Abstract
Two new drimane sesquiterpene lactones and one new tricarboxylic acid derivative were isolated from the Berkeley Pit extremophilic fungus Penicillium solitum. The structures of these compounds were deduced by spectroscopic analysis. Berkedrimanes A and B inhibited the signal transduction enzymes caspase-1 and caspase-3 and mitigated the production of interleukin 1-β in the induced THP-1 (promonocytic leukemia cell line) assay.
Microorganisms isolated from the acidic, metal-rich waters of the Berkeley Pit Lake System in Butte, Montana have been a rich source of interesting and biologically active compounds.1,2a–f We recently isolated a filamentous fungus from surface waters of the Pit Lake which was identified as Penicillium solitum. The organic extracts of broth cultures of this fungus exhibited potent activity in caspase-1, caspase-3 and matrix metalloproteinase-3 enzyme inhibition assays. These enzyme assays were used to guide the isolation of two new drimane sesquiterpenes, berkedrimane A and B. Their isolation, characterization, inhibition of the caspases and mitigation of interleukin-1β production in induced THP-1 cells will be described in this report.
The role of caspase-1 in inflammation is complex. Caspase-1 is activated upon binding to the inflammasome, a multiprotein complex that plays a key role in innate immunity by activating the proinflammatory cytokines interleukin 1-β and IL-18.3 There is a strong correlation between dysregulated inflammasome activity and both inherited and acquired inflammatory diseases.3 Caspase-1 inhibitors have shown promise in either delaying the onset or in mitigating the effects of a number of diseases, including Huntington’s disease,4 amyotropic lateral sclerosis,5 stroke 6 and multiple sclerosis.7, 8 Caspase-1 inhibitors have also been proposed as potential therapies for osteoarthritis and rheumatoid arthritis,9 Alzheimer’s disease,10 and brain and nerve trauma.6, 8
There is a complex relationship between different members of the caspase enzyme class. Caspase-3 is an executioner caspase and an important component of apoptosis, which can be activated by either extrinsic or intrinsic factors.11 High levels of caspase-3 have been associated with Alzheimer’s disease, particularly when coupled with high levels of caspase-1. The pathological hallmarks of Alzheimer’s disease include neuronal loss, extracellular senile plaques containing the peptide amyloid-β, and neurofibrillary tangles (NFTs).10 Although amyloid plaques and NFTs have been largely regarded as independent neuropathologic phenomena, they may be functionally linked and caspases, particularly caspase-3 and 1, may play an important role in their development.10 We have begun exploring the implications of compounds with activity against both of these caspases. The development of new enzyme inhibitors will not only provide potential chemotherapeutics or pharmacophore models but will also provide tools for the investigation of the intricacies of signal transduction.
RESULTS AND DISCUSSION
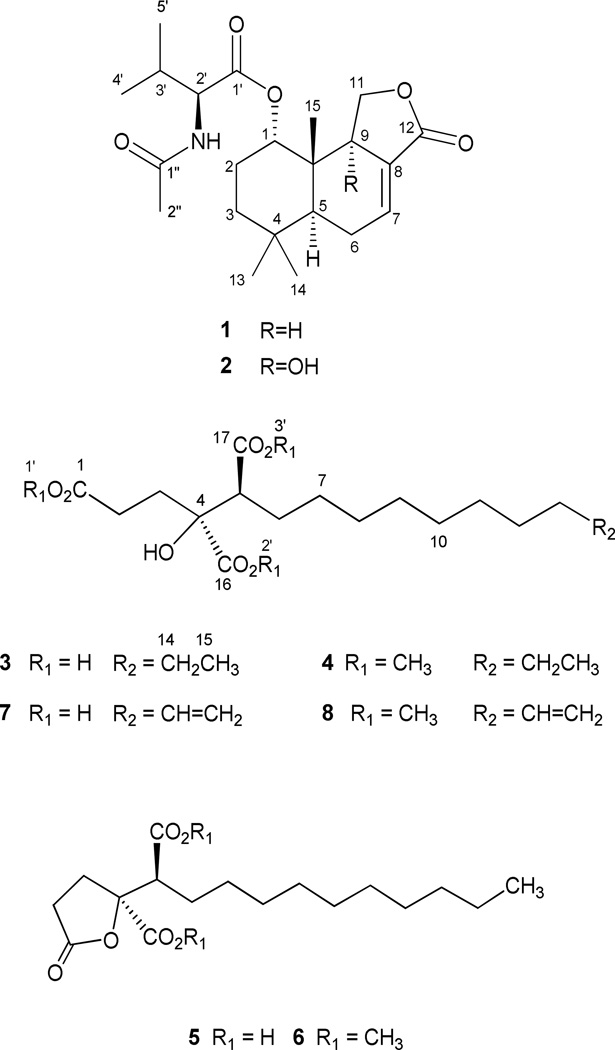
Penicillium solitum, a surface isolate of acidic Berkeley Pit Lake (pH 2.7), was grown in mycological broth for 22 days. At time of harvest, the mycelium was removed by filtration and the filtrate was extracted with CHCl3. When compared to other fungal extracts from Pit microbes in our study, the organic extract of P. solitum demonstrated excellent inhibitory activity in the signal transduction assays. Flash silica gel chromatography followed by silica gel HPLC yielded the two new drimane derivatives 1 and 2, that contributed to the observed enzyme inhibitory activity. A known tricarboxylic acid (3) and its lactone (5) and the new tricarboxylic acid derivative, compound 7 were also isolated from this extract.
The major drimane derivative, 1, isolated as a colorless oil, had a molecular formula of C22H33NO5 as deduced by HRCIMS [M+H]+, with seven degrees of unsaturation. The IR spectrum indicated the presence of an α,β unsaturated γ-butyrolactone, and both ester and amide functionalities (1756, 1733 and 1675 cm−1 respectively).12 All 22 carbons were observed in the 13C NMR spectrum and analysis of this spectrum along with the DEPT and HMQC spectra indicated the presence of three ester/amide carbonyls (δC 171.5, 170.2, 169.7), one trisubstituted double bond (δC 135.7, 126.9), two oxygen-bearing carbons (δC 75.5, 66.7), one nitrogen-bearing carbon (δC 57.4), two quaternary carbons (δC 37.5, 33.0), three methine carbons (δC 44.3, 42.9, 31.1), three methylene carbons (δC 34.7, 25.1, 22.7) and six methyl carbons (δC 32.7, 23.3, 21.5, 19.5, 17.3, 13.6). An N-acetyl valine residue was deduced from the following data: the 1H-1H COSY spectrum indicated the presence of the spin system NH-CH-CH-(CH3)2 and the HMBC spectrum showed a correlation from both NH and H-2′ to carbonyl C-1′ (δC 171.5), and from NH and methyl singlet H3-2″ (δH 2.08) to carbonyl C-1″ (δC 170.2). These assignments accommodated C7H12NO2 and both the amide and the ester functionalities. There remained to assign C15H21O3 which suggested a sesquiterpene which included three singlet methyls (δH 1.00, 0.99, 0.92) and the α,β unsaturated γ-butyrolactone, which accommodated three degrees of unsaturation. The remaining two degrees required two additional rings suggesting a fused tricyclic carbon skeleton.
The 1H-1H COSY spectrum indicated three isolated spin systems: OCH2-CH, =CH-CH2-CH, and OCH-CH2-CH2, The HMBC spectrum provided key correlations to connect these systems and to assemble the molecule. The α,β unsaturated γ-butyrolactone provided an appropriate starting point. Oxygen-bearing methylene H-11α (δH 4.23) exhibited three-bond correlations to olefinic C-8 (δC 126.9) and carbonyl C-12 (δC 169.7), and H-11β (δH 4.02) had a similar correlation to C-10 (δC 37.5). Both protons showed two-bond correlations to C-9 (δC 42.9). H-9 (δH 3.18) showed three-bond correlations to methyl C-15 (δC 13.6) and olefinic C-7 (δC 135.7). H-7 (δH 6.88) exhibited three-bond correlations to C-9, -12, and -5 (δC 44.3) connecting the B and C rings to the A ring. H-5 (δH 1.74) exhibited three-bond correlations to C-1 (δC 75.5), C-15 (δC 13.6) and C-9 (δC 42.9). Oxygen-bearing methine H-1 (δH 4.78) exhibited three-bond correlations to both valine C-1′ (δC 171.5) and C-3 (δC 34.7) linking the tricyclic moiety to valine. These data fit the drimane skeleton and generated the structure of berkedrimane A (1) as shown.
Berkedrimane A (1), is isomeric with the known compound, purpuride, isolated from Penicillium purpurogenum13 (the double bond is Δ8 rather than Δ7) and has the same sesquiterpene skeleton as nebularilactone B isolated from Lepista nebularis.14 Comparison of the NMR data of berkedrimane A with that of nebularilactone B provided additional support for the structure of compound 1 as shown.
The relative configuration of 1 was established by consideration of coupling constants and from 1-D difference NOE studies. H-1 appeared as a broad singlet, indicating that it occupied an equatorial position in the cyclohexane ring and that the ester substituent must therefore be axial. Irradiation of H-9 (δH 3.18) enhanced the intensity of H-11α (4.23) as well as that of H-5 (δH 1.74) indicating a 1,3-syn-diaxial relationship between H5 and -9. In C6D6 the methyl singlets of H-13, H-14 and H-15 shifted from δH 1.00, 0.99 and 0.92 ppm respectively to δH 0.55, 0.66 and 0.27 ppm. This greater signal dispersion in C6D6 allowed selective irradiation of the methyl resonances to observe the resultant NOE enhancements. A large NOE enhancement was observed between methyl H-14 and methyl H-15, indicating a 1,3-diaxial relationship. Irradiation of H-15 also enhanced the absorption of methine H-1. These data were sufficient to generate the relative configuration of berkedrimane A 1, which was consistent with that of purpuride and nebularilactone B.
The molecular formula of 2 was C22H33NO6, established by HRCIMS [M+H]+. The IR spectrum again indicated the presence of α,β unsaturated γ-butyrolactone, ester and amide functionalities as in compound 1, but compound 2 also showed an additional -OH stretch at 3436 cm−1. The 1H NMR and 13C NMR spectra (Table 1) for 1 and 2 were very similar, with the following key exceptions: methine C-9 of compound 1 (δC 42.9) was replaced with a quaternary oxygen-bearing carbon in compound 2 (δC 78.0); methylene H2-11 in compound 1 appeared as two triplets (δH 4.23 and 4.02, J = 9.2 Hz) whereas the analogous protons appeared as geminally coupled doublets at (δH 4.40 and 4.25, J = 10.1 Hz) in compound 2. These data suggested that the hydroxy moiety in 2 was at position C-9. All of the HMBC correlations were similar to those of 1 and were in accord with this proposed structure. Key NOE correlations were also similar to those of 1, indicating the same relative stereoconfiguration and the structure of berkedrimane B (2) as shown.
Table 1.
13C and 1H NMR Data for Compounds 1, 2, and 8 in CDCl3.a
| 1 | 2 | 8 | ||||
|---|---|---|---|---|---|---|
| no. | δC, mult.d | δH, mult. (J in Hz)b | δC, mult. d | δH, mult. (J in Hz)b | δC, mult. d | δH, mult. (J in Hz)c |
| 1 | 75.5, CH | 4.78, bs | 76.3, CH | 5.01, bs | 173.4, C | |
| 2 | 22.7, CH2 | 1.85, m | 23.5, CH2 | 1.92, dt (14.5, 3.1) | 28.6, CH2 | 2.46, m |
| 1.76, m | 1.76, dq (14.5, 3.5) | 2.11, m | ||||
| 3 | 34.7, CH2 | 1.69, m | 34.8, CH2 | 1.61, td (13.7, 3.4) | 32.3, CH2 | 2.17 |
| 1.32, dt (13.7, 3.2) | 1.35, dt (13.7, 3.7) | 1.97, | ||||
| 4 | 33.0, C | 32.6, C | 77.7, C | |||
| 5 | 44.3, CH | 1.74, dd (11.5, 6.0) | 37.9, CH | 2.51, dd (15.3, 5.8) | 53.8 | 2.76 |
| 6 | 25.1, CH2 | 2.50, m | 25.5, CH2 | 2.58, ddd (20.1, 5.7, 3.9) | 27.7, CH2 | 1.82, m |
| 2.21, m | 2.24, m | 1.35, m | ||||
| 7 | 135.7, CH | 6.88, bq (3.5) | 139.9, CH | 7.01, t (3.5) | 27.9, CH2 | 1.22, bs |
| 8 | 126.9, C | 130.4, C | 29.5, CH2 | 1.22, bs | ||
| 9 | 42.9, CH | 3.18, m | 78.0, C | 29.3, CH2 | 1.22, bs | |
| 10 | 37.5, C | 41.0, C | 29.5, CH2 | 1.22, bs | ||
| 11 | 66.7, CH2 | α 4.23, t (9.2) | 73.9, CH2 | 4.40, d (10.1) | 29.3, CH2 | 1.22, bs |
| Β 4.02, t (9.2) | 4.25, d (10.1) | |||||
| 12 | 169.7, C | 168.5, C | 31.9, CH2 | 1.22, bs | ||
| 13 | 32.7, CH3 | 1.00, s | 33.0, CH3 | 1.04, s | 33.8, CH2 | 2.03, bq |
| 1.95, m | ||||||
| 14 | 21.5, CH3 | 0.98, s | 21.6, CH3 | 1.03, s | 139.2, CH | 5.78, ddt (17.1, 10.2, 6.8) |
| 15 | 13.6, CH3 | 0.92, s | 18.6, CH3 | 0.92, s | 114.1, CH2 | 4.91, dt (10.2, 1.6) |
| 4.97, dq (17.1, 1.6) | ||||||
| 16 | 174.4, C | |||||
| 17 | 174.4, C | |||||
| 1′ | 171.5, C | 170.9, C | 51.9, CH3 | 3.64, s | ||
| 2′ | 57.4, CH | 4.62, m | 58.9, CH | 4.36, dd (7.4, 5.8) | 51.8, CH3 | 3.79, s |
| 3′ | 31.1, CH | 2.21, m | 29.9, CH | 2.20, m | 53.1 CH3 | 3.70, s |
| 4′ | 19.5, CH3 | 1.02, d (6.8) | 19.4, CH3 | 1.02, d (7.0) | ||
| 5′ | 17.3, CH3 | 0.93, d (7.0) | 17.9, CH3 | 0.98, d (6.8) | ||
| Ac | 170.2, C | 169.9, C | ||||
| Ac CH3 |
23.3, CH3 | 2.08, s | 23.2, CH3 | 2.07, s | ||
| NH | 5.90, bd (8.4) | 5.94, bd (6.8) |
All assignments are based on COSY, HSQC, HMBC and NOE experiments.
300 MHz for 1H NMR, HMBC, COSY.
500 MHz for 1H NMR, HMBC, COSY.
75 MHz for 13C NMR
The magnitude of the scalar coupling constants (3J ) associated with olefinic H-7 also supported the assigned configuration at C-9. In the energy minimized model (Molecular Dynamics, MM2 force field calculations) of berkedrimane B as shown, the dihedral angles of H-7 to Hα-6 and Hβ-6 were −55° and 58°, respectively, which would indicate that H-7 was a doublet of doublets (J = 4.0, 3.9). In the MM2 energy minimized model of 9-epi-berkedrimane, the dihedral angles of H-7 were 100° relative to Hβ-6 and −7.5° relative to Hα-6. If this were the case, H-7 would be a doublet of doublets (J = 12, 1). In fact, H-7 is a triplet (J = 3.5) which supports the structure of compound 2 as shown. Similar results were obtained when the minimum energy geometries were determined using Hartree-Fock (3-21G) calculations for compound 2 and its C-9 epimer.
The absolute configuration of the valine residue in berkedrimane B (2) was established by Marfey’s analysis.15 Compound 2 was hydrolyzed in boiling 6M HCl and treated with 2,4,-dinitro-5-fluoro-L-alanamide (Marfey’s reagent). The resulting derivative was analyzed by LC-MS. Authentic samples of D and L-valine were derivatized under identical conditions. The valine derivative from the hydrolysis of 2 was identical to the L-valine stereoisomer.
We also isolated two tricarboxylic acids and a tricarboxylic acid lactone along with the two drimane derivatives. These acids were difficult to purify, but were easily separated following methylation. A mixture of acids 3, 5 and 7 was treated with diazomethane and separated by HPLC to yield the methyl esters 4, 6, and 8. Analysis of NMR and mass spectral data indicated that compound 4 was the methyl ester of secospiculisporic acid (3),16 and compound 6 was the methyl ester of the tricarboxylic acid lactone, spiculisporic acid (5).17 Both of these acids have been previously described as metabolites of Penicillium spiculisporum.17
The HRCIMS [M+H]+ of the trimethyl ester 8 established a molecular formula of C20H34O7. The 1H and 13C NMR spectra of compounds 4 and 8 were almost identical, with one key exception; the 1H NMR of trimethylester 4 indicated the presence of a terminal methyl group, and this methyl triplet was not present in the 1H NMR spectrum of compound 8. Instead, both the 1H and 13C NMR spectra indicated the presence of a terminal monosubstituted double bond [δC 114.1, δH 4.97 (ddt, J = 17.1, 2.0, 1.6), 4.91 (ddt, J = 10.2, 2.0, 1.2) and δC 139.2, δH 5.78 (ddt, J = 17.1, 10.1, 6.7)]. This placed the double bond at the end of the straight chain and established the structure of triacid 7 as the previously undescribed 4-hydroxy-4,5-dicarboxy-9-pentadecenoic acid.
Secospiculisporic acid 3 and spiculisporic acid 5 have received much attention because of their amphiphilic nature and their ability to aggregate as liposome-like vesicles. They act as biosoaps and biosurfactants and can associate with metals in solution and precipitate these metal salts.18 According to the NCBI PubChem website, spiculosporic acid was evaluated in 63 different biological assays, including several enzyme assays, and was inactive in each test.19
Berkedrimane A and B were evaluated for their ability to inhibit caspase-1 and caspase-3 in vitro in fluorometric assays. Berkedrimane A (1) exhibited IC50 value of 100 µM against caspase-1 and 200 µM against caspase-3. It maintained moderate inhibition of caspase-3 with an IC25 value of 15 µM. Berkedrimane B (2) was slightly more potent with IC50 values of 90 µM against caspase-1 and 50 µM against caspase-3. It maintained moderate inhibition of caspase-3 with an IC25 value of 10 µM. The three methyl esters were inactive against the two caspases and MMP-3.
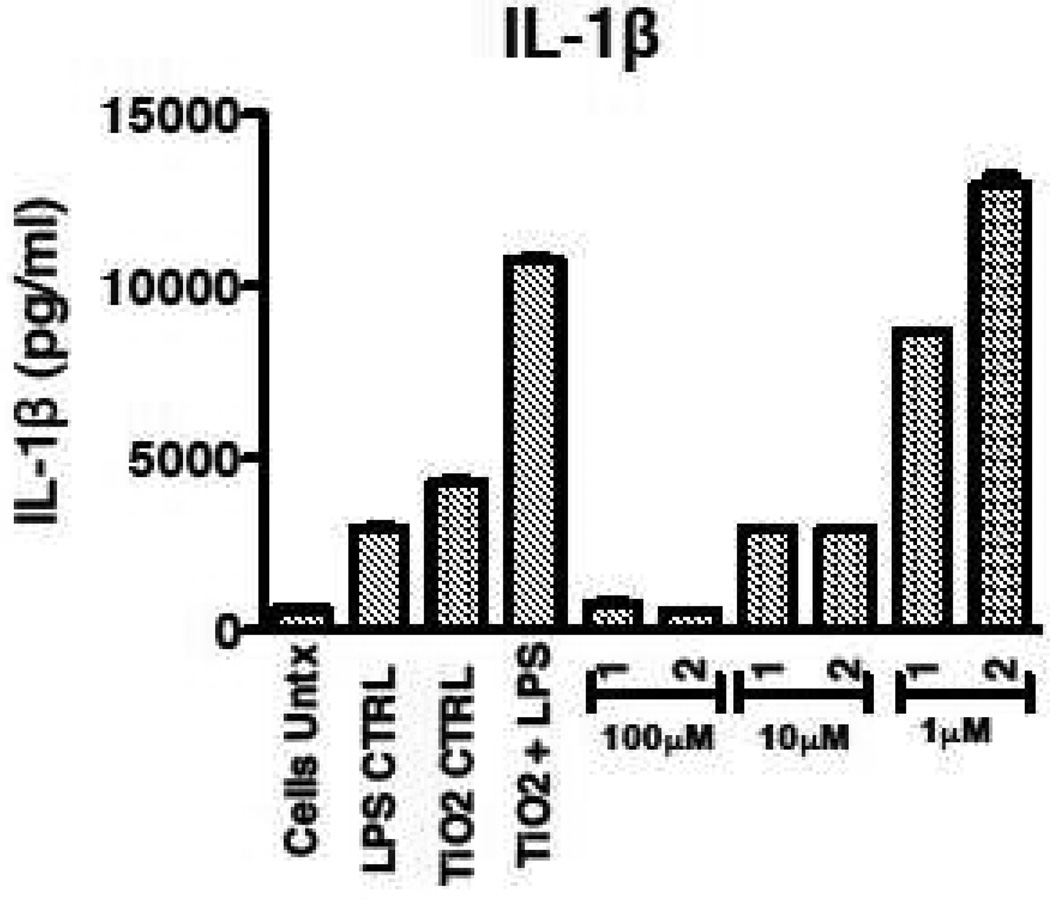
When induced with titanium nanowires and bacterial lipopolysaccharide (LPS), THP-1 cells produce large numbers of inflammasomes, which in turn activate pro-IL-1B to the active form, IL-1β. Induced THP-1 cells were exposed to compounds 1 and 2 at concentrations of 100, 10 and 1 µM and the concentrations of IL-1β post-exposure were determined (Figure 2). Both berkedrimanes effectively inhibited the production of IL-1β by intact inflammasomes at low micromolar concentrations showing greater potency in the cell line assay than in the enzyme assay. Further work is underway to determine the relevance of these observations.
Figure 2.
Production of IL-1β in THP-1 cells induced with TiO2 and bacterial LPS following treatment with compounds 1 and 2.
The inflammasome is becoming an important therapeutic target as studies continue to link inflammation and disease. In mice, potential therapeutic agents that target the inflammasome and mitigate the production of IL-1β have been shown to reduce the inflammatory response following thromboembolic stroke20 and to improve histopathology following traumatic brain injury.21 The biological activity of the berkedrimanes is similar to that of parthenolide, an anti-inflammatory sesquiterpene lactone isolated from feverfew, Tanacetum parthenium.22
EXPERIMENTAL SECTION
General Experimental Procedures
Optical rotations were recorded on a Perkin Elmer 241 Polarimeter using a 1 mL cell. IR spectra were recorded on a Nicolet NEXUS 670 FT-IR spectrometer. 1H-NMR and 13C-NMR spectra were obtained on Bruker DPX-300 or Varian 500 NMR spectrometers. 1H-NMR spectra were recorded at 300 or 500 MHz and the 13C-NMR spectra were recorded at 75 MHz unless otherwise noted. All of the chemical shifts were recorded with respect to the deuterated solvent shift (CDCl3, δH 7.24, δC 77.0 or C6D6, δH 7.18, δC 128.0). Mass spectral data were provided by the University of Montana Mass Spectrometer facility. All solvents used were spectral grade or distilled prior to use.
Collection, Extraction, and Isolation Procedures
The collection and isolation of Berkeley Pit fungi have previously been described.2e Penicillium solitum was grown in potato dextrose broth (8 flasks with 500 mL broth per flask, in 1L Erlenmeyer flasks) for 22 days (still culture). At time of harvest MeOH (50 mL per flask) was added to each flask, the mycelia was removed by filtration and the filtrate was extracted with CHCl3 (3 × 1 L). Removal of the CHCl3 in vacuo yielded 0.4128 g of total crude extract. This extract showed potent enzyme inhibition in the MMP-3, caspase-1, and caspase-3 assays. It was chromatographed on a flash Si gel column in a gradient fashion, starting with hexane, then with increasing amounts of IPA. The column was washed with 100% IPA, and finally with MeOH. The fraction that eluted with 50% IPA/hexane was active in both the caspase-1 and caspase-3 enzyme inhibition assays. It was further purified with Si gel HPLC in an isocratic mode using 20% IPA/hexane to yield berkedrimane A 1 (11.6 mg) and B 2 (5.1 mg). The two berkedrimanes which were 2.8% and 1.2% of the crude CHCl3 respectively, were active in the enzyme inhibition assays, although several of the other column fractions exhibited activity as well. The fraction that eluted with 20% IPA (180.2 mg) was a mixture of acids (3, 5, and 7) that was difficult to separate. The acid fraction was methylated with diazomethane and then chromatographed on Sigel HPLC to give methyl esters of the known 4-hydroxy-4,5-dicarboxypentadecanoic acid 3 (40.3 mg), the previously undescribed 4-hydroxy-4,5-dicarboxy-9-pentadecenoic acid 7 (75.3 mg), and spiculisporic acid 5 (60.2 mg).
Berkedrimane 1
colorless oil, [α]25D +31.4°(c .0014, CHCl3); IR (CHCl3) νmax 3436, 2966, 1756, 1733, 1675, 1506, 1373, 1016, 892cm−1; 1H NMR and 13C NMR (CDCl3) see Table 1 ; 1H NMR (C6D6, 500 MHz) δ 0.27 (s, 3 H, H-15 ) 0.55 (s, 3 H, H-13) 0.66 (s, 3 H, H-14) 0.82 (d, J=6.73 Hz, 6 H, H-4’,5’) 1.50 (s, 3 H, OAc) 1.89 - 1.99 (m, 1 H) 3.11 (m, 1 H, H-9) 3.60 (m, 1 H, H-11) 4.19 (m, 1 H, H-11) 4.56 (m, 1 H, H-1) 4.66 (m, 1 H, H-2’) 5.32 (bd, 1 H, NH) 6.61 (m, 1 H, H-7): HRCIMS [M+H]+ m/z 392.2466 (calcd for C22H34NO5, 392.2437).
Berkedrimane 2
colorless oil, [α]25D -15.3° (c .0145, CHCl3); IR (CHCl3) νmax 3446, 3081, 2966, 1751, 1730, 1670, 1238, 1022, 729 cm−1; 1H NMR (CDCl3) see Table 1; 13C NMR (CDCl3) see Table 1 ; EIMS m/z [M]+ 407 (2), 390 (10), 230 (55), 78 (100); HRCIMS [M+H]+ m/z 408.2361 (calcd for C22H33NO6, 408.2386).
Determination of the Absolute Configuration of Valine
Drimane 2 (1 mg) was suspended in freshly distilled constant boiling 6 M HCl (100 µL) and heated in a Teflon sealed glass vial at 110 °C for 15 h. The solution was then concentrated to dryness under a stream of N2. This solid was treated with a solution of 2,4,-dinitro-5-fluoro-L-alanamide (100 µL, 1% w/v in acetone), followed by 1.0 M NaHCO3 (25 µL) and heated in a Teflon sealed glass vial at 80 °C for 10 min. The mixture was cooled and quenched with 1.0 M HCl (25 µL). Authentic L- and D-valine standards were derivatized under identical conditions.15
LC-MS Analysis
The Marfey’s derivatives and the standards derivatives (see above) were analyzed by LC-MS using a ZORBAX Extend C-18 (2.1 X 50mm, 3.5 µM) column, chromatography parameters are as follows: initial, 80% solvent A (H2O + 0.1% formic acid), 20 % solvent B (acetonitrile), at 9 min to 60% A, at 10 min to 100% B. PDA parameters were: channel A 340 nm, channel B 254 nm. MS parameters were: APCI source. Retention time for the authentic L-valine derivative was 10.6 min, retention time for the authentic D-valine was 12.6 min. The Marfey’s derivative of the hydrolysis of 2 had a retention time of 10.6 min and gave a mass spectrum identical to the authentic valine samples.
Methylation of carboxylic acid mixture
The mixture of acids (10.3 mg) was dissolved in diethyl ether (2 mL) and a solution of CH2N2 dissolved in ether was added dropwise until the yellow color remained. After 1 minute the solvent was removed in vacuo and the mixture subjected to HPLC using 5% EtOAc/Hexane. The trimethyl ester of the known tricarboxylic acid, secospiculisporic acid (4-hydroxy-4,5-dicarboxypentadecanoic acid) 4 (5.2 mg) and the dimethyl ester of spiculisporic acid 6, (3.2 mg) were isolated along with the trimethyl ester of the previously unreported 4-hydroxy-4,5-dicarboxy-9-pentadecenoic 8, (2.2 mg).
Trimethyl 4-hydroxy-4,5-dicarboxy-9-pentadecenoate (8)
colorless oil, [α]25D -5.0° (c 0.0022, CHCl3)1H NMR (CDCl3, 500 MHz) δ 1.22 (bs, 12 H) 1.35 (m, 2 H) 1.83 (m, 1 H) 1.98 (m, 2 H) 2.14 (m, 2 H) 2.46 (m, 1 H) 2.76 (dd, J=11.8, 3.3 Hz, 1 H) 3.64 (s, 3 H) 3.70 (s, 3 H) 3.79 (s, 3 H) 4.91 (dt, J=10.2, 1.0 Hz, 1 H) 4.97 (dq, J=17.1, 1.8 Hz, 1 H) 5.78 (dd, J=17.0, 10.1 Hz, 1 H); 13C NMR (CDCl3, 125 MHz) δ 27.7, 27.9, 28.6, 29.3(3C), 29.5, 31.9, 32.3, 33.8, 51.8, 51.9, 53.1, 53.8, 77.7, 114.1, 139.2, 173.4, 174.1, 174.4; HRCIMS [M+H]+ m/z 387.2344 (calcd for C20H35O7, 387.2383).
Signal Transduction Assays
The signal transduction Drug Discovery Kits for caspase-1, caspase-3, and matrix metalloproteinase-3 enzymes were purchased from Enzo Life Sciences. The positive controls (enzyme inhibitors) used in these assays are Ac-YVAD-CHO for the caspase-1 assay, Ac-DEVD-CHO for the caspase-3 assay, and N-Isobutyl-N-(4-methoxyphenylsulfonyl)-glycylhydroxamic acid (NNGH) for the MMP-3 assay.
In Vitro THP-1 Assay
Human monocyte cell line THP-1 was purchased from ATCC (#TIB-202). The cells were suspended at (2–4) × 105 viable cells/mL in RPMI media supplemented with 10% fetal bovine serum, 0.05 mM 2-mercaptoethanol, sodium pyruvate, and an antimycotic/antibiotic cocktail containing penicillin, streptomycin and amphotericin B (Mediatech, VWR). The cells were differentiated into macrophage-like cells by the phorbol ester, PMA (1 µg/mL, Sigma), 24 hour prior to experimentation. The transformed cells were removed from the flask by scraping, and centrifuged at 450 g for 5 min. The resulting cell pellet was suspended at 1.0 × 106 cells/mL and exposed to caspase-1 inhibitors at concentrations described below (0.5%–0.005%), LPS [20ng/mL] and TiO2 nanowires (100 µg/mL). Experiments were conducted in 96-well plates for 24 h in 37 °C water-jacketed CO2 incubators (ThermoForma).
Toxicity Assay
Cell viability was determined by MTS reagent using the CellTiter96 assay (Promega), according to the manufacturer’s protocol. The plate was read at 490 nm.
Cytokine Assays
Human IL-1β DuoSet was obtained from R&D Systems and ELISA assays performed according to the manufacturer’s protocol. The plate was read at 490 nm.
Supplementary Material
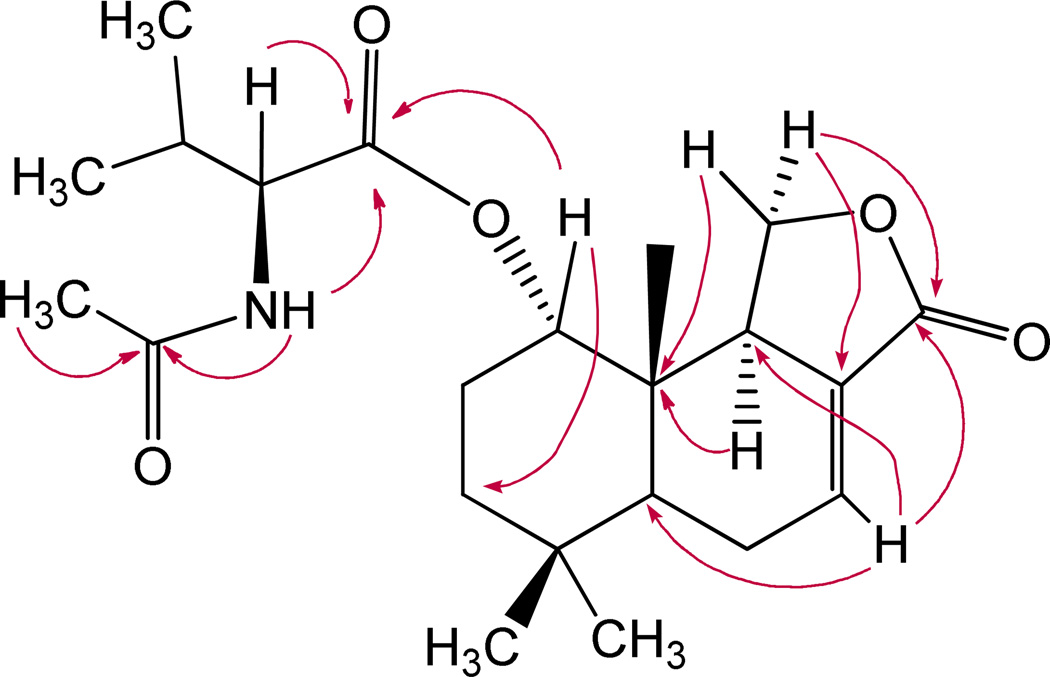
Figure 1.
Key HMBC correlations for berkedrimane 1.
Figure 3.
ACKNOWLEDGEMENTS
We thank B. Parker (University of Montana) for high resolution mass spectrometry data. We thank NSF grant #CHE-9977213 for acquisition of an NMR spectrometer and the MJ Murdock Charitable Trust Ref # 99009:JVZ:11/18/99 for acquisition of the mass spectrometer. The project was supported by NIH grants R01CA139159, P20RR16455-04, P20RR017670, 5P30NS055022, and RC2ES018742.
Footnotes
ASSOCIATED CONTENT
Supporting Information
Experimental details including the 1H NMR spectrum of berkedrimane A (1) in both CDCl3 and C6D6, and of berkedrimane B (2) in CDCl3, the 13C NMR, COSY, HMBC and HSQC spectra for both compounds in CDCl3, and the 1H NMR and 13C NMR spectra of trimethylester 8 in CDCl3. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Wilson ZE, Brimble MA. Nat. Prod. Rep. 2009;26:44–71. doi: 10.1039/b800164m. [DOI] [PubMed] [Google Scholar]
- 2.For the previous isolation of secondary metabolites from this lake see: Stierle AA, Stierle DB, Parker K, Goldstein E, Bugni T, Baarson C, Gress J, Blake D. J. Nat. Prod. 2003;66:1097–1100. doi: 10.1021/np030044w. Stierle DB, Stierle AA, Hobbs JD, Stokken J, Clardy J. Org. Lett. 2004;6:1049–1052. doi: 10.1021/ol049852k. Stierle AA, Stierle DB, Kemp K. J. Nat. Prod. 2004;67:1392–1395. doi: 10.1021/np049975d. Stierle AA, Stierle DB, Kelly K. J. Org. Chem. 2006;71:5357–5360. doi: 10.1021/jo060018d. Stierle DB, Stierle AA, Patacini B. J. Nat. Prod. 2007;70:1820–1823. doi: 10.1021/np070329z. Stierle AA, Stierle DB, Patacini B. J. Nat. Prod. 2008;71:856–860. doi: 10.1021/np0705054.
- 3.Franchi L, Eigenbrod T, Muñoz-Planillo R, Nuñez G. Nat. Immunol. 2009;10:241–256. doi: 10.1038/ni.1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ona VO, Li M, Vonsattel JPG, Andrews LJ, Khan SQ, Chung WM, Frey AS, Menon AS, Li XJ, Stieg PE, Yuan J, Penney JB, Young AB, Cha JHJ, Friedlander RM. Nature. 1999;399:263–267. doi: 10.1038/20446. [DOI] [PubMed] [Google Scholar]
- 5.Li M, Ona VO, Guegan C, Chen M, Jackson V, Andrews LJ, Olszewski AJ, Stieg PE, Lee J, Przedborski S, Friedlander RM. Science. 2000;288:335–339. doi: 10.1126/science.288.5464.335. [DOI] [PubMed] [Google Scholar]
- 6.Rabuffetti M, Sciorati C, Tarozzo G, Clementi E, Manfredi AA, Beltramo M. J. Neurosci. 2000;20:4398–4404. doi: 10.1523/JNEUROSCI.20-12-04398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ming X, Li W, Maeda Y, Blumberg B, Raval S, Cook SD, Dowling PC. J. Neurol. Sci. 2002;197:9–18. doi: 10.1016/s0022-510x(02)00030-8. [DOI] [PubMed] [Google Scholar]
- 8.Furlan R, Martino G, Galbiati F, Poliani PL, Smiroldo S, Bergami A, Desina G, Comi G, Flavell R, Su MS, Adorini L. J. Immunol. 1999;163:2403–2409. [PubMed] [Google Scholar]
- 9.Gamblin TC, Chen F, Zambrano A, Abraha A, Lagalwar S, Guillozet AL, Lu M, Fu Y, Garcia-Sierra F, LaPointe N, Miller R, Berry RW, Binder LI, Cryns VL. Proc. Nat. Acad. Sci. U.S.A. 2003;100:10032–10037. doi: 10.1073/pnas.1630428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rudolphi K, Gerwin N, Verzijl N, van der Kraan P, van den Berg W. Osteoarthritis Cartilage. 2003;11:738–746. doi: 10.1016/s1063-4584(03)00153-5. [DOI] [PubMed] [Google Scholar]
- 11.Walters J, Pop C, Scott FL, Drag M, Swartz P, Mattos C, Salvesen GS, Clark AC. Biochem. J. 2009;424:335–345. doi: 10.1042/BJ20090825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavia DL, Lampman GM, Kriz GS. Introduction to Spectroscopy. 3rd edition. Orlando, FL: Harcourt College; 2001. p. 53. [Google Scholar]
- 13.King TJ, Roberts JC, Thompson DJ. J. Chem. Soc. Perkin Trans. 1973:78–80. doi: 10.1039/p19730000078. [DOI] [PubMed] [Google Scholar]
- 14.Wangun HV, Dorfelt H, Hertweck C. Eur. J. Org. Chem. 2006:1643–1646. [Google Scholar]
- 15.Marfey P. Carlsberg Res. Comm. 1984:591–596. [Google Scholar]
- 16.Turner WB, Aldridge DC. Fungal Metabolites II. Academic Press; 1983. p. 377. [Google Scholar]
- 17.Brandaenge S. Chem. Scand. Ser. B. 1984:837–844. [Google Scholar]
- 18.Hong J, Yang S, Choi YK, Lee CH. J. Coll. Interface Science. 1995;173:92–103. [Google Scholar]
- 19. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?q=sids&reqid=6542588476838064189.
- 20.Abulafia DP, de Rivero Vaccari JP, Lozano JD, Lotocki G, Keane RW, Dietrich WD. J. Cereb. Blood Flow Metab. 2009;29:534–544. doi: 10.1038/jcbfm.2008.143. [DOI] [PubMed] [Google Scholar]
- 21.de Rivero Vaccari JP, Lotocki G, Alonso OF, Bramlett HM, Dietrich WD, Keane RW. J. Cereb. Blood Flow Metab. 2009;29:1251–1261. doi: 10.1038/jcbfm.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juliana C, Fernandes-Alnemri T, Wu J, Datta P, Solorzano L, Yu J, Meng R, Quong AA, Latz E, Scott CP, Alnemri ES. J. Biol. Chem. 2010;285:9792–9802. doi: 10.1074/jbc.M109.082305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.