Abstract
Members of the fibroblast growth factor (FGF) family play a critical role in embryonic lung development and adult lung physiology. The in vivo investigation of the role FGFs play in the adult lung has been hampered because the constitutive pulmonary expression of these factors often has deleterious effects and frequently results in neonatal lethality. To circumvent these shortcomings, we expressed FGF-3 in the lungs under the control of the progesterone antagonist-responsive binary transgenic system. Four binary transgenic lines were obtained that showed ligand-dependent induction of FGF-3 with induced levels of FGF-3 expression dependent on the levels of expression of the GLp65 regulator as well as the dose of the progesterone antagonist, RU486, administered. FGF-3 expression in the adult mouse lung resulted in two phenotypes depending on the levels of induction of FGF-3. Low levels of FGF-3 expression resulted in massive free alveolar macrophage infiltration. High levels of FGF-3 expression resulted in diffuse alveolar type II cell hyperplasia. Both phenotypes were reversible after the withdrawal of RU486. This system will be a valuable means of investigating the diverse roles of FGFs in the adult lung.
The primary function of the lung is to ensure aeration of the blood. To achieve this, the lung has a complex architecture composed of airways and alveoli lined with a variety of cell types. Perturbation of this pulmonary architecture results in debilitating and/or lethal diseases. Transgenic and knockout technologies have elucidated the role of many factors involved in lung development (1). Transgenic and gene ablation experiments demonstrated that members of the fibroblast growth factor (FGF) family are critical for the regulation of lung development and epithelial cell differentiation (2–4).
The FGFs constitute a family of homologous glycoproteins controlling normal growth and differentiation of mesenchymal, epithelial, and neuroectodermal cell types (5, 6). Deregulation of the expression of these growth factors can result in pathological situations such as tumorigenesis (7–9). FGFs signal through their cognate tyrosine kinase receptors to effect their varying activities on cells. Most FGFs bind to one or more of four known FGF receptors, FGFRs 1–4 (10). The complexity of FGF signaling is increased through the process of alternative splicing, generating many variants of each of these receptors including both soluble and membrane-bound forms (11). All four FGFRs are expressed in the lung, (12, 13) and of these, FGFR2 has been shown to be critical for lung development (2).
FGFR2 is expressed preferentially in epithelial cells during development, and disruption of FGFR2iiib signaling by the expression of a dominant negative form of the receptor results in failure of branching morphogenesis in the lung (2). The FGFs that bind this receptor are FGF-3 (int-2), FGF-7 (keratinocyte growth factor), and FGF-10 (4, 10). FGF-10 was found to be a key regulator of lung branching and morphogenesis because FGF-10-deficient mice were nonviable because of a lack of lung development (2). FGF-7 has also been implicated in regulating pulmonary function in the adult lung by regulating alveolar type II cell differentiation (14) and protecting alveoli from hyperoxic damage (15). FGF-3 is located on human chromosome 11q13 (16). This chromosomal region was shown to be amplified in some head, neck, and pulmonary cancers (17–19). The above observations demonstrate that the FGFR2 signaling axis is important for lung development. However, an in vivo transgenic analysis of the role of this signaling axis in the adult has been hampered because transgenic manipulation of the FGF signaling axis results in neonatal lethality (2). Establishment of a regulatable system for FGF transgene expression in the lungs would allow the investigation of the role these factors play in the adult lung.
To investigate the phenotypic consequences of the activation of FGFR2 expression in the adult lung, FGF-3 was expressed in the lungs of transgenic mice by using the GLp65 bitransgenic regulatory system. This bitransgenic system is based on a previously established ligand-inducible regulator, the Gal-4 DNA-binding domain (GLVP), the transcriptional activity of which is modulated by a progesterone antagonist, RU486 (20). The GLp65 regulator consists of the mutated progesterone receptor ligand binding domain, the DNA binding domain of the yeast Gal4 transcription factor, and the p65 activation domain of the transcription factor NFκB. GLVP and GLp65 regulators have been demonstrated to regulate the expression of target transgenes containing the upstream activating sequences for the yeast Gal4 gene (UASG) after germ line (21, 22) and somatic gene delivery (23, 24), respectively. Transgenic mice were generated with the GLp65 regulator and placed under the transcriptional control of the human surfactant protein-C (SP-C) gene, which targets transgene expression to the alveolar type II cells and distal Clara cells of the mouse lungs (25). The UASG-FGF-3 target transgenic line of mice used were generated previously by Ornitz et al. (26) in the establishment of a Gal4-based binary transgenic system for the expression of FGF-3 in the mammary glands of transgenic mice. Establishment of the UASG-FGF-3/SP-C-GLp65 bitransgene has allowed development of a system for temporal and quantitative regulation of FGF-3 in the adult lung.
Materials and Methods
Generation of SP-C-GLp65 Transgenic Mice.
The regulator transgene was generated by ligating the 3.7-kb SP-C promoter fragment (J. Whitsett, University of Cincinnati, Cincinnati, OH) into the PAP CMV-GL914p65′SV plasmid (24) by blunt-end ligation. The 3.7-kb NdeI–SalI fragment was inserted into the Asp718–AscI site of the PAP CMV-GL914p65′SV (24). The resulting construct was denoted pSP-C-GLp65. For the generation of SP-C-GLp65 transgenic mice, a 6.6-kb DNA fragment was freed from the pSP-C-GLp65 by digesting with NotI and PmeI and was isolated by using Geneclean (Bio 101). The fragment was injected into either FvB/N or C57BL/6J fertilized eggs at a concentration of 2 ng/μl. Transgenic mice were generated according to the standard procedure described by Hogan et al. (27). Founder mice were analyzed by PCR and Southern hybridization.
Heterozygous SP-C-GLp65 mice were bred with homozygous UASG-FGF-3 monotransgenic mice (Philip Leder, Harvard University, Cambridge, MA). The UASG-FGF-3 target mice were initially described by Ornitz et al. (26) as the UASG-int-2 transgenic mice.
Administration of RU486 to Mice.
RU486 (Mifepristone) was obtained from Roussel-UCLAF. For administration to bitransgenic mice, RU486 pellets at dosages of 60, 120, 180, and 450 μg per pellet were designed for release of RU486 at a constant daily dose of 66, 130, 200, and 500 μg/kg body weight for 30 days, respectively (Innovative Research of America). The pellets were implanted s.c. Placebo pellets containing an equal amount of formative components were used in each experiment to serve as a vehicle control. RU486 was given also to both male and nonpregnant female nontransgenic mice. No toxicity from RU486 was seen.
Ribonuclease Protection Assay (RPA).
Total RNA was extracted from mouse lungs by using Trizol reagent (GIBCO/BRL). Expression of the mRNAs for the GLp65 regulator transgene, FGF-3 target transgene, cyclins, and cytokines was accomplished by RNase protection using [32P]UTP (ICN) and the RPA II kit (Ambion, Austin, TX). The template for the GLp65 mRNA was generated by inserting a 295-bp SalI and BamHI fragment containing the GaL4 DNA binding domain into pBluescript (Stratagene). An antisense riboprobe for RPA was generated by digesting the plasmid with XbaI, and T3 RNA polymerase was used for in vitro RNA synthesis. A template for FGF-3 expression was generated by inserting a 1.0-kb EcoRI–PstI fragment of the FGF-3 cDNA into the EcoRI and BamHI sites of pBluescript. An antisense riboprobe for RPA was generated by digestion of this plasmid with BamHI and by using T7 RNA polymerase for in vitro RNA synthesis.
Multiprobe templates for the expression of the cytokines IL-7, granulocyte/macrophage colony-stimulating factors (GM-CSFs), were made by using the mCK4 multiprobe template set (PharMingen). The mCyc-1 multiprobe template set was used for RPA analysis of Cyclins A1, A2, B1, C, D1, and D2.
S1 Nuclease Protection Assay.
Expression of mRNAs for the SP-A, -B, and -C, Clara cell secretory protein (CCSP), and cytochrome p450 2F2 was accomplished by S1 nuclease protection as described (28, 29). The ribosomal protein L32 was used as an internal control.
Western Blot Analysis.
FGF-3 protein levels were assayed by Western blot analysis. Mouse lungs were homogenized, and protein levels were quantified by Bradford assay (Bio-Rad). Then 60 μg of total protein from each sample was separated on a 10% SDS/PAGE gel and then electroblotted to a membrane (Millipore). The membranes then were probed with goat anti-mouse FGF-3 polyclonal IgG (Santa Cruz Biotechnology) and horseradish peroxidase-conjugated anti-goat IgG (Santa Cruz Biotechnology). Finally, the signals were visualized by ECL+plus (Amersham Pharmacia).
Immunohistochemistry.
Lung tissue was fixed with buffered formalin for histological analysis by gravity perfusion through the canulated trachea. Immunohistochemistry for Mac-3 was accomplished by using rat anti-mouse Mac-3 antibodies (PharMingen). Immunohistochemistry for NKx2.1 was accomplished by using antibodies for recombinant human NKx2.1 generated in rabbits. Biotin-conjugated rabbit anti-rat IgG and goat anti-rabbit IgG (Vector Laboratories) were used as secondary antibodies for Mac-3 and TTF-1 staining, respectively. Immunoperoxidase staining was accomplished as described by Sepulveda et al.(30). Nontransgenic mouse lung sections were analyzed concurrently as normal controls as well as to control for specific protein staining. Standard methods were used for routine processing and hematoxylin and eosin (H&E) staining of tissues.
Results
Ligand-Inducible Expression of FGF-3 in Bitransgenic Mice.
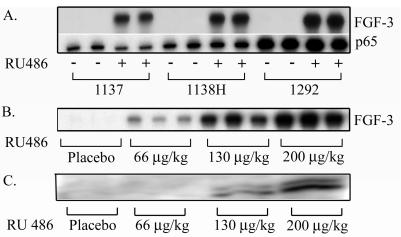
Twelve regulator mouse lines carrying the GLp65 transgene were identified by Southern blot analysis and PCR on tail biopsies of the F0 generation. In an initial test of the functional properties of our regulatory system, we injected RU486 intraperitoneally at a concentration of 500 μg/kg body weight for 5 days. Controls received sesame oil alone. RU486-dependent expression of FGF-3 was observed in 4 of 12 GLp65/UASG-FGF-3 bitransgenic lines. To assess the inducibility for each line further, we implanted the RU486 pellets s.c. with a dosage of 500 μg/kg for 1 week. This dose ensured a constant release of RU486 at an equal concentration in all the mice. Placebo pellets lacking RU486 served as a vehicle control. The degree of induction as well as the basal levels of FGF-3 depended on the level of expression of the GLp65 regulator (Fig. 1A). Mouse line 1137 showed the lowest level of GLp65 and the lowest level of induction of FGF-3 after the administration of RU486 (Fig. 1A). Mouse line 1138H showed an intermediate level of GLp65 expression and an intermediate induced level of FGF-3 and has no detectable expression of FGF-3 in the placebo-treated group. Mouse line 1292 displayed the highest level of GLp65 mRNA and had the highest level of FGF-3 after the administration of RU486. However, basal expression of FGF-3 was also detectable in placebo-treated binary mice. One mouse line expressed very low levels of GLp65 as well as a low level of FGF-3 expression after the administration of RU486 (data not shown), and six other founders demonstrated either leaky basal expression of FGF-3 or FGF-3 expression that could not be induced. The variable inducibility and leakiness of transgenes in different founders is most likely a consequence of the site of transgene integration. More importantly, RU486 has no effect on monogenic FGF-3, monogenic GLp65, or wild-type mice (data not shown). In fact, RU486, at the doses used in this report, showed no effects on monogenic FGF-3, monogenic GLp65, or wild-type mice when compared with animals that were administered placebo in any of the experiments presented to validate this system (data not shown).
Figure 1.
(A) RPA of FGF-3 and GLp65 expression in the lungs after administration of RU486 at a dosage of 500 μg/kg for 1 week (+) on transgenic lines 1292, 1138H, and 1137. Bitransgenic littermates were administered placebo pellets as control (−). RPA of FGF-3 mRNA (B) and Western blot analysis of FGF-3 protein expression (C) in the lungs of mice from line 1138H after administration of placebo or RU486 at a dosage of 66, 130, and 200 μg/kg for 1 month are shown. Each lane represents an independent mRNA and protein from an individual mouse.
The levels of induction of the FGF-3 target transgene not only correlated with the levels of expression of the regulator but also could be modulated by varying the dose of RU486 administered. To assess this, RU486 was administered at dosages of 0, 66, 130, and 200 μg/kg body weight to the 1138H intermediate-expressing line for 1 month. This line was chosen because it demonstrated induction without any background expression of FGF-3. Fig. 1, B and C, shows that levels of FGF-3 mRNA and protein expression, respectively, correlated with the dosage of RU486 administered.
Phenotypic Consequences of Induction of FGF-3 in the Adult Mouse Lung.
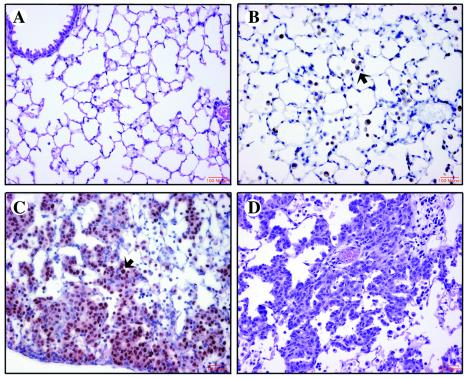
The phenotypic consequences of induction of FGF-3 were assayed after a 2-month induction with 500 μg/kg RU486 pellets. Monogenic SP-C-GLp65 and UASG-FGF-3 as well as nontransgenic mice were also administered RU486 to serve as controls. Placebo pellets were also implanted as a vehicle control. Three mice were in each control and treatment group. Each binary transgenic mouse line produced a characteristic histologic pattern that depended on the degree and duration of induced FGF-3 expression. There were no significant histopathologic alterations in any of the monogenic mice treated with RU486 or placebo or in the placebo-treated binary transgenic mice (Fig. 2A). Phenotypic analysis was conducted at 6 weeks for line 1137 (low induction), 2 weeks for line 1138H (intermediate induction), and 1 week for 1292 (highest induction). The time course chosen to examine the phenotypic consequences was determined by killing mice at the first signs of distress. FGF-3 induction in line 1137 showed a characteristic pattern of massive FAM infiltration. The cellular infiltration was confirmed to be FAMs by immunohistochemical staining with anti-Mac-3 antibody (Fig. 2B). Minimal type II cell hyperplasia was observed in the periphery of the lungs. The 1138H line with the intermediate level of expression of FGF-3 showed a distinct morphology with a mixture of diffuse type II cell hyperplasia and FAM infiltration after a 2-week treatment (Fig. 2C). The hyperplastic cells were identified as pulmonary epithelial cells by immunohistochemical staining of the lungs with an antibody to NKx 2.1. NKx2.1 is expressed in the alveolar type II cells and Clara cells of the adult mouse lung (Fig. 2C; ref. 31). No other type of acute or chronic inflammatory cell infiltrate was identified in any line of mice.
Figure 2.
Histological changes in the lungs of GLp65/FGF-3 bitransgenic mice treated with placebo or 500 μg/kg RU486. (A) H&E-stained section of lung from placebo-treated mouse. (B) Immunohistochemical staining for Mac-3 of lung from 1137 bitransgenic mouse treated with RU486 for 6 weeks. Free alveolar macrophages (FAMs) are abundant in the alveoli as indicated by the arrow. (C) Immunohistochemical staining for Nkx2.1 of lung from the 1138H bitransgenic mouse treated with 500 μg/kg RU486 for 2 weeks. Type II epithelial cells are increased strikingly along the septa and in alveoli as indicated by the arrow. (D) H&E section of lung from the 1292 bitransgenic mouse treated with 500 μg/kg RU486 for 1 week.
Histological analysis of the lungs from the 1292 line (highest degree of induction) revealed a diffuse intense type II pneumocyte cell hyperplasia after 1 week of RU486 administration. The hyperplastic cells were arranged in nodular aggregates of type II cells centered on the intersection of alveolar walls with adjacent bronchoalveolar spread (Fig. 2D).
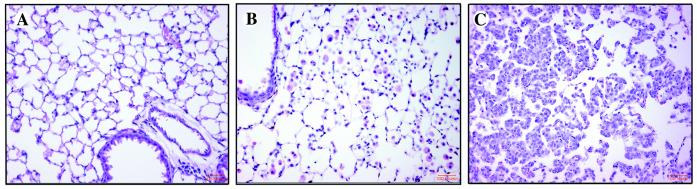
Because the above phenotypic consequences correlated with the level of FGF-3 expression, we determined whether these two phenotypes could be induced in a single line by varying the dosage of RU486. RU486 at dosages of 0, 66, 130, and 200 μg/kg body weight was administered to line 1138H (intermediate) for 1 month, because this line demonstrated induction without any background expression of FGF-3. At the dosage of 66 μg/kg of RU486, no microscopically apparent phenotype was observed, although the level of FGF-3 was elevated (data not shown). The induction of FGF-3 at a dose of 130 μg/kg of RU486 showed only macrophage infiltration (Fig. 3B), and both macrophage infiltration and type II cell proliferation could be achieved with induction of FGF-3 at a dosage of 200 μg/kg of RU486 (Fig. 3C). Interpretation of these results supports the hypothesis that the two phenotypes are caused by the dose response of the lungs to FGF-3.
Figure 3.
The response of 1138H bitransgenic mouse lungs to the induction of FGF-3 for 4 weeks with varying doses of RU486. (A) H&E-stained section of lung from placebo-treated 1138H bitransgenic mouse. (B) H&E-stained section of lungs from 1138H bitransgenic mouse treated with 130 μg/kg RU486. FAMs are abundant. (C) H&E-stained section of lungs from 1138H bitransgenic mouse treated with 200 μg/kg RU486. Type II cell hyperplasia is apparent.
The Phenotypic Effect of FGF-3 Expression Is Reversible and Repeatable.
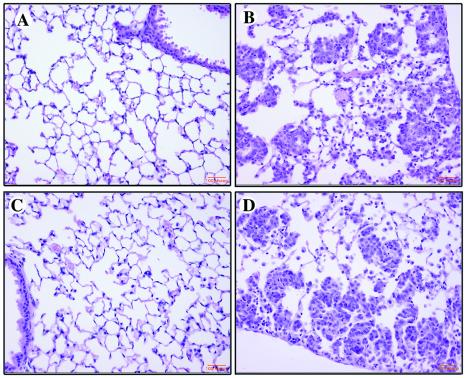
The value of an inducible bitransgenic system is the ability to turn the expression of the transgene on and off and to assay the phenotypic consequences. The reversibility and repeatability of the induction of the FGF-3 target transgene was assessed by s.c. implantation of RU486 pellets at a dosage of 500 μg/kg in a group of 15 1138H GLp65/FGF-3 bitransgenic mice. Three mice were killed after 1 week, three more were killed after 2 weeks, and the pellets were removed from the surviving mice. Two groups of three mice were killed at 1-week intervals after RU486 withdrawal. The three mice surviving were then reimplanted with RU486 and killed after 2 weeks. RNA was extracted from each group of mice, and RPA and H&E staining were performed. As observed in the initial characterization of the 1138H line, we observed an intense type II cell hyperplasia, most pronounced in the lung periphery, mixed with FAMs in animals treated with RU486 for 2 weeks (Fig. 4B) as compared with mice that were administered a placebo (Fig. 4A). These histological changes underwent nearly complete regression when RU486 was removed for 2 weeks (Fig. 4C). In animals that subsequently had RU486 reimplanted, the same histologic changes recurred (Fig. 4D).
Figure 4.
The response of 1138H bitransgenic mouse lungs after the administration and withdrawal of RU486 at 2-week intervals. H&E staining of lung sections from 1138H bitransgenic mice: treated with placebo (A); treated with 500 μg/kg RU486 for 2 weeks (B); 2 weeks after the withdrawal of RU486 (C); and re-treated with 500 μg/kg RU486 for 2 weeks after withdrawal (D).
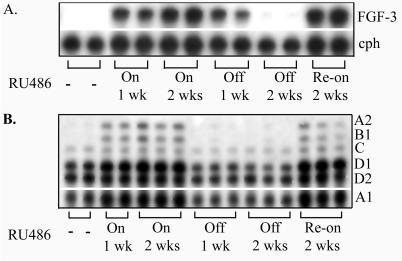
The molecular regulation of the hyperplasia was assayed by measuring the expression of FGF-3 and the cyclin genes with the induction and withdrawal of RU486. Expression of the target transgene, FGF-3, paralleled the administration of RU486 (Fig. 5A). By using multiriboprobes for cyclins A–D, we detected an overall increased level of cyclins A1, A2, B1, C, D1, and D2 (Fig. 5B). RPA of cyclin expression demonstrated a tight correlation between FGF-3 gene expression and expression of the cyclin genes. This data strongly demonstrates that our inducible system produces lines of mice with reversible and repeatable induction of transgene expression and phenotypic consequences.
Figure 5.
RPA of FGF-3 (A) and cyclin mRNAs (B) in the lungs of 1138H bitransgenic mouse lungs after the administration and withdrawal of 500 μg/kg RU486. −, RNA from mice treated with placebo; On 1 wk, mice treated with RU486 for 1 week; On 2 wks, mice treated with RU486 for 2 weeks; Off 1 wk, mice after withdrawal of RU486 for 1 week; Off 2 wks, mice after the withdrawal of RU486 for 2 weeks after treatment with RU486; Re-on 2 wks, mice treated with RU486 after withdrawal of RU486 for 2 weeks. Expression of cyclophillin (cph) served as a control for RNA-loading analysis of FGF-3 expression. Expression of L32 served as a control for RNA loading for cyclin gene expression (data not shown). Each lane represents an independent mRNA from an individual mouse.
The Effects of FGF-3 Induction on the Expression of Cytokines and Pulmonary Epithelial Markers.
The marked FAM migration in the RU486-treated binary transgenic mice is an unexpected finding. To investigate the mechanisms of this phenomenon, RPA was performed on IL-7 and GM-CSF in bitransgenic mice with the 1137 activator line. Although no increase in IL-7 and GM-CSF was observed after 1 week of FGF-3 induction, significant increases in mRNA levels of IL-7 and GM-CSF were seen after 6 weeks of induction (Fig. 6A). This increase was concurrent with the increase in FAMs in the airway. The late induction of these cytokines indicates that their increase may not be the primary response to FGF-3 induction but a response to an alteration in pulmonary gene expression. SP-A has been found to promote macrophage phagocytosis (32) and chemotaxis (33). Induction of SP-A by FGF-3 may play a role in the FAM phenotype. Therefore, analysis of the expression of pulmonary epithelial markers was conducted.
Figure 6.
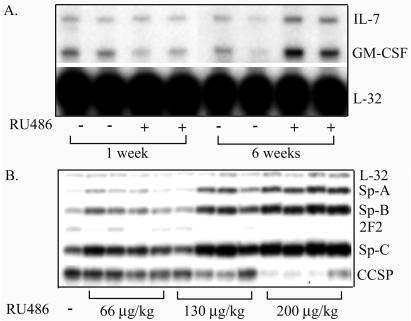
The effect of FGF-3 induction on IL-7, CM-CSF, and pulmonary epithelial marker gene expression. (A) RPA of the expression of IL-7 and GM-CSF in lungs from 1137 mice treated with 500 μg/kg RU486 for either 1 or 6 weeks. L32 was used as internal control for RNA loading in the RPA. (B) S1 nuclease protection analysis of the expression of epithelial markers, SP-A, SP-B, and SP-C, CCSP, and cytochrome p450 2F2 in bitransgenic mice from line 1138H were administered RU486 for 1 month at a dose of 0, 66, 130, and 200 μg/kg. L32 was used as internal control for RNA loading in the S1 nuclease protection analysis. Control samples were assayed in triplicate (data not shown).
The expression of pulmonary epithelial-specific genes was investigated to determine whether FGF-3 induction exhibited an effect on pulmonary epithelial cell differentiation. Expression of surfactant protein genes, SP-A, SP-B, and SP-C, as well as Clara cell-specific markers, CCSP, and cytochrome P450 2F2 were assayed in line 1138H by S1 nuclease protection analysis with varying doses of RU486 administered for 1 month. RU486 had no effect on the expression of any of these markers in control mice (data not shown). As Fig. 6B shows, the degree of induction of the surfactant protein genes, SP-C, a marker for alveolar type II cells, SP-A, and SP-B, a marker for airway epithelial cells, as well as type II cells correlated with the induction of FGF-3. Interestingly, genes expressed specifically in the Clara cells, CCSP and cytochrome p450 2F2 of the airway were reduced at the higher doses. This reduction in proximal airway gene expression may reflect three possibilities. First, the chronic high level of the expression of FGF-3 could alter the differentiation of Clara cells, which could be manifested in a reduction in the expression of these cell-specific markers. Second, inflammatory cytokines may have affected expression of these genes directly and/or the phenotype of airway epithelial cells (32, 33). Finally, and more likely, the increase in the number of alveolar type II cells could result in the dilution of Clara cell-specific mRNA, because the proportion of airway cells in the lung could decrease with the increase in distal cells. The effect of FGF-3 expression on these proximal airway genes requires further investigation.
Discussion
The GLp65 bitransgenic regulatory system has allowed the establishment of an inducible system for FGF-3 expression in the murine lung. By using this system, the effects of FGF-3 expression can be evaluated in a fully developed mature lung without the developmental defects that may result from constitutive transgene expression during development. The GLp65/UASG-FGF-3 bitransgenic system demonstrated the following characteristics. First, there are no phenotypic consequences observed in the monogenic GLp65 regulator, UASG-FGF-3 target mice, or mice given the above doses of RU486. Second, there are either no detectable or low levels of target gene expression in the bitransgenic mice before the induction of target transgene expression with RU486. Third, the level of induction of the target transgene depends on the dose of RU486 administered and is at a level that it can exert a phenotypic consequence. Fourth, the expression and the phenotypic consequences of the target transgene are reversible after withdrawal of the ligand. The most interesting facet of the establishment of this system is that the absolute level of induction of FGF-3 in the mouse lung depended on the degree of expression of the regulator. Thus, lines of bitransgenic mice have been established in which a variety of levels of FGF-3 expression could be achieved, and interestingly, the phenotype observed depended on the level of FGF-3 expression.
The phenotypic consequences of FGF-3 expression in the lung depend on the level of FGF-3 induction. At maximal levels of FGF-3 expression, the predominant phenotype was alveolar type II cell hyperplasia. At lower levels of FGF-3 expression, the epithelial cell hyperplasia was minimal, and the predominant phenotype was macrophage invasion into the alveoli. The first phenotype of cellular hyperplasia was not unexpected, because deregulated expression of FGF-3 in other tissues such as mammary and coagulating glands caused profound epithelial cell hyperplasia (34). FGF-7, which acts through the same receptor as FGF-3, has also been shown to cause pulmonary hyperplasia in the embryo and adult mouse lung (35, 36). The hyperplastic phenotype may be a recapitulation of the role of FGF signaling during pulmonary development. During development, FGF-10 acting through its receptor, FGFR2iiib, was shown to regulate branching morphogenesis in the lungs of mice (3, 4). The high levels of FGF-3 may result in the activation of pathways that reflect the role of FGF signaling during development of the airways.
The proliferative phenotype observed after the induction of high levels of FGF-3 was rapid and reversible. Examination of the expression of the cyclin genes during administration and withdrawal of RU486 demonstrated that proliferative marker gene expression correlated tightly with the induction and withdrawal of FGF expression. These results indicate that this system can be exploited to identify alterations in gene expression associated with the induction and withdrawal of FGF-3. The exploitation of this system will allow for identification of the genes that are primary in the regulation of alveolar type II cell proliferation and are responsible for the remodeling of the airways after the withdrawal of growth factor stimulation. Also, exploiting the system will allow for the identification of processes that are crucial for regulation of homeostatic growth of the distal airway epithelial cells and will shed light on processes disrupted during the development of pulmonary diseases such as lung cancer.
The second phenotype, the recruitment of macrophages into the airways, was not expected. This phenotype may reflect the role of FGFR2 signaling in the adult lung. Examination of FGFR2 signaling in the adult has been demonstrated by examining the role of FGF-7 on alveolar type II cells cultured in vitro. FGF-7 has also been demonstrated to be responsible for the maintenance of the cellular functions of alveolar type II cells in vitro (14). Administration of FGF-7 to type II cells in culture is responsible for maintenance of the expression of surfactant proteins, which are lost in culture. Therefore, the phenotype observed with low levels of FGF-3 may reflect the role of FGF in regulating the expression of surfactant proteins in the adult lung. An increase in surfactant protein mRNA was correlated best with the induction of FGF-3. FGF-3 stimulation of surfactant protein production may result directly in the recruitment of macrophages into the lung (32, 33). Alternatively, the FAM recruitment may be an indirect result of increased surfactant production by the alveolar Type II cells. Evidence for this comes from examining the role of GM-CSF in surfactant metabolism.
GM-CSF has been shown to be essential for the regulation of pulmonary surfactant homeostasis. Mice homozygous for a targeted disruption of the GM-CSF gene develop a progressive accumulation of surfactant lipids and proteins in the alveolar space because of the inability of the lung to recruit macrophages into the airways (37). Transgene expression of GM-CSF in the lungs has been shown to regulate macrophage recruitment (38), surfactant metabolism, and alveolar type II cell hyperplasia (39). In this report, mice with low levels of FGF-3 expression showed an increase in GM-GSF with time, which correlated with the FAM phenotype. In this model, FGF-3-stimulated FAM infiltration in the lungs could be a result of increased GM-CSF production in response to altered surfactant production and metabolism by the type II cells.
In summary, the animal models generated in this report will be useful in dissecting the role of FGF signaling in the regulation of pulmonary epithelial physiology and proliferation. The mice expressing high levels of FGF-3 can be used to determine the molecular mechanism regulating epithelial cell proliferation. The role of FGF may be important in studies of the regulation of lung development and the establishment of animal models to investigate the progression of lung cancer. The lines of mice showing low expression of FGF-3 will be important to investigate the molecular mechanisms by which FGF signaling regulates alveolar type II cell biology as well as the communication of the pulmonary epithelium with the cells that contribute to immune surveillance and maintenance of airway integrity. These animals will allow us to assess how signaling pathways that regulate development participate in adult physiology.
Acknowledgments
Jie Wang, Janet L. DeMayo, M. S., Mei Rong Gu, and Louise Hadsell contributed technical expertise. John Ellsworth and Norio Takamoto aided in the preparation of this manuscript, and Fred Pereira aided in the critical review of this manuscript. This research was supported by American Cancer Society Grants RPG-95-020-04 and RO1 HL61406 (to F.J.D.), P01 HL59314 (Project 3) and DAMD 179919077 (to S.Y.T.), and RO1 HL64888 and RO1 ES08964 (to S.D.R. and B.R.S.).
Abbreviations
- FGF
fibroblast growth factor
- FGFR
FGF receptor
- GLVP
Gal-4 DNA-binding domain, ligand-binding domain of the progesterone receptor and the VP-16 activation domain
- UASG
upstream activating sequences for the yeast Gal4 gene
- SP-A
surfactant protein-A
- SP-B
surfactant protein-B
- SP-C
surfactant protein-C
- RPA
ribonuclease protection assay
- GM-CSF
granulocyte/macrophage colony-stimulating factor
- CCSP
Clara cell secretory protein
- H&E
hematoxylin and eosin
- FAM
free alveolar macrophage
References
- 1.Korfhagen T R, Whitsett J A. Chest. 1997;111:S83–S88. [Google Scholar]
- 2.Peters K, Werner S, Liao X, Wert S, Whitsett J, Williams L. EMBO J. 1994;13:3296–3301. doi: 10.1002/j.1460-2075.1994.tb06631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 4.Kato S, Sekine K. Cell Mol Biol. 1999;45:631–638. [PubMed] [Google Scholar]
- 5.Naski M C, Ornitz D M. Front Biosci. 1998;3:D781–D794. doi: 10.2741/a321. [DOI] [PubMed] [Google Scholar]
- 6.Cuevas P. Surg Neurol. 1998;50:571–573. doi: 10.1016/s0090-3019(97)00375-3. [DOI] [PubMed] [Google Scholar]
- 7.Daphna-Iken D, Shankar D B, Lawshe A, Ornitz D M, Shackleford G M, MacArthur C A. Oncogene. 1998;17:2711–2717. doi: 10.1038/sj.onc.1202212. [DOI] [PubMed] [Google Scholar]
- 8.Galdemard C, Brison O, Lavialle C. Oncogene. 1995;10:2331–2342. [PubMed] [Google Scholar]
- 9.Rosen A, Sevelda P, Klein M, Dobianer K, Hruza C, Czerwenka K, Hanak H, Vavra N, Salzer H, Leodolter S, et al. Br J Cancer. 1993;67:1122–1125. doi: 10.1038/bjc.1993.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ornitz D M, Xu J, Colvin J S, McEwen D G, MacArthur C A, Coulier F, Gao G, Goldfarb M. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 11.Johnson D E, Williams L T. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez A M, Hill D J, Logan A, Maher P A, Baird A. Pediatr Res. 1996;39:375–385. doi: 10.1203/00006450-199603000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Weinstein M, Xu X, Ohyama K, Deng C X. Development (Cambridge, UK) 1998;125:3615–3623. doi: 10.1242/dev.125.18.3615. [DOI] [PubMed] [Google Scholar]
- 14.Shannon J M, Gebb S A, Nielsen L D. Development (Cambridge, UK) 1999;126:1675–1688. doi: 10.1242/dev.126.8.1675. [DOI] [PubMed] [Google Scholar]
- 15.Barazzone C, Donati Y R, Rochat A F, Vesin C, Kan C D, Pache J C, Piguet P F. Am J Pathol. 1999;154:1479–1487. doi: 10.1016/S0002-9440(10)65402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huebner K, Ferrari A C, Delli Bovi P, Croce C M, Basilico C. Oncog Res. 1988;3:263–270. [PubMed] [Google Scholar]
- 17.Brookes S, Lammie G A, Schuuring E, de Boer C, Michalides R, Dickson C, Peters G. Genes Chromosomes Cancer. 1993;6:222–231. doi: 10.1002/gcc.2870060406. [DOI] [PubMed] [Google Scholar]
- 18.Rubin J S, Qiu L, Etkind P. J Laryngol Otol. 1995;109:72–76. doi: 10.1017/s0022215100129305. [DOI] [PubMed] [Google Scholar]
- 19.Finkelstein S D, Hasegawa T, Colby T, Yousem S A. Am J Pathol. 1999;155:633–640. doi: 10.1016/S0002-9440(10)65159-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai S Y, O'Malley B W, DeMayo F J, Wang Y, Chua S S. Adv Drug Delivery Rev. 1998;30:23–31. doi: 10.1016/s0169-409x(97)00104-x. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, DeMayo F J, Tsai S Y, O'Malley B W. Nat Biotechnol. 1997;15:239–243. doi: 10.1038/nbt0397-239. [DOI] [PubMed] [Google Scholar]
- 22.Pierson T M, Wang Y, DeMayo F J, Matzuk M M, Tsai S Y, Omalley B W. Mol Endocrinol. 2000;14:1075–1085. doi: 10.1210/mend.14.7.0478. [DOI] [PubMed] [Google Scholar]
- 23.Burcin M M, O'Malley B W, Tsai S Y. Front Biosci. 1998;3:c1–c7. doi: 10.2741/a258. [DOI] [PubMed] [Google Scholar]
- 24.Burcin M M, Schiedner G, Kochanek S, Tsai S Y, O'Malley B W. Proc Natl Acad Sci USA. 1999;96:355–360. doi: 10.1073/pnas.96.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glasser S W, Korfhagen T R, Wert S E, Bruno M D, McWilliams K M, Vorbroker D K, Whitsett J A. Am J Physiol. 1991;261:L349–L356. doi: 10.1152/ajplung.1991.261.4.L349. [DOI] [PubMed] [Google Scholar]
- 26.Ornitz D M, Moreadith R W, Leder P. Proc Natl Acad Sci USA. 1991;88:698–702. doi: 10.1073/pnas.88.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hogan B, Costantini F, Lacy E. Manipulating the Mouse Embryo, A Laboratory Manual. Plainview, New York: Cold Spring Harbor Lab. Press; 1986. pp. 151–197. [Google Scholar]
- 28.Mango G W, Johnston C J, Reynolds S D, Finkelstein J N, Plopper C G, Stripp B R. Am J Physiol. 1998;275:L348–L356. doi: 10.1152/ajplung.1998.275.2.L348. [DOI] [PubMed] [Google Scholar]
- 29.Stripp B R, Lund J, Mango G W, Doyen K C, Johnston C, Hultenby K, Nord M, Whitsett J A. Am J Physiol. 1996;271:L656–L664. doi: 10.1152/ajplung.1996.271.4.L656. [DOI] [PubMed] [Google Scholar]
- 30.Sepulveda A R, Finegold M J, Smith B, Slagle B L, DeMayo J L, Shen R F, Woo S L, Butel J S. Cancer Res. 1989;49:6108–6117. [PubMed] [Google Scholar]
- 31.Zhou L, Lim L, Costa R H, Whitsett J A. J Histochem Cytochem. 1996;44:1183–1193. doi: 10.1177/44.10.8813084. [DOI] [PubMed] [Google Scholar]
- 32.Wright J R, Youmans D C. Am J Physiol. 1993;264:L338–L344. doi: 10.1152/ajplung.1993.264.4.L338. [DOI] [PubMed] [Google Scholar]
- 33.Weikert L F, Lopez J P, Abdolrasulnia R, Chroneos Z C, Shepherd V L. Am J Physiol. 2000;279:L216–L223. doi: 10.1152/ajplung.2000.279.2.L216. [DOI] [PubMed] [Google Scholar]
- 34.Muller W J, Lee F S, Dickson C, Peters G, Pattengale P, Leder P. EMBO J. 1990;9:907–913. doi: 10.1002/j.1460-2075.1990.tb08188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simonet W S, DeRose M L, Bucay N, Nguyen H Q, Wert S E, Zhou L, Ulich T R, Thomason A, Danilenko D M, Whitsett J A. Proc Natl Acad Sci USA. 1995;92:12461–12465. doi: 10.1073/pnas.92.26.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tichelaar J W, Lu W, Whitsett J A. J Biol Chem. 2000;275:11858–11864. doi: 10.1074/jbc.275.16.11858. [DOI] [PubMed] [Google Scholar]
- 37.Dranoff G, Crawford A D, Sadelain M, Ream B, Rashid A, Bronson R T, Dickersin G R, Bachurski C J, Mark E L, Whitsett J A, et al. Science. 1994;264:713–716. doi: 10.1126/science.8171324. [DOI] [PubMed] [Google Scholar]
- 38.Huffman Reed J A, Rice W R, Zsengeller Z K, Wert S E, Dranoff G, Whitsett J A. Am J Physiol. 1997;273:L715–L725. doi: 10.1152/ajplung.1997.273.4.L715. [DOI] [PubMed] [Google Scholar]
- 39.Reed J A, Whitsett J A. Proc Assoc Am Physicians. 1998;110:321–332. [PubMed] [Google Scholar]