s002130051128. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Broadbent J, Burkhart-Kasch S, Henderson C, Wenger CD, McMullin C, McKinnon CS, Cunningham CL. Genetic correlational analyses of ethanol reward and aversion phenotypes in short-term selected mouse lines bred for ethanol drinking or ethanol-induced conditioned taste aversion. Behav Neurosci. 2005;119:892–910. doi: 10.1037/0735-7044.119.4.892. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Reed C, Burkhart-Kasch S, Li N, Hitzemann R, Yu C-H, Brown LL, Helms ML, Crabbe JC, Belknap JK. A method for mapping intralocus interactions influencing excessive alcohol drinking. Mamm Genome. 2010;21:39–51. doi: 10.1007/s00335-009-9239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintanilla ME, Israel Y, Sapag A, Tampier L. The UChA and UChB rat lines: metabolic and genetic differences influencing ethanol intake. Addict Biol. 2006;11:310–323. doi: 10.1111/j.1369-1600.2006.00030.x. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Best K, Belknap JK, Finn DA, Crabbe JC. Evaluation of a simple model of ethanol drinking to intoxication in C57BL/6J mice. Physiol Behav. 2005;84:53–63. doi: 10.1016/j.physbeh.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Rhodes JS, Ford MM, Yu C-H, Brown LL, Finn DA, Garland T, Jr, Crabbe JC. Mouse inbred strain differences in ethanol drinking to intoxication. Genes Brain Behav. 2007;6:1–18. doi: 10.1111/j.1601-183X.2006.00210.x. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Bell RL, Kuc KA, Murphy JM, Lumeng L, McBride WJ. Effects of concurrent access to multiple ethanol concentrations and repeated deprivations on alcohol intake of high-alcohol-drinking (HAD) rats. Addict Biol. 2009;14:152–164. doi: 10.1111/j.1369-1600.2008.00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers DA, McClearn GE. Sucrose versus ethanol appetite in inbred strains of mice. Quart Jf Stud Alcohol. 1964;25(1):26–35. [PubMed] [Google Scholar]
- Rodgers DA, McClearn GE, Bennett EL, Herbert M. Alcohol preference as a function of its caloric utility in mice. J Comp Physiol Psychol. 1963;56(4):666–672. doi: 10.1037/h0040350. [DOI] [PubMed] [Google Scholar]
- Ryabinin AE, Yoneyama N, Tanchuck MA, Mark GP, Finn DA. Urocortin 1 microinjection into the mouse lateral septum regulates the acquisition and expression of alcohol consumption. Neuroscience. 2008;151:780–790. doi: 10.1016/j.neuroscience.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer W, Hyytia P, Kiianmaa K. The alcohol-preferring AA and alcohol-avoiding ANA rats: neurobiology of the regulation of alcohol drinking. Addict Biol. 2006;11:289–309. doi: 10.1111/j.1369-1600.2006.00037.x. [DOI] [PubMed] [Google Scholar]
- Song MJ, Lewis CK, Lance ER, Chesler EJ, Yordanova RK, Langston MA, Lodowski KH, Bergeson SE. Reconstructing generalized logical networks of transcriptional regulation in mouse brain from temporal gene expression data. EURASIP J Bioinform Syst Biol. 2009:545176. doi: 10.1155/2009/545176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanagel R. Alcoholism: a systems approach from molecular physiology to addictive behavior. Physiol Rev. 2009;89:649–705. doi: 10.1152/physrev.00013.2008. [DOI] [PubMed] [Google Scholar]
- Sparta DR, Sparrow AM, Lowery EG, Fee JR, Knapp DJ, Thiele TE. Blockade of the corticotropin releasing factor type 1 receptor attenuates elevated ethanol drinking associated with drinking in the dark procedures. Alcohol Clin Exp Res. 2008;32:259–265. doi: 10.1111/j.1530-0277.2007.00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JP, Liang T, Liu L, Johnson PL, Foroud T, Carr LG, Shekhar A. From QTL to candidate gene: a genetic approach to alcoholism research. Curr Drug Abuse Rev. 2009;2:127–134. doi: 10.2174/1874473710902020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Kechris K, Hu W, Bhave SV, Finn DA, Grahame NJ, Hoffman PL. The genomic determinants of alcohol preference in mice. Mamm Genome. 2008;19:352–365. doi: 10.1007/s00335-008-9115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenina-Rigaldie E, Jones BJ, Mormede P. Pleiotropic effect of a locus on chromosome 4 influencing alcohol drinking and emotional reactivity in rats. Genes Brain Behav. 2003a;2:125–131. doi: 10.1034/j.1601-183x.2003.00018.x. [DOI] [PubMed] [Google Scholar]
- Terenina-Rigaldie E, Moisan MP, Colas A, Beauge F, Shah KV, Jones BC, Mormede P. Genetics of behaviour: phenotypic and molecular study of rats derived from high- and low-alcohol consuming lines. Pharmacogenetics. 2003b;13:543–554. doi: 10.1097/01.fpc.0000054120.14659.8c. [DOI] [PubMed] [Google Scholar]
- Toth LA, Gardiner TW. Food and water restriction protocols: physiological and behavioral considerations. Contemp Top Lab Anim Sci. 2000;39:9–17. [PubMed] [Google Scholar]
- Treadwell JA, Pagniello KB, Singh SM. Genetic segregation of brain gene expression identifies retinaldehyde binding protein 1 and syntaxin 12 as potential contributors to ethanol preference in mice. Behav Genet. 2004;34:425–439. doi: 10.1023/B:BEGE.0000023648.78190.ee. [DOI] [PubMed] [Google Scholar]
- Vadasz C, Saito M, Gyetvai BM, Oros M, Szakall I, Kovacs KM, Prasad VV, Morahan G, Toth R. Mapping of QTLs for oral alcohol self-administration in B6.C and B6.I quasi-congenic RQI strains. Neurochem Res. 2007;32:1099–1112. doi: 10.1007/s11064-006-9234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlsten D, Bachmanov A, Finn DA, Crabbe JC. Stability of inbred mouse strain differences in behavior and brain size between laboratories and across decades. Proc Natl Acad Sci USA. 2006;103:16364–16369. doi: 10.1073/pnas.0605342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J, Symons MN, Singh SM. Ethanol-responsive genes (Crtam, Zbtb16, and Mobp) located in the alcohol-QTL region of chromosome 9 are associated with alcohol preference in mice. Alcohol Clin Exp Res. 2009a;33:1409–1416. doi: 10.1111/j.1530-0277.2009.00971.x. [DOI] [PubMed] [Google Scholar]
- Weng J, Symons MN, Singh SM. Studies on Syntaxin 12 and alcohol preference involving C57BL/6J and DBA/2J strains of mice. Behav Genet. 2009b;39:183–191. doi: 10.1007/s10519-008-9249-5. [DOI] [PubMed] [Google Scholar]
- Whatley VJ, Johnson TE, Erwin VG. Identification and confirmation of quantitative trait loci regulating alcohol consumption in congenic strains of mice. Alcohol Clin Exp Res. 1999;23:1262–1271. doi: 10.1111/j.1530-0277.1999.tb04287.x. [DOI] [PubMed] [Google Scholar]
- Worst TJ, Tan JC, Robertson DJ, Freeman WM, Hyytia P, Kiianmaa K, Vrana KE. Transcriptome analysis of frontal cortex in alcohol-preferring and nonpreferring rats. J Neurosci Res. 2005;80:529–538. doi: 10.1002/jnr.20496. [DOI] [PubMed] [Google Scholar]
- Yoneyama N, Crabbe JC, Ford MM, Murillo A, Finn DA. Voluntary ethanol consumption in 22 inbred mouse strains. Alcohol. 2008;42:149–160. doi: 10.1016/j.alcohol.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]