Abstract
Uncovering the cis-regulatory logic of developmental enhancers is critical to understanding the role of non-coding DNA in development. However, it is cumbersome to identify functional motifs within enhancers, and thus few vertebrate enhancers have their core functional motifs revealed. Here we report a combined experimental and computational approach for discovering regulatory motifs in developmental enhancers. Making use of the zebrafish gene expression database, we computationally identified conserved non-coding elements (CNEs) likely to have a desired tissue-specificity based on the expression of nearby genes. Through a high throughput and robust enhancer assay, we tested the activity of ~100 such CNEs and efficiently uncovered developmental enhancers with desired spatial and temporal expression patterns in the zebrafish brain. Application of de novo motif prediction algorithms on a group of forebrain enhancers identified five top-ranked motifs, all of which were experimentally validated as critical for forebrain enhancer activity. These results demonstrate a systematic approach to discover important regulatory motifs in vertebrate developmental enhancers. Moreover, this dataset provides a useful resource for further dissection of vertebrate brain development and function.
Keywords: enhancers, motifs, conserved non-coding elements, zebrafish, brain development
Introduction
The development of an organism is dictated by the precise patterns of gene expression orchestrated in space and over time. One type of regulatory non-coding DNA, known as enhancers (Blackwood and Kadonaga, 1998; Khoury and Gruss, 1983; Levine and Tijan, 2003), is critical for driving tissue-specific and time-dependent gene expression during embryonic development, thus modulating distinct epigenetic states in the given cell type. Understanding the cis-regulatory codes of developmental enhancers is crucial to uncovering the function of non-coding DNA in cell fate specification and tissue or organ patterning, and shall also facilitate single-nucleotide-polymorphism (SNP)-based association studies of human developmental disorders.
Traditionally, enhancers have been identified through empirical deletion analysis and in vitro footprinting of selected gene loci (Davidson, 2001; Small et al., 1992). In recent years, comparative genomic analyses have suggested that at least 5% of the sequences in the human genome are under negative or purifying selection and are hence functionally important (Pheasant and Mattick, 2007; Waterston et al., 2002). Even distantly related species, such as human and the puffer fish Fugu rubripes or the teleost zebrafish Danio rerio, share many thousands of such conserved non-coding elements (CNEs), far beyond what would be expected in the absence of selective pressure. The majority of these sequences are outside known protein-coding regions, making them potential candidates for enhancers (Pennacchio and Rubin, 2001). Indeed, randomly selected CNEs have been tested experimentally in mice (Pennacchio et al., 2006; Visel et al., 2008) and in zebrafish (Navratilova et al., 2009; Shin et al., 2005; Woolfe et al., 2007; Woolfe et al., 2005), confirming that CNEs are a good source for potential enhancers. Recently, chromatin immunoprecipitation with the enhancer-associated protein P300 followed by massively parallel sequencing has also been shown to identify tissue-specific enhancers (Visel et al., 2009).
Despite these advancements in enhancer detection, only a few vertebrate tissue-specific enhancers have been analyzed to uncover novel functionally critical motifs de novo (Rastegar et al., 2008)(Pennacchio et al., 2006). Indeed, one challenging goal toward understanding the function of the genome is to unveil the regulatory logic embedded in DNA sequences. The classical experimental approach of deletion/mutation analyses targeting random nucleotides in an enhancer is too laborious and often not practical. Computational searches of transcription factor binding sites using methods such as TRANSFAC (Reuter and Schacherer, 2000) are biased by our existing knowledge and moreover, predict too many sites to be validated experimentally in an effective way. Although there are now several hundred human enhancers whose activity has been verified in mouse, de novo motif detection has not been substantially investigated for them. For example, in an extensive experimental study of human enhancers (Pennacchio et al., 2006), de novo motif prediction was performed only on one set of four forebrain-specific enhancers, and the predicted motifs were not experimentally validated.
Here we describe a systematic approach, employing existing experimental and bioinformatic methodologies and the vertebrate model organism zebrafish, to discover novel functional motifs within tissue-specific enhancers. As an example, we focused our analysis on the developing anterior brain (fore- or mid-brain regions). The establishment of the vertebrate anterior brain character requires suppression of the activity of posteriorizing signals including BMP, Wnts, Fgfs, Nodal, and retinoic acids (Wilson and Houart, 2004). In addition, a number of evolutionarily conserved transcription factors are expressed in specific regions along the anterior-posterior neural axis (Bally-Cuif and Boncinelli, 1997). For example, otx2 (Simeone et al., 1992) and too few/fezl (Guo et al., 1999; Hashimoto et al., 2000; Levkowitz et al., 2003) are specifically expressed in the anterior brain at early somitic stages. A set of Hox genes and Krox-20 are specifically expressed in some hindbrain rhombomeres (Lumsden and Krumlauf, 1996). An elaborate gene regulatory network is likely needed to translate the complex extrinsic signals into distinct anterior-posterior identity in neural progenitor cells. However, little information on such regulatory network is currently available.
In this study, we selected a set of 101 CNEs near genes expressed either in the anterior or posterior (hind-) brain regions. Subsequently, we tested their ability to drive expression of a cis-reporter gene using an improved transient transgenesis method, which significantly alleviates the problem of mosaic expression. We found that 25% of tested CNEs exhibited the desired anterior brain enhancer activity. Application of de novo motif prediction algorithms on a group of 13 forebrain enhancers uncovered five top-ranked 6-nucleotide motifs that were significantly enriched in these enhancers. Experimental analyses of these motifs in zebrafish revealed that all five are functionally critical for anterior brain enhancer activity (hence a validation rate of 100%). Finally, we built an online resource (zebrafishcne.org) to store information on these and future experiments into the coding logic of developmental enhancers.
These findings demonstrate a practical way to uncover functional motifs of vertebrate developmental enhancers. The data resources we have developed provide important tools for further dissection of vertebrate brain development and function.
Materials and methods
Bioinformatic identification of expression pattern-associated CNEs
Based on literature and gene expression database in zfin (http://www.zfin.org), groups of anterior brain specific/enriched or posterior brain specific/enriched genes were chosen as candidates for selection of nearby CNEs (Table 1 and Fig. S1). CNEs were then selected from amongst those with a minimum 60% identity and 100 bp conservation between zebrafish (zv6) and human (hg18), which are straightforward constraints relevant to our experimental organism and human. Most CNEs were chosen using cneViewer (cneviewer.zebrafishcne.org)(Persampieri et al., 2008), a tool that we have created to make use of publicly available zebrafish tissue and temporal gene expression data. cneViewer allows users to specify an anatomy and developmental timing and retrieve CNEs near genes expressed with that specificity. cneViewer was supplemented by individual inspection of CNEs using tools such as the ECRbrowser (Ovcharenko et al., 2004) and UCSC genome browser (Kent et al., 2002). Other groups have used assorted thresholds to identify highly conserved noncoding elements for experimental study (Nobrega et al., 2003; Pennacchio et al., 2006; Woolfe et al., 2005), though they are generally comparable to the length and identity criteria we have used here.
Table 1.
Predicted and validated pattern-associated CNEs near brain-expressed genes.
| Strong expression in the anterior CNS | Strong expression in anterior CNS and other regions | Strong expression in the posterior CNS | |
|---|---|---|---|
| Genes | 20 fezl, titf1b, six3a, arx, six3b, vax1, emx2, arl3l1, elov4, foxh1, bhlhb5, arr3l, zgc 103611, barhl2, otx1, dlx1a/dlx2a, dlx5a/dlx6a, sox5 , stka, calrl2 | 18 lmo1, bcat, sb:cb648, sp8l, isl1, sb:cb306, atp6v1ba, six1, ckb, fxr1, pax2a, prox1, fbp1l, foxp1b, LOC797593, LOC797945, zgc: 66439, gli3 | 11 hoxa2b, hoxa13b, hoxa3a, hoxa13a, egr2b, eng1b, eng2b, hoxb1b, tall, irx4a, hprt1l |
| #of computationally predicted CNEs (-500kb-500kb 100bp, 60%) | 282 | 199 | 46 |
| # of experimentally validated CNEs | 49 | 30 | 22 |
Molecular cloning, plasmids, and site-directed mutagenesis
The enhancer activity detection plasmid, termed pT2KXIGQ, was derived from pT2KXIG with minor modification: First, pT2KXIG was digested with BglII and NruI, then self-ligated after T4 fill-in. EF-1α promoter was replaced with the E1B minimal promoter. For functional assays, each individual CNE or motif was cloned into XhoI and BglII sites upstream of the E1B minimal promoter.
Site-directed mutagenesis was carried out by a PCR strategy. Mutagenic primers and flanking primers were used to generate two intermediate PCR products with the overlapping ends that also contain the desired nucleotide changes. The intermediate products were denatured, re-annealed at their overlapping complementary regions, and used as templates for a second round of PCR. The resulting fusion product is further amplified using the flanking primers. Final product was cloned between Xho I and Bgl II sites of pT2KXIGQ. Deletions were generated as described above, except that the internal primers contain the desired deletions. Plasmids were subjected to sequencing to ensure that only the desired mutations have been introduced into the constructs.
Animal husbandry and transgenesis
Wild type zebrafish were maintained at 28.5°C according to standard protocols (Westerfield, 1995). Fertilized eggs were collected and then micro-injected with enhancer activity detection constructs at one-cell stage. After injection, embryos were raised at 28.5°C, and staged according to published methods ((Kimmel et al., 1995), in Daneau's solution (30× stock: 174mM NaCl, 21mM KCl, 12mM MgSO4, 18mM Ca(NO3)2, 15 mM HEPES, pH7.6). To prevent pigment formation, 0.003% phenylthiocarbamide was added at around the tailbud stage.
At least 50 embryos were injected for each CNE construct. At least 20 embryos were evaluated for each CNE activity. The reported expression pattern for each CNE was observed in at least 50% of embryos. For the generation of stable zebrafish transgenic lines, embryos injected with CNE constructs were raised to adulthood. Founders that transmitted the transgene through the germline were kept, and the next generations were raised to adulthood. The reported stable transgenic pattern of each CNE construct was observed in at least three independent transgenic lines.
De novo motif prediction and transcription factor binding site analysis
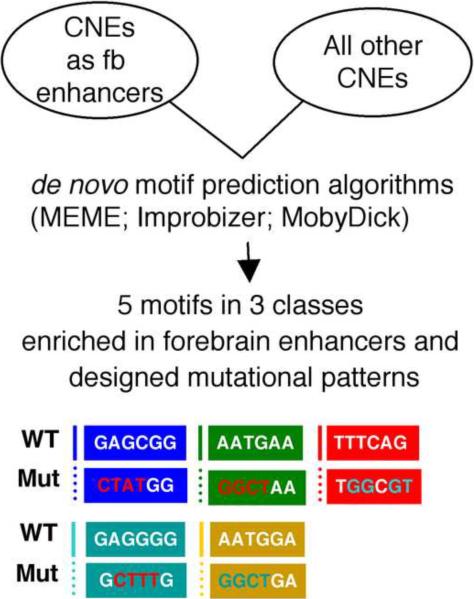
We applied several computational tests to identify motifs important to the activity of forebrain enhancers. To maximize the quality of predicted motifs, we focused on motifs with consistent evidence across multiple motif detection algorithms, a pragmatic approach that has been advocated in the motif detection literature (Tompa et al., 2005). We first searched for common sequence motifs in a set of 13 experimentally validated forebrain-expressed zebrafish CNEs [the first 13 forebrain CNEs that we discovered, 5010 nucleotides (nt) total]. We then applied the programs MEME (settings mod=oops, nmotifs=6, minw=6, maxw=6, revcomp and other settings default) and Improbizer (motif length=6, ignore location, reverse complement, 3 occurrences per sequence, right align, restrain expansionist tendencies and other settings default) to the data. We found 3 classes of motifs robustly predicted by both methods: (gagcgg~gagggg,, tttcag, aatgaa~aatgga). These motifs were strong enough to be found whether or not the reverse complement option was employed. They were not found when the bases in each CNE were shuffled, as expected (this shuffling check was performed 5 times).
We ran a variety of additional algorithms to further test the quality of the motifs. Ao et al (Ao et al., 2004) described a method in which biologically active motifs were found by applying Improbizer to sequences near tissue-specific genes and removing motifs found near other types of genes. To parallel this approach, we applied MEME and Improbizer to a set of 38 CNEs that did not drive reporter gene expression in forebrain (the first 38 CNEs found to not drive forebrain gene expression). Motifs in this control set did not overlap those found for the anterior brain set, showing that the 5 motifs meet the standard of (Ao et al., 2004). As another verification, we applied the MobyDick maximum-likelihood motif detection algorithm (Bussemaker et al., 2000) to the 538 CNEs (>60% human-zebrafish identity, >100 bp) within 500kb of a curated set of 34 forebrain specific genes. Two of the motifs (gagcgg~gagggg: MobyDick gagggg p=1×10−3 and tttcag: MobyDick ggtttcag p=1×10−12) were found significant by Moby Dick, using the set of all remaining CNEs in the genome (Persampieri et al., 2008) for contrast. When the full experimental CNE set was completed, we counted the number of copies of the 5 motif strings in all experimental CNEs (on both the forward and reverse strands). We found significant enrichment of the 5 strings (ccgctc~cccctc, ctgaaa, ttcatt~tccat) in CNEs driving forebrain expression over those not driving forebrain expression (7.5 motif copies/1000 bp in a dataset of 11852 bp vs 5.0 copies/1000 bp in a dataset of 19995 bp, R prop.test P = 0.0061). We also ran the motif-prediction algorithms AlignAce and Weeder on the 13 forebrain CNEs. The gagcgg motif was returned as the one “interesting motif” by Weeder. The gagcgg motif was also found as one of the top motifs in AlignACE (MAP score 1.7) though the other motifs were not found. Although not all algorithms gave identical results, the relative consistency across algorithms suggested these motifs were worth testing experimentally.
Matching of motifs to transcription factor binding sites was done using the motif position-specific scoring matrices from MEME and Improbizer. The motif matching program STAMP (default settings) was used with a file of human and zebrafish transcription factor binding matrices obtained from TRANSFAC Professional (Mahony and Benos, 2007; Wingender et al., 2000). Output from STAMP was manually reviewed and the lowest E-value candidate transcription factors were chosen.
Imaging analysis
Live zebrafish embryos injected with CNE reporter constructs were photographed using a Zeiss epi-fluorescent compound microscope connected with a CCD camera. For high resolution imaging analysis, embryos were immuno-labeled with chicken anti-GFP (Abcam), mouse anti-Hu (Invitrogen) antibodies, DNA dyes Hoechst 34580 (Invitrogen), and subjected to photography on a confocal microscope.
Results
Identification of CNEs near genes expressed in specific regions of the developing zebrafish brain
The developing vertebrate brain shares a conserved structure and can be crudely divided into the forebrain, midbrain, and hindbrain along the anteroposterior (AP) axis. Such regionalization occurs early during embryonic development (Kiecker and Lumsden, 2005; Lumsden and Krumlauf, 1996; Redies and Puelles, 2001; Rubenstein et al., 1998; Stern, 2001; Wilson and Houart, 2004). Further subdivisions along both AP and dorsoventral (DV) axes give rise to structures including the telencephalon, thalamus, hypothalamus, tectum, tegmentum, and hindbrain rhombomeres r1 to r7. Subsequently, distinct cell types populate these brain subdivisions.
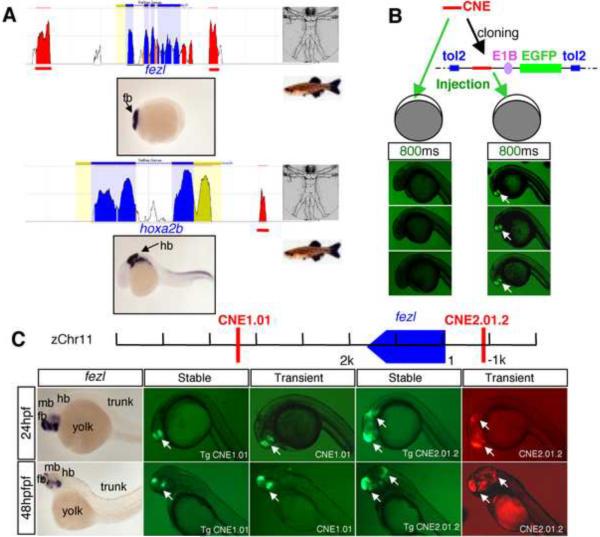
In this study, we focused on a set of genes that display enriched expression in either the anterior (fore- or mid-) or the posterior (hind-) brain during somitogenesis stages of zebrafish embryos (from tailbud to 24 hours post fertilization, -hpf)(Thisse and Thisse, 2005)(Table 1, and Fig. S1). CNEs were computationally identified within 500 kilobases (kb) on both sides of these genes, using the criteria of a minimal 60% identity and greater than 100 base pairs (bps) in length between human and zebrafish orthologous genes. These criteria identify sequences that are far more conserved than would be expected for any neutrally evolving DNA (Fig. 1A, and Table 1). From a total number of 527 computationally predicted CNEs, 101 were selected as an experimental training set, based on combined criteria of proximity to the gene start site and diversity: closer CNEs were generally preferred, and broader coverage of genes in our list was targeted (Table 1).
Figure 1.
A Tol2-based system provides robust and high throughput in vivo enhancer detection in zebrafish
To fully evaluate the efficiency and sensitivity of enhancer detection methodologies, two transient transgenesis methods in zebrafish were compared (Fig. 1B). In the first method, which has been reported to work effectively in zebrafish (Woolfe et al., 2005), each CNE (~20 tested) was PCR amplified and the purified PCR product was micro-injected directly into zebrafish embryos together with the reporter PCR product composed of EGFP under the control of a minimal promoter. Both E1B and the mouse beta-globin basal promoters were tested. This method is supposedly of high efficiency because it obviates the need for cloning each CNE into a plasmid vector. However in our hands, for most CNEs tested, the GFP signal was too weak to be viewed in live transgenic embryos (Fig. 1B), hence requiring additional immunocytochemistry (with an anti-EGFP antibody) for pattern visualization. We therefore tested a second method, in which each CNE was cloned into a plasmid containing the E1B minimal promoter and a fluorescent reporter gene (GFP or mCherry) with the Tol2 transposon backbone (Kawakami and Shima, 1999), and co-injected with Tol2 transposase mRNA into zebrafish embryos. This method yielded robust signals, and moreover, there was little mosaicism in GFP patterns, possibly due to early integration of the transposon facilitated by the Tol2 transposase (Fig. 1B). For each CNE enhancer, at least 20 embryos were examined, greater than 50% of which showed robust and consistent patterns. With this method, we also established stable transgenic lines for 2 CNEs. A comparison of the reporter patterns in transient versus stable transgenics revealed a good match (Fig. 1C). Based on these results, we decided that the Tol2-based method represents a more reliable and effective system for our CNE analysis in zebrafish.
Analysis of expression pattern-associated CNE training sets reveals many distinct spatial and temporal enhancers
Using the transposon-based method described above, we functionally tested the activity of our 101 selected CNEs in zebrafish. Given our research interest in the anterior brain, 79 were selected near anterior brain-expressed genes (including genes with detectable strong expression only in the anterior brain, and genes with strong expression in the anterior brain and elsewhere), whereas 22 were selected from near posterior brain-expressed genes (Table 1). We examined the CNE-driven reporter expression pattern at two developmental stages, 24 hpf and 48 hpf. The results of our analysis are summarized in Table 2, Fig. 2, and Fig. S2. The chromosomal location of each CNE shown in Fig. 2, as well as nearby gene expression patterns, can be found in Fig. S2. For each CNE analyzed, at least 50 embryos were injected, out of which at least 20 embryos were evaluated for CNE activity, and the reported expression pattern was observed in at least 50% of evaluated embryos.
Table 2.
Summary of the activity of experimentally validated CNE training sets.
| CNE category | CNEs near genes expressed in the anterior CNS (79) | CNEs near genes expressed in the posterior CNS (22) |
|---|---|---|
| CNE activity | ||
| Anterior brain enhancers | 20 | 1 |
| Posterior brain enhancers | 4 | 3 |
| Other tissue enhancers | 10 | 2 |
| Temporal enhancers | 14 | 2 |
| Broad pattern enhancers | 29 | 6 |
| No enhancer activity | 15 | 10 |
Note: Certain CNEs have enhancer activity in multiple regions, and thus have been included in multiple categories.
Figure 2.
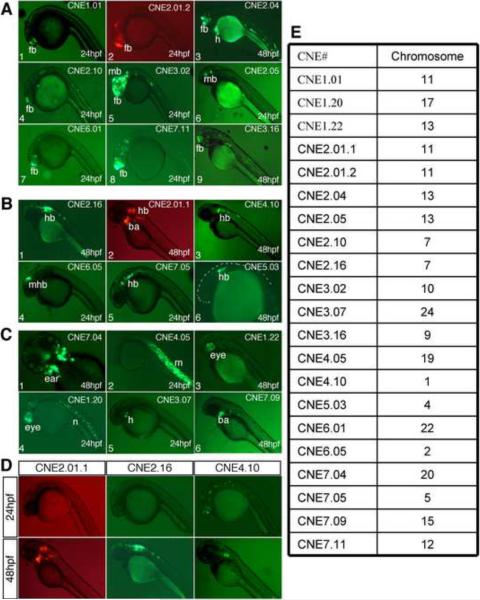
Overall, we found that 76/101 of the CNEs were enhancers, either in specific tissues or broadly in the embryo, providing support for the general concept of CNEs as transcriptional enhancers. Moreover, our approach resulted in the identification of a substantial number of CNEs able to drive expression in the desired tissue of zebrafish. For example, 20/79 CNEs (25%), chosen based on their proximity to genes expressed in the anterior brain, displayed enriched activity in the developing anterior brain (Fig. 2A, and Table 2). Some of these CNEs drove even more finely subdivided expression patterns. For example, CNE 2.10 drove reporter expression in the telencephalon, and CNE2.05 drove reporter expression in the midbrain tegmentum (Fig. 2A, panels 4 and 6). 3/22 (14%) posterior brain-associated CNEs displayed enriched activity in the developing hindbrain (Fig. 2B, and Table 2). Finally, 12/101 CNEs displayed enhancer activity specific for all other tissues combined (Fig. 2C, and Table 2).
One major advantage of zebrafish is that their development can be followed in real time, thus allowing the observation of temporal enhancer activity. A significant fraction of CNEs also exhibited temporally restricted activity (Fig. 2D, and Table 2). 16/76 (21%) CNEs that drove reporter expression showed region-specific expression that clearly differed in at least one anatomical location between the two time points that we examined, 24 hpf and 48 hpf. This result indicates that temporal specificity of CNEs may be a common theme.
High-resolution analysis reveals brain subdivision-specific activity of anterior brain enhancers
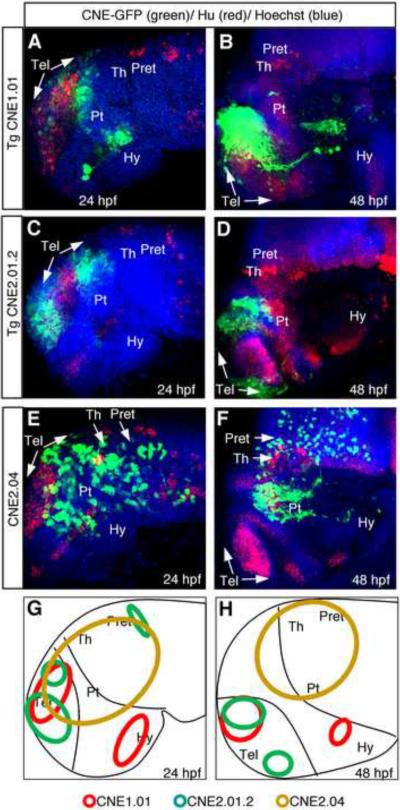
Based on anatomical features, the anterior brain can be further sub-divided into multiple regions including telencephalon, hypothalamus, prethalamus, thalamus, pretectum, tectum and tegmentum (Wilson and Houart, 2004). To gain a better understanding of the region-specific activity of anterior brain enhancers, we carried out high-resolution analyses of embryos expressing reporter constructs driven by a subset of identified anterior brain enhancers (Fig. 3). Three CNEs, CNE1.01, CNE2.01.2, and CNE2.04, and two developmental stages, 24 hpf and 48 hpf, were analyzed. Embryos expressing CNE driven reporters were processed by triple labeling with anti-GFP antibody, anti-Hu antibody (labeling new born neurons), and Hoescht dye (labeling DNA), and visualized through confocal microscopy. Our analyses revealed distinct brain subdivision-specific activity of individual CNEs at these developmental stages. CNE1.01 displayed largely restricted activity in posterior-ventral telencephalon and in distinct cell clusters in the hypothalamus (Fig. 3A–B). CNE2.01.2 drove reporter expression in dorsal-anterior and posterior telencephalon (but not in the medial telencephalon) as well as in a small region in the prethalamus at 24 hpf (Fig. 3C). At 48 hpf, medial telencephalon remained devoid of reporter expression, which was detected in subdivisions of telencephalon and prethalamus (Fig. 3D). CNE2.04 exhibited activity in the posterior telencephalon, prethalamus, thalamus, and pretectum regions (Fig. 3E–F). Together, these analyses reveal distinct activity of CNEs in various brain sub-divisions at the developmental stages analyzed.
Figure 3.
De novo motif prediction reveals short motifs enriched in anterior brain enhancers
We next applied de novo motif prediction algorithms to uncover potentially functional motifs within the identified anterior brain enhancers (Fig. 4). To identify motifs with the best chance of functional activity, we searched for motifs with consistent evidence across multiple prediction algorithms, a practical and stringent approach that has been espoused in the motif detection literature (Tompa et al., 2005). 13 CNEs (5010 nucleotides total) with forebrain enhancer activity (Table 3) were used for this analysis. We applied the programs MEME (Bailey and Elkan, 1994) and Improbizer (Ao et al., 2004)(The Improbizer algorithm is available online at www.cse.ucsc.edu/~kent/improbizer/improbizer.html) to the data. We searched for motifs of 6 base pairs (bps) in length, since this is approximately the length of known transcription factor binding sites, and because longer motifs are too rare at background occurrence rates to be evaluated accurately in a dataset of this size. Five 6-base pair motifs were identified robustly by both methods, and we categorized these into 3 classes: 1) GAGCGG~GAGGGG, 2) TTTCAG, 3) AATGAA~AATGGA (Fig. 4). The locations of these motifs in the 13 CNEs are delineated in Table 3.
Figure 4.
Table 3.
Anterior brain CNEs used for de novo motif prediction
| CNE1.01 | 142_aatgga | 230_ccgctc | 262_aatgaa | 369_ctgaaa | 400_aatgga | 440_ccgctc | 509_aatgaa | 577_gagcgg | 657_gagggg |
| CNE1.06 | 163_ttcatt | 181_gagggg | 193_gagcgg | 204_aatgaa | 233_ttcatt | 286_cccctc | 309_ttcatt | ||
| CNE1.16 | 3_tttcag | 38_tccatt | 192_cccctc | 222_aatgga | 335_gagggg | 410_ccgctc | 429_aatgaa | 592_aatgga | |
| CNE1.20 | 37_cccctc | 253_ctgaaa | 285_tccatt | ||||||
| CNE2.01.2 | 125_aatgaa | 243_ttcatt | 308_tttcag | 553_tttcag | 580_aatgaa | ||||
| CNE2.04 | 8_tttcag | 166_ctgaaa | 247_ttcatt | 274_gagcgg | 481_cccctc | 491_ccgctc | 499_gagggg | 582_gagggg | |
| CNE2.05 | 67_ccgctc | 105_tccatt | |||||||
| CNE2.10 | 5_gagggg | ||||||||
| CNE2.17 | 185_ccgctc | 228_ctgaaa | 282_tccatt | 398_aatgga | 439_gagcgg | 492_aatgaa | 499_tttcag | ||
| CNE3.05 | 99_tccatt | ||||||||
| CNE3.06 | 253_cccctc | 312_aatgaa | |||||||
| CNE6.01 | 96_ctgaaa | 99_tttcag | 320_ttcatt | 380_ctgaaa | |||||
| CNE7.11 | 38_ccgctc | 67_tttcag |
Note: The numbers indicate the motif location in the CNEs. Both orientations of the motifs are indicated.
Several other computational tests provided support for these motifs (details in Methods), including application of the Mobydick, AlignAce, and Weeder algorithms, as well as analysis of motifs found in CNEs not driving anterior brain expression and analysis of motifs in anterior-driving CNEs with the bases shuffled. After manual analysis of these multiple computational results, we selected five best motifs to be tested experimentally.
Mutating each of the five predicted motifs impairs the forebrain enhancer activity of CNE2.01.2 and CNE1.01
To determine whether these five predicted motifs are critical for anterior brain enhancer activity, mutagenesis was carried out in selected CNEs, followed by in vivo reporter assays in zebrafish. To be consistent, we mutated 4 nucleotides in each motif. We chose to mutate the nucleotides that are conserved between zebrafish and human. In cases where fewer than four conserved nucleotides are found in a given motif, we randomly chose additional non-conserved nucleotides and mutated them to different ones (Fig. 4). CNE 2.01.2 and CNE1.01, which are located 5' and 3' respectively to the fezl (also known as fezf2) gene, were selected for analysis (See Fig. 1C). fezl is critical for forebrain patterning and neurogenesis in vertebrates (Chen et al., 2005; Hirata et al., 2006; Hirata et al., 2004; Jeong et al., 2007; Jeong et al., 2006; Levkowitz et al., 2003; Molyneaux et al., 2005).
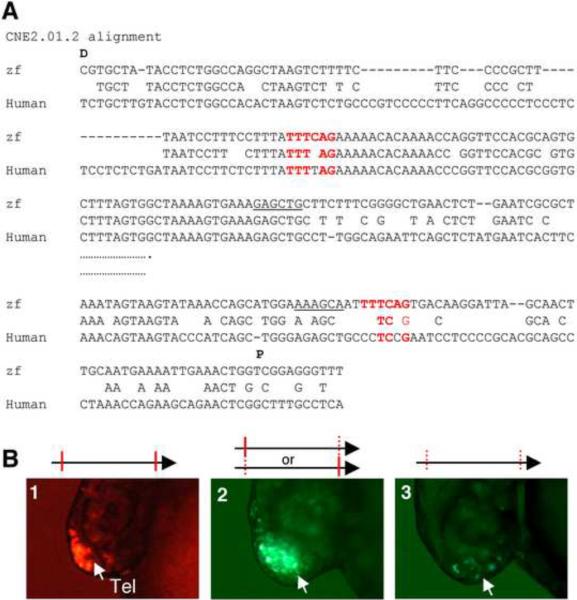
CNE2.01.2 is ~400 bp in length and nested in the CNE2.01. It contains 2 copies of the de novo predicted motif TTTCAG (243 nucleotides apart): one is located in a conserved stretch of sequence (with 5/6 nucleotides being identical between the zebrafish and human motifs) while the other is not (with 3/6 nucleotides being identical between the zebrafish and human motifs) (Fig. 5A). CNE2.01.2 exhibits a strong enhancer activity in the telencephalon (Fig. 5B, panel 1, 95%, n=100). Mutations in either the proximal (19% with reduced reporter expression and the remaining 81% with normal expression, n=68) or the distal copy (19% with reduced reporter expression and the remaining 81% with normal expression, n=57) of the motif partially impair the activity of CNE2.01.2 (Fig. 5B, panel 2). When both copies of the motif were mutated, 82% embryos lost reporter expression and the 18% embryos had significantly reduced reporter expression (Fig. 5B, panel 3, n=34). Together, these results indicate that TTTCAG is a critical code for the forebrain enhancer activity of CNE2.01.2, and the two copies of TTTCAG motif carry out partially redundant functions.
Figure 5.
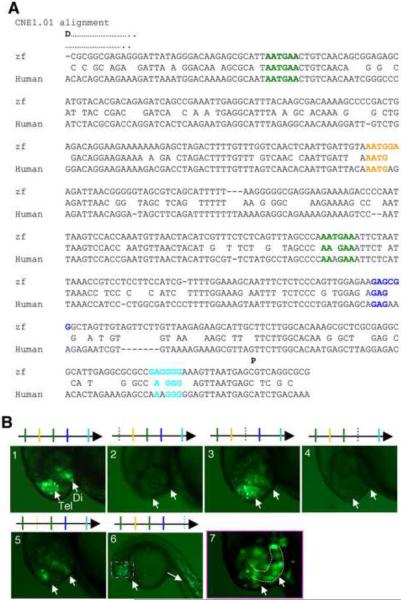
We next analyzed CNE1.01, which is ~500 bp in length (Fig. 6A): It contains four de novo predicted motifs (2 copies of AATGAA, one copy each of AATGGA, GAGCGG, GAGGGG), and the relative conservation of these motifs between zebrafish and human are shown in Fig. 6A. CNE1.01 exhibits enhancer activity in both the telencephalon and diencephalon (Fig. 6B, panel 1, 96%, n=100). When the distal copy of AATGAA was mutated, the activity of CNE1.01 was abolished (Fig. 6B, panel 2, 91%, n=43). When the proximal copy of AATGAA was mutated, 30% embryos showed no reporter expression, while the rest displayed reduced reporter expression, particularly in the diencephalon (Fig. 6B, panel 3, 70%, n=44). Likewise, mutating the motif GAGCGG or AATGGA also significantly impaired the CNE enhancer activity (Fig. 6B, panel 4, 62%, n=34, and panel 5, 71%, n=42). Interestingly, mutating the motif GAGGGG led to ectopic enhancer activity in the muscle and eyes (Fig. 6B, panels 6 and 7, 80%, n=44), suggesting that this motif mediates an inhibition of gene expression in the eyes and muscle, or alternatively, the introduced mutations led to a gain-of-function effect in these tissues.
Figure 6.
To determine whether mutating any residue in a CNE may abolish their enhancer activity, we mutated two randomly chosen stretches of nucleotide sequences in CNE 2.01.2, one in a highly conserved region, and the other in a less conserved region (Fig. 5A, underlined). Alone or in combination, we did not see any effect on the enhancer activity (data not shown). Taken together, these experimental analyses have validated the computational prediction by demonstrating that all five de novo predicted short motifs (100% validation rate) represent critical codes for forebrain enhancer activity.
A Database for Functional CNE Analysis in Zebrafish
We have placed all experimentally validated CNEs and the motif analysis in a public database, available at http://zebrafishcne.org. The site will serve as a data repository for current and future experiments from our laboratory and from other laboratories that wish to share information on the analysis of zebrafish CNEs. The database allows direct submission of CNE images and sequences by users, and provides other annotations including genome coordinates, experiment type, specifications of timing and anatomical location via ZFIN-defined terms. This ZFIN-based anatomical organization ties CNE regulatory patterns to the controlled language that is the standard for the thousands of gene expression measurements that have been compiled for zebrafish genes (Sprague et al., 2006). CNEs can be searched based on characteristics including ZFIN stage or anatomy, sequence, chromosome, user, institute, user comments and broad-based expression (positive/negative) characteristics. Outside users will be able to store their data on zebrafish CNE experiments here as well, to improve data sharing among research groups.
The zebrafishCNE database is well-suited for use with cneViewer (cneviewer.zebrafishcne.org) (Persampieri et al., 2008), a companion website to prioritize candidate zebrafish CNE sequences for experimental testing based on their proximity to genes of a desired tissue- and stage-specific expression, as well as other characteristics such as sequence identity, length, and synteny. Together the cneViewer website and the zebrafishCNE database create a workflow for the experimental researcher by simplifying experimental design, data storage, and analysis for zebrafish CNEs.
Discussion
In this study, we have identified vertebrate brain region-specific enhancers through a high throughput analysis of expression-pattern associated CNEs in zebrafish. These data provide a basis for our subsequent identification of functional motifs critical for the activity of these brain-specific enhancers, employing bioinformatic motif prediction algorithms followed by functional validation in vivo. These findings lay an important foundation for future dissection of gene regulatory networks involved in vertebrate brain development, as well as demonstrate a practical way to uncover functional motifs in vertebrate developmental enhancers.
Many of the CNEs we analyzed have specific spatial or temporal enhancer activity, making them versatile tools for engineering desired patterns of gene expression in vivo. Consistent with the previously observed modular nature of enhancer activity, CNE-driven activity was often nested in sub-groups of cells where the endogenous genes are expressed. For example, the zebrafish islet1 gene is expressed in the eyes, forebrain, midbrain, hindbrain cranial motor neurons, cranial sensory neurons, and spinal motor neurons (Appel et al., 1995; Inoue et al., 1994; Korzh et al., 1993; Thisse and Thisse, 2005; Tokumoto et al., 1995). Moreover, a GFP transgenic line driven by the ~15 kb islet1 5' regulatory sequences displays reporter expression in cranial motor neurons and cranial sensory neurons (Higashijima et al., 2000). Our result showed that the CNE 7.05 ~8 kb distal to the islet1 gene drove reporter expression only in hindbrain motor neurons at 24 hpf and mid/hind-brain motor neurons at 48 hpf (Fig. S3A). Conversely, when two unrelated CNEs with distinct enhancer activity were combined, an additional pattern of reporter expression was derived (Fig. S3B), suggesting combinatorial use of CNEs can direct new and desired patterns of gene expression.
Zebrafish CNE sequences that have enhancer activity have essentially the same average cross-species conservation as those that do not (72% identity vs. 70% identity). Base composition is also nearly identical among these sets (43.9% GC vs 43.4% GC). Interestingly, CNEs that drive expression are on average shorter than those that do not (365 bp vs 476, P=0.04), and are also slightly closer to the nearest gene (47kb vs 70kb, P=0.16). These characteristics could provide useful rules of thumb for predicting enhancer-coding CNEs, though their roughness supports the idea that finer structures such as motifs are important. Our study supports a pragmatic approach to motif detection in which one searches for motifs with robust evidence across prediction algorithms. Although the outputs of de novo motif detection algorithms are not always consistent(Tompa et al., 2005), the experimental validations indicate that such algorithms can be effectively used for enhancer motif studies, as was also found in another study using different prediction algorithms but a similar basic approach (Rastegar et al., 2008). The mechanisms by which motifs act in enhancers have been either computationally or experimentally characterized in only a relatively small number of cases (Markstein et al., 2004; Rastegar et al., 2008; Stathopoulos et al., 2002). It is often assumed that most enhancers operate by recruitment of transcription factors; however, in some cases, they can interact with regulatory non-coding RNA (Petruk et al., 2006; Sanchez-Elsner et al., 2006). Our functionally validated motifs match TRANSFAC binding profiles (Mahony and Benos, 2007; Wingender et al., 2000) for transcription factors involved in fundamental developmental/regulatory roles, including the developing central nervous system (Table 4 and Supplementary online text). For example, the transcription factor YY1 associates with the motif TCCATT. YY1 is known to regulate Otx2, an early developmental gene involved in vertebrate head formation, and the regulation occurs via binding of YY1 to an upstream enhancer (Takasaki et al., 2007). The involvement of microRNAs in enhancing transcription has also been reported recently (Place et al., 2008), suggesting that regulatory RNAs could interact with these motifs. Interestingly, there are 8 zebrafish miRNAs in the Sanger Zebrafish miRNA database with sequences complementary to the identified motifs (data not shown). Experimental identification of the trans-regulatory factors, either proteins or miRNAs, which interact with these motifs is an important future goal. Curiously, the functional motif instances do not always have every base conserved across species, suggesting that motif degeneracy and/or species-specific behavior (Hare et al., 2008)are important even in these highly conserved sequences.
Table 4.
Transcription Factors Associated with Motifs
| MOTIF | TRANSCRIPTION FACTOR | Function |
|---|---|---|
| TTCATT | ICSBP/IRF | Regulate viral response in developing vertebrate embryos and inflammatory cytokines (Levraud et al., 2007) |
| TCCATT | YY1 | Regulate Otx2 and vertebrate head formation (Takasaki et al., 2007) |
| CGTAAA | FOXP3 | Forkhead developmental transcription factor (Curiel, 2007) |
| CCCCTC | MZF1 | Kruppel-family zinc-finger, active in cell differentiation (Bieker, 2001) |
| CCGCTC | PAX1 | paired-box transcription factor family, neural tube/column development, active in segmentation (Kafri et al., 2006) |
Motifs were matched to transcription factors in the TRANSFAC database using the STAMP software package (Mahony and Benos, 2007).
About one-third of expression pattern-associated CNEs that we analyzed have no detectable enhancer activity in any tissues at the stages analyzed, contesting the paradigm of CNEs functioning solely as transcriptional enhancers. While such sequences could still have gene regulatory function (e.g. as enhancers at different time points, or as silencing elements), they may also have alternative functions. For example, recent reports suggest that a large fraction of vertebrate genomes may be transcribed (Birney et al., 2007) and some CNEs are likely to encode non-protein-coding regulatory RNAs. Future studies of CNEs for various functional roles will be crucial to understanding the full complexity of the genomic landscape.
Conclusion
Our analyses have delineated a systematic approach, using expression-pattern associated CNEs to reveal tissue-specific enhancers and moreover their core functional motifs. Such an approach can be applied to any tissue/organ of interest, since gene expression profiles for many tissues/organs are available either from gene expression databases in zebrafish (Thisse and Thisse, 2005) and mice (Gray et al., 2004)(and the Allen mouse brain atlas at http://www.brain-map.org), or expression array profiling data (Su et al., 2002). In addition, we have established a central public database of tissue-specific enhancers and motifs in zebrafish to house data on zebrafish CNEs, which we envision will significantly facilitate the use of zebrafish as a model organism for understanding development and modeling human diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Munchua for zebrafish maintenance, Nadav Ahituv and members of Chuang and Guo laboratories for discussions and helpful comments on the manuscript. This work is supported by Boston College Department of Biology startup fund to J.C., NIH grants (HD 051835 and NS 042626) to S.G., and NIH grant (GM 70808) and Packard Fellowship in Science & Engineering to H.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA Supplemental data accompany this manuscript.
REFERENCES
- Ao W, Gaudet J, Kent WJ, Muttumu S, Mango SE. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science. 2004;305:1743–1746. doi: 10.1126/science.1102216. [DOI] [PubMed] [Google Scholar]
- Appel B, Korzh V, Glasgow E, Thor S, Edlund T, Dawid IB, Eisen JS. Motoneuron fate specification revealed by patterned LIM homeobox gene expression in embryonic zebrafish. Development. 1995;121:4117–25. doi: 10.1242/dev.121.12.4117. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology; Menlo Park, California: AAAI Press; 1994. pp. 28–36. [PubMed] [Google Scholar]
- Bally-Cuif L, Boncinelli E. Transcription factors and head formation in vertebrates. Bioessays. 1997;19:127–135. doi: 10.1002/bies.950190207. [DOI] [PubMed] [Google Scholar]
- Bieker JJ. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 2001;276:34355. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281:60–3. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- Bussemaker HJ, Li H, Siggia ED. Building a Dictionary for Genomes: Identification of Presumptive Regulatory Sites by Statistical Analysis. Proc. Natl. Acad. Sci. 2000;97:10096. doi: 10.1073/pnas.180265397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proc. Natl. Acad. Sci. 2005;102:17184–17189. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curiel TJ. Regulatory T-cell development: is Foxp3 the decider? Nat. Med. 2007;13:250. doi: 10.1038/nm0307-250. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Genomic regulatory systems: development and evolution. Academic Press; San Diego: 2001. [Google Scholar]
- Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–7. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- Guo S, Wilson SW, Cooke S, Chitnis AB, Driever W, Rosenthal A. Mutations in the zebrafish unmask shared regulatory pathways controlling the development of catecholaminergic neurons. Dev. Biol. 1999;208:473–487. doi: 10.1006/dbio.1999.9204. [DOI] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet. 2008;4:e1000106. doi: 10.1371/journal.pgen.1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, et al. Expression of the zinc finger gene fez-like in zebrafish forebrain. Mech. Dev. 2000;97:191–195. doi: 10.1016/s0925-4773(00)00418-4. [DOI] [PubMed] [Google Scholar]
- Higashijima S, Hotta Y, Okamoto H. Visualization of cranial motor neurons in live transgenic zebrafish expressing green fluorescent protein under the control of the islet-1 promoter/enhancer. Journal of Neuroscience. 2000;20:206–218. doi: 10.1523/JNEUROSCI.20-01-00206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata T, Nakazawa M, Yoshihara S, Miyachi H, Kitamura K, Yoshihara Y, Hibi M. Zinc-finger gene Fez in the olfactory sensory neurons regulates development of the olfactory bulb non-cell-autonomously. Development. 2006;133:1433–1443. doi: 10.1242/dev.02329. [DOI] [PubMed] [Google Scholar]
- Hirata T, Suda Y, Nakao K, Narimatsu M, Hirano T, Hibi M. Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Dev. Dyn. 2004;230:546–556. doi: 10.1002/dvdy.20068. [DOI] [PubMed] [Google Scholar]
- Inoue A, Takahashi M, Hatta K, Hotta Y, Okamoto H. Developmental regulation of islet-1 mRNA expression during neuronal differentiation in embryonic zebrafish. Dev. Dyn. 1994;199:1–11. doi: 10.1002/aja.1001990102. [DOI] [PubMed] [Google Scholar]
- Jeong J, Einhorn Z, Mathur P, Chen L, Lee S, Kawakami K, Guo S. Patterning the zebrafish diencephalon by the conserved zinc finger protein Fezl. Development. 2007;134:127–136. doi: 10.1242/dev.02705. [DOI] [PubMed] [Google Scholar]
- Jeong J, et al. Neurogenin1 is a determinant of zebrafish basal forebrain dopaminergic neurons and is regulated by the conserved zinc finger protein Tof/Fezl. Proc. Natl. Acad. Sci. 2006;103:5143–5148. doi: 10.1073/pnas.0600337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafri R, Levy M, Pilpel Y. The regulatory utilization of genetic redundancy through responsive backup circuits. Proc. Natl. Acad. Sci. 2006;103:11653–8. doi: 10.1073/pnas.0604883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Shima A. Identification of the Tol2 transposase of the medaka fish Oryzias latipes that catalyzes excision of a nonautonomous Tol2 element in zebrafish Danio rerio. Gene. 1999;240:239–244. doi: 10.1016/s0378-1119(99)00444-8. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G, Gruss P. Enhancer Elements. Cell. 1983;33:313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Kiecker C, Lumsden A. Compartments and their boundaries in vertebrate brain development. Nat. Rev. Neurosci. 2005;6:553–564. doi: 10.1038/nrn1702. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Korzh V, Edlund T, Thor S. Zebrafish primary neurons initiate expression of the LIM homeodomain protein Isl-1 at the end of gastrulation. Development. 1993;118:417–25. doi: 10.1242/dev.118.2.417. [DOI] [PubMed] [Google Scholar]
- Levine M, Tijan R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Levkowitz G, et al. Zinc finger protein too few controls the development of monoaminergic neurons. Nat. Neurosci. 2003;6:28–33. doi: 10.1038/nn979. [DOI] [PubMed] [Google Scholar]
- Levraud JP, Boudinot P, Colin I, Benmansour A, Peyrieras N, Herbomel P, Lutfalla G. Identification of the zebrafish IFN receptor: implications for the origin of the vertebrate IFN system. J. Immunol. 2007;178:4385–94. doi: 10.4049/jimmunol.178.7.4385. [DOI] [PubMed] [Google Scholar]
- Lumsden A, Krumlauf R. Patterning the vertebrate neuraxis. Science. 1996;274:1109–1114. doi: 10.1126/science.274.5290.1109. [DOI] [PubMed] [Google Scholar]
- Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucl. acids Res. 2007;35:W253. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine M. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2387–94. doi: 10.1242/dev.01124. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Navratilova P, Fredman D, Hawkins TA, Turner K, Lenhard B, Becker TS. Systematic human/zebrafish comparative identification of cis-regulatory activity around vertebrate developmental transcription factor genes. Dev. Biol. 2009;327:526–40. doi: 10.1016/j.ydbio.2008.10.044. [DOI] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucl. Acids Res. 2004;32:W280–286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio LA, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Rubin EM. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- Persampieri J, Ritter DI, Lees D, Lehoczky J, Li Q, Guo S, Chuang JH. cneViewer: A Database of Conserved Noncoding Elements for Studies of Tissue-Specific Gene Regulation. Bioinformatics. 2008;24:2418–9. doi: 10.1093/bioinformatics/btn443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, et al. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–21. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pheasant M, Mattick JS. Raising the estimate of functional human sequences. Genome Res. 2007;17:1245–53. doi: 10.1101/gr.6406307. [DOI] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc. Natl. Acad. Sci. 2008;105:1608–13. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegar S, et al. The words of the regulatory code are arranged in a variable manner in highly conserved enhancers. Dev. Biol. 2008;318:366–77. doi: 10.1016/j.ydbio.2008.03.034. [DOI] [PubMed] [Google Scholar]
- Redies C, Puelles L. Modularity in vertebrate brain development and evolution. Bioessays. 2001;23:1100–11. doi: 10.1002/bies.10014. [DOI] [PubMed] [Google Scholar]
- Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucl. Acids Res. 2000;28:316–19. doi: 10.1093/nar/28.1.316. 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Shimamura K, Martinez S, Puelles L. Regionalization of the prosencephalic neural plate. Annu. Rev. Neurosci. 1998;21:445–477. doi: 10.1146/annurev.neuro.21.1.445. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Shin JT, Priest JR, Ovcharenko I, Ronco A, Moore RK, Burns CG, MacRae CA. Human-zebrafish non-coding conserved elements act in vivo to regulate transcription. Nucleic Acids Res. 2005;33:5437–45. doi: 10.1093/nar/gki853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nested expression domains of four homeobox genes in developing brain. Nature. 1992;358:687–690. doi: 10.1038/358687a0. [DOI] [PubMed] [Google Scholar]
- Small S, Blair A, Levine M. Regulation of even-skipped strip 2 in the Drosophila embryo. EMBO J. 1992;11:4047–57. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, et al. The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res. 2006;34:D581–6. doi: 10.1093/nar/gkj086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos A, Van Drenth M, Erives A, Markstein M, Levine M. Whole-genome analysis of dorsal-ventral patterning in the Drosophila embryo. Cell. 2002;111:687–701. doi: 10.1016/s0092-8674(02)01087-5. [DOI] [PubMed] [Google Scholar]
- Stern CD. Initial patterning of the central nervous system: how many organizers? Nat. Rev. Neurosci. 2001;2:92–98. doi: 10.1038/35053563. [DOI] [PubMed] [Google Scholar]
- Su AI, et al. Large-scale analysis of the human and mouse transcriptomes. Proc. Natl. Acad. Sci. 2002;99:4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki N, Kurokawa D, Nakayama R, Nakayama J, Aizawa S. Acetylated YY1 regulates Otx2 expression in anterior neuroectoderm at two cis-sites 90 kb apart. EMBO J. 2007;26:1649–59. doi: 10.1038/sj.emboj.7601619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission. 2005 http://zfin.org.
- Tokumoto M, Gong Z, Tsubokawa T, Hew CL, Uyemura K, Hotta Y, Okamoto H. Molecular heterogeneity among primary motoneurons and within myotomes revealed by the differential mRNA expression of novel islet-1 homologs in embryonic zebrafish. Dev. Biol. 1995;171:578–89. doi: 10.1006/dbio.1995.1306. [DOI] [PubMed] [Google Scholar]
- Tompa M, et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–44. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, et al. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat. Genet. 2008;40:158–60. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book: a guide for the laboratory use of zebrafish, Brachydanio rerio. The University of Oregon Press; Eugene, OR: 1995. [Google Scholar]
- Wilson SW, Houart C. Early steps in the development of the forebrain. Dev. Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, et al. TRANSFAC: an integrated system for gene expression regulation. Nucl. Acids Res. 2000;28:316–9. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, et al. CONDOR: a database resource of developmentally associated conserved non-coding elements. BMC Dev. Biol. 2007;7:100. doi: 10.1186/1471-213X-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A, et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:1–15. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.