Abstract
Domestic animals have played a key role in human history. Despite their importance, however, the origins of most domestic species remain poorly understood. We assessed the phylogenetic history and population structure of domestic goats by sequencing a hypervariable segment (481 bp) of the mtDNA control region from 406 goats representing 88 breeds distributed across the Old World. Phylogeographic analysis revealed three highly divergent goat lineages (estimated divergence >200,000 years ago), with one lineage occurring only in eastern and southern Asia. A remarkably similar pattern exists in cattle, sheep, and pigs. These results, combined with recent archaeological findings, suggest that goats and other farm animals have multiple maternal origins with a possible center of origin in Asia, as well as in the Fertile Crescent. The pattern of goat mtDNA diversity suggests that all three lineages have undergone population expansions, but that the expansion was relatively recent for two of the lineages (including the Asian lineage). Goat populations are surprisingly less genetically structured than cattle populations. In goats only ≈10% of the mtDNA variation is partitioned among continents. In cattle the amount is ≥50%. This weak structuring suggests extensive intercontinental transportation of goats and has intriguing implications about the importance of goats in historical human migrations and commerce.
Keywords: animal domestication, mitochondrial DNA, conservation genetics, Capra taxonomy and evolution
Domestic goats (Capra hircus) might have played a central role in the Neolithic agricultural revolution and the spread of human civilizations around the globe (1, 2). The origins of domestic goats remain uncertain and controversial, but archaeological evidence suggests that they were probably first domesticated in the Fertile Crescent region of the Near East ≈10,000 years ago (YA) (1–5). Some studies hint that a second domestication in Pakistan could have given rise to the cashmere breeds (2, 6). Others suggest that at least two wild species of Capra (7) could have contributed to the gene pool of domestic goats (8). This seems possible in light of the enormous morphological diversity among the more than 300 goat breeds (2). Goats are the most adaptable and geographically wide spread livestock species, ranging from the mountains of Siberia to the deserts and tropics of Africa. They are the main economic resource in numerous developing countries, and are growing in economic importance in western countries (2).
To help understand the phylogenetic history of goats, we sequenced the first hypervariable segment (HVI) of the mtDNA control region of 406 goats originating from 44 countries throughout Europe, Asia, Africa, and the Middle/Near East. To estimate the time since divergence of the mtDNA lineages, we sequenced the entire cytochrome b gene in six goats. We then compared the geographic patterns of HVI variation in goats to the patterns in cattle by using published data (9, 10). Our objectives were to (i) assess the maternal origins of domestic goats and (ii) estimate levels of gene flow among goat populations to infer the importance of goats in historical human commerce and population movements. These objectives are addressed in light of recent publications in both archeozoology and molecular genetics of the four main livestock species [cattle (9, 10), sheep (11), pigs (12), and goats].
Methods
Sampling and DNA Extraction.
Our extensive sampling spanned most of the Old World distribution of goats from Nigeria to Iceland and Mongolia to Malaysia, including potential centers of domestication [e.g., Turkey, Egypt, Jordan, Iraq, and Pakistan (see Table 3, which is published as supplemental data on the PNAS web site, www.pnas.org)]. We sampled only pure indigenous goats from small remote villages and excluded research centers, large cities, and coastal harbors where recent international shipping of goats is possible. Fourteen wild Capra individuals were sampled, including representatives of all major taxa, to use as outgroups. Blood or skin biopsies were collected and stored for 6–12 months at −20°C or in 95% ethanol (respectively) before DNA extraction. DNA was extracted by using standard commercial kits (Qiagen tissue and blood kits, Chatsworth, CA).
Sequencing.
We sequenced the HVI of the mtDNA control region because it has proven informative for inferring the history of humans and domestic animals (9–11, 13, 14). The primers CAP-F (5′-CGTGTATGCAAGTACATAC-3′) and CAP-R (5′-CTGATTAGTCATTAGTCCATC-3′) were used to amplify a 579-bp DNA fragment (this value excludes insertions/deletions). PCR amplifications were conducted in a 25-μl volume containing 2.5 mM MgCl2, 200 μM of each dNTP, 1 μM of each primer, and 1 unit of AmpliTaq Gold Polymerase (Applied Biosystems). The PCR mixture underwent 35 cycles of 30 s at 95°C, 30 s at 50°C, and 1 min at 72°C. PCR products were purified by using the Quaquick PCR columns (Qiagen). We then sequenced a 481-bp segment of the PCR products by using two “internal” primers CAP-FI (5′-TCCATATAACGCGGACATAC-3′) and CAP-RI (5′ATGGCCCTGAAGAAAGAAC-3′). All sequences were obtained for both DNA strands by using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) in a 20-μl volume containing 40–50 ng of purified DNA, and 3.2 pmol of primer. Sequencing reactions underwent 25 cycles of 30 sec at 96°C, 30 sec at 58°C, and 4 min at 72°C on a thermocycler (PE 2400, PE 9600, or PE 9700; Perkin–Elmer). Excess dye terminators were removed by spin-column purification. Sequencing reactions were electrophoresed for 6 h on an ABI 377 PRISM DNA sequencer (Applied Biosystems) in a 5% Long Ranger gel (FMC).
The 1,140 bp of cytochrome b (mtDNA) was sequenced for two goats arbitrarily chosen (with different control region sequences) from each divergent lineage (C. hircus lineages A–C) by using the primers L14724V (3′-ATGATATGAAAAACCATCGTTG-5′) and H15915V (3′-TCTCCTTCTCTGGTTTACAAGAC-5′). PCR was conducted as for HVI except the same primers were used for the initial PCR and sequencing under the following reaction conditions: 35 cycles (or 25 for sequencing) of 30 sec at 95°C, 30 sec at 55°C, and 2 min at 72°C. The two domestic sheep (Ovis aries) sequences were obtained from two independent studies listed in GenBank (accession nos. AF034730 and X56284).
Data Analysis.
Neighbor-joining trees, as well as UPGMA (unweighted pair group method using arithmetic averages) and maximum likelihood trees, were constructed by using PAUP* software, Version 0.64d. Following Wakeley (15), heterogeneity in substitution rates among branches was modeled for goat HVI by using a gamma distribution. The alpha shape parameter of the gamma distribution was estimated by a maximum-likelihood method from a set of 35 wild and domestic goats by using paml software, Version 1.3b (16), under the Kimura 2-parameter substitution model (17). We observed substantial heterogeneity in substitution rates among nucleotide sites (alpha 0.29), as has been observed for HVI in other mammals (10, 13, 14). The hierarchical components of mtDNA variation were computed under the AMOVA (Analysis of Molecular Variance) framework (18) by using arlequin software, Version 1.1 (http://anthropologie.unige.ch/arlequin). The AMOVA procedure incorporates both the estimated divergence between sequences and the frequencies. Between the domestic and wild Capra (used as out groups), we detected only four and two insertions/deletions of one bp and two bp, respectively; the remaining polymorphisms were single nucleotide substitutions.
Phylogenetic trees (maximum parsimony and neighbor joining) group the goat cytochrome b sequences into the same three clusters (C. hircus A–C) identified by the control region HVI sequences in Fig. 1. A molecular-clock likelihood-ratio test (for heterogeneity in substitution rates) was carried out by using the six goat and two sheep cytochrome b sequences analyzed for the 380 nucleotides at third codon positions (i.e., synonymous positions unlikely to be under selection). The test was not significant (P > 0.05), allowing us to use the amount of divergence between sheep and goat sequences to estimate the approximate time to the most recent ancestor (TMRCA) of the three goat lineages. Using five or seven million YA as the divergence between sheep and goats (19, 20), we estimate a TMRCA for domestic goat genes of roughly 201,380 years or 281,932 years. All sequences were deposited in GenBank (accession nos. AJ317533–AJ317875).
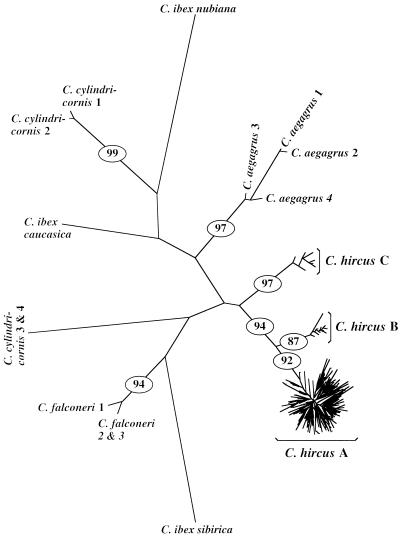
Figure 1.
Neighbor-joining tree of mtDNA types from 406 domestic goats and 14 wild Capra. Trees constructed by using other methods (e.g., UPGMA or neighbor-joining with alpha = 0.20–0.40) were nearly identical in shape. The large star-shaped cluster (C. hircus A) contains 316 mtDNA types (found in 370 individuals and in all breeds). The two smaller lineages (C. hircus B and C) contain only eight and seven mtDNA types (found in 25 and 11 individuals, respectively). C. hircus B was detected only in eastern and southern Asia. C. hircus C was found in Mongolia, Switzerland, and Slovenia (Fig. 2). Numbers on branches are the percent of 2,000 bootstrap trees with the same branch structure. Only bootstrap values >70 are given. The wild taxon with sequences most similar to domestic goats is Capra aegagrus (61.3 substitutions, on average, using the gamma-corrected distance). The second most similar taxon is Capra cylindricornis (84.5 substitutions; see Table 4, which is published as supplemental data on the PNAS web site, www.pnas.org). It is not surprising that some wild taxa appear to be paraphyletic because (i) the taxonomy of Capra is very poorly understood and erroneous taxonomic classifications are possible (7), (ii) paraphyly has been reported (33, 34), and (iii) intertaxon hybridization is possible (23) and is thought to occur in Daghestan where our (paraphyletic) samples originated.
Results and Discussion
The control region sequences were highly polymorphic: 160 variable sites defined 331 mitochondrial types in domestic goats. Most mutations were single nucleotide substitutions. Only three insertions/deletions of one, two, and 76 nucleotides were detected in three, two, and two different sequences, respectively. The ratio of transitions to transversions was high (17/1), as has been observed for the HVI segment of mtDNA in cattle and humans (9, 10, 13).
Goat Origins and mtDNA.
Phylogenetic analysis revealed three highly divergent goat mtDNA lineages comprising 316, eight, and seven mtDNA types, respectively (Fig. 1, C. hircus A–C). The three distinct lineages could be interpreted as evidence for either (i) three separate maternal origins from genetically distinct populations, or (ii) one origin from an extremely large population containing three highly divergent lineages.
An argument for a single origin is that the three goat lineages appear to be monophyletic (Fig. 1). If three distinct wild Capra populations had been domesticated, we might expect a paraphyletic tree in which some of the sequences from wild Capra are clustered between the three domestic goat lineages (C. hircus A–C). However, this argument is not convincing, in part because many of the likely progenitor populations of wild Capra are now extinct (7), as for cattle, and thus cannot be sampled. Furthermore, it is unlikely that such divergent lineages have evolved from a single ancestral population, because the population would have had to be extremely large to maintain lineages that were divergent enough to have given rise to the highly divergent lineages found in goats today. The size of the ancestral population that would be necessary to have given rise to the three divergent lineages is ≈38,000–82,000 reproductive females, assuming a 2–3 year generation length and assuming that domestication occurred ≈10,000 YA (see Supplemental Text, which is published as supplemental data on the PNAS web site, www.pnas.org). Goat domestication is highly unlikely to have been much before 10,000 YA according to the comprehensive archeological record (see below and Table 1).
Table 1.
Some archaeological sites from which claims for evidence of livestock domestication have been made
| Area and country | Site | Predomestic/domestic species | Date calibrated | Refs. |
|---|---|---|---|---|
| Taurus, Southeastern Turkey* | Nevali Çori | Goat, sheep, pig | 8,500–8,000 B.C. | 4 |
| Cayönü | Pig, goat?, sheep? | 8,500–8,000 B.C. | 28 | |
| Euphrates valley (N. Syria) | Tell Halula | Goat, cattle? | 8,000–7,800 B.C. | 4 |
| Tell Abu Hureyra | Goat, sheep | 8,000–7,800 B.C. | 29 | |
| Central Anatolia, Turkey† | Asikli Höyük | Goat, sheep | 8,000–7,800 B.C. | 3 |
| Cyprus | Shillourokambos | Goat, sheep, pig, cattle | 8,500–8,000 B.C. | 30 |
| Damas basin, Syria† | Choraifé | Goat?, sheep | 7,600–7,500 B.C. | 21 |
| Aswad | Goat?, sheep | 7,600–7,500 B.C. | 21 | |
| South Levant, Israel | Jericho | Goat, sheep | 7,500 B.C. | 21 |
| South Levant, West Jordan | Ain Ghazal | Goat, cattle? | 7,600–7,500 B.C. | 21 |
| Basta | Goat, sheep, cattle | 7,500–7,000 B.C. | 21 | |
| West Zagros, Iran* | Ganj Dareh | Goat | 8,000–7,800 B.C. | 5 |
| Tepe Guran | Goat, sheep | 7,500–7,000 B.C. | 31, 32 | |
| Baluchistan, Pakistan* | Mehrgarh | Goat, sheep, cattle | 7,000 B.C. | 6 |
All dates are calibrated and approximate. Some dates are not yet firmly established and are not derived from directly-dated fossil material. ?, Not definitively domestic.
Probable local domestication.
Possible local domestication.
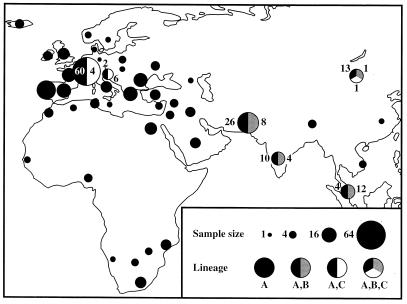
On the other hand, four lines of evidence support the hypothesis of multiple maternal origins. First, the C. hircus B lineage (Fig. 1) was detected only in eastern and southern Asia (Mongolia, Pakistan, India, and Malaysia; Fig. 2), and not in Europe, Africa, or the Middle/Near East where sampling was extensive (including 314 individuals from 63 breeds and 36 countries). This lineage is likely to have arisen in Asia. It is unlikely that all three lineages had a single geographic origin (say, in the Fertile Crescent) and that the B lineage spread to eastern Asia, leaving no trace in other parts of the world. It is indeed unlikely that this lineage would have disappeared via chance (drift) from the Fertile Crescent region, because early domestic populations were expanding, and drift is limited during population expansions. It is therefore more likely to have arisen in Asia.
Figure 2.
Geographic distribution of samples and of the three mtDNA lineages. The size of each circle is proportional to the sample size (1 to 62) from each of 44 countries. The presence of each lineage in a country is represented by a different color (black, lineage A; gray, lineage B; white, lineage C). Thus, the Asian B lineage occurs in Pakistan, India, Malaysia, and Mongolia. The numbers beside each circle on the map show the number of individuals from each lineage. The complete list of breeds and number of individuals sampled per breed and country are shown in Table 3, which is published a supplemental data.
Second, the time since divergence among the three domestic goat lineages (C. hircus A–C) vastly predates the time of domestication suggested from the fossil record. We estimated the divergence time of the goat lineages by sequencing the entire mtDNA cytochrome b gene from two goats from within each lineage and then by calibrating the rate of substitution for the gene (at the third position of codons) by comparing the goat and sheep sequences. Sheep and goats diverged ≈5–7 million YA, according to the ungulate fossil record (19, 20). The estimated distance between goat and sheep sequences at the third codon position was 0.536 substitutions per site, giving an estimated rate of 0.038–0.054 substitutions per site per million years, when using 7 and 5 million years, respectively, as the divergence time. When using this calibration, the most recent common ancestor (i.e., origin) of the domestic goat lineages dates to between ≈201,380 and 281,932 YA. This is long before the estimated domestication time (≈9,000–13,000 YA) suggested by the fossil records for all farm animals and plants that have been domesticated in the Old World (1–5). This estimated divergence time is similar to that (≈200,000 YA) estimated for the two cattle mtDNA lineages that are generally accepted to have originated from two independent domestications (9, 10). Therefore, these data suggest that the three goat lineages arose from genetically discrete populations rather than from a single wild population.
Third, recent mtDNA studies in each of the other major farm animals (cattle, sheep, and pigs), have also revealed multiple highly divergent lineages, with one existing only or primarily in southern or eastern Asia (9–12). This implies a possible Asian center of origin, in addition to the center(s) in the Fertile Crescent (1–5). These results lend support to the hypothesis of multiple genetic origins of goats, and they suggest that multiple maternal origins are a general theme among domestic livestock species.
Fourth, most recent archaeological data are consistent with at least two (and possibly up to five) distinct places for domestication of goats (Table 1). Goats were actually kept in captivity in the southern Turkish region of the Euphrates valley ≈11,000 YA, as revealed at Nevali Çori (4). The appearance of domestic goats ≈10,000 YA in the Zagros (Ganj Dareh, Iran), more than 800 km toward the east, indicates a second and probably independent place for domestication of goats (5). The appearance of domestic goats (together with sheep, pigs, and cattle) ≈9,000 YA in the Indus Basin (Mehrgarh, Eastern Baluchistan) suggests a possible third independent local domestication (6). Finally, the questions concerning local domestication of goats ≈9,000–10,000 YA in the Southern Levant (21) and central Anatolia (3) are still under debate (Table 1).
In light of the above evidence from both archaeology and molecular genetics of livestock, the most parsimonious interpretation of our data is that goats have multiple maternal origins, possibly arising through multiple independent domestications. However, it is still possible that the multiple maternal lineages in goats (and all livestock species) originated via introgression, and not through separate domestication events. Introgression is possible because all domestic livestock species (goats, cattle, sheep, and pigs) can interbreed with wild related species (9, 11, 12, 22–24). It is worth noting that mtDNA is less sensitive to introgression from wild species than is nuclear DNA because mtDNA in transmitted only from females. Thus, mtDNA is more useful for studying domestication because it would detect only introgression from females, which is less likely than introgressions from wild males (24).
Population Expansions.
Mismatch distribution analysis revealed a genetic signature of a population expansion with a different expansion date for each of the three goat mtDNA lineages (Fig. 3). Analyses of mismatch distributions (i.e., distributions of all pairwise sequence differences), by using mtDNA control region sequences, have often been used to detect and date historical human population expansions (13, 14, 25–27). For goats, the bell-shaped mismatch distributions and the star-shaped phylogenies (Figs. 1 and 3) are consistent with a demographic population expansion, such as would be expected following the origin of each lineage from a limited number of founder individuals (25–27).
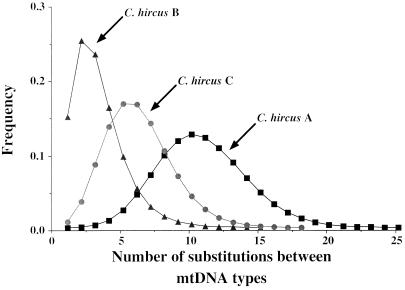
Figure 3.
Mismatch distributions (i.e., pairwise sequence-difference distributions) for mtDNA types from the major lineage of goat sequences, C. hircus A, and from the two smaller lineages, C. hircus B and C. A signature of population growth (i.e., a bell-shaped distribution) is clearly evident in the distribution for all three clusters of sequences, as would be expected for populations expanding after the domestication of relatively few founder-individuals (25). The means of the distributions are different, suggesting different expansion dates for each of the three goat lineages. The Asian lineage (C. hircus B) displays the most recent expansion date.
We can speculate about the approximate relative dates of expansion of each goat lineage by comparing the amount of sequence variation within each lineage and by assuming a similar (limited) amount of mtDNA diversity within each founder lineage. The data suggest that the lineage C. hircus A underwent a relatively ancient population expansion, whereas the other two lineages (B and C) experienced a relatively recent expansion. The diversity within the large lineage C. hircus A (≈10.9 pairwise differences, on average) is far higher than the diversity within each of the two smaller lineages (≈2.3–5.6 differences; Fig. 3). The smaller lineages are widespread geographically (e.g., C. hircus B is found from Mongolia to Pakistan and Malaysia; see below) probably because of a relatively recent spread of these mtDNA lineages. If the two smaller lineages had originated and spread 8,000–10,000 YA, shortly after the estimated date of the initial domestication (1–5), we would expect more divergent lineages to have evolved, as in the larger cluster of mtDNAs (C. hircus A). The relative dating of the population expansions is informative because the expansion dates should roughly correspond to the time that each goat lineage began to become numerous and thus to become important in historical human societies and economies.
We can roughly estimate the relative dates of expansion of each lineage by using the parameters of a stepwise population expansion (estimated from the mismatch distributions), and by taking into account heterogeneity of mutation rates following a gamma distribution with an estimated parameter α = 0.29 (26). The expansion times expressed in mutation units (τ = 2ut, where u is the mutation rate for the 481 base pairs, and t is time in generations) are found to equal 10.8 (CI90% = 9.37 − 11.27) for C. hircus A, 2.3 (CI90% = 0.95 − 3.89) for C. hircus B, and 6.6 (CI90% = 3.67 − 8.94) for C. hircus C. Assuming that the large lineage (C. hircus A) corresponds to an initial domestication that occurred ≈10,000 YA, we find dates of ≈2,130 and ≈6,110 YA, respectively, for the expansions of C. hircus B and C. We can estimate a minimum and maximum value for these latter two dates based on the 90% confidence intervals for the expansion dates of C. hircus B and C expressed in mutational units. These minimal and maximal dates are 841–4,151 YA for C. hircus B, and 3,253–9,536 YA for C. hircus C. These time ranges do not overlap with the primary expansion time (10,000 YA) and thus provide some support for the hypothesis that both C. hircus B and C represent secondary expansions after the initial relatively ancient expansion (C. hircus C). This analysis employs relative dates and thus does not depend on the accuracy of the estimated absolute date of the initial domestication (i.e., ≈10,000 YA).
Population Structure.
Geographic structuring was surprisingly weak among goat populations, as revealed by a phylogeographic analysis of the 406 control region sequences. For example, mtDNA types from the main lineage (C. hircus A) were found in all countries and breeds. The mtDNAs from C. hircus B were found across much of Asia, including Pakistan (three breeds), India (two breeds), Malaysia, and Mongolia. Representatives of C. hircus C were detected as far away as Slovenia, Switzerland, and Mongolia (Fig. 2 and Table 3, which is published as supplemental data). Closely related mtDNA types (differing by 1–4 substitutions) were found in distant locations—e.g., Denmark and Portugal, Mongolia and Ukraine, and Algeria and Turkey. Furthermore, highly divergent mtDNA types (differing by >18 substitutions) were found within breeds and geographic regions. We quantified the degree of structuring by computing the components of mtDNA variation (within breeds vs. between breeds vs. between continental groups of breeds) under the hierarchical AMOVA framework (18). This computation revealed that a large percentage (78.72%) of the total mtDNA variation in C. hircus is distributed within breeds. A smaller but significant percentage exists among continental groups (10.58%; P < 0.001) and among breeds (10.70%; P < 0.001) (Table 2).
Table 2.
Hierarchical distribution of mtDNA (HVI) diversity within and among breeds (and continental groups of breeds) for goats and cattle as computed under the AMOVA framework (15)
| Species | Distribution
of sequence variation, % of total
|
||
|---|---|---|---|
| Within breeds | Among breeds within groups | Among continental groups of breeds | |
| Goats (i) | 78.7 | 10.6 | 10.7 (Northern Europe/Southern Europe/Northern Africa/ Southern Africa/Asia/Middle and Near East) |
| Goats (ii) | 74.0 | 12.1 | 13.9 (Europe/Africa/Asia) |
| Goats (iii) | 72.7 | 12.2 | 15.1 (Europe/Africa) |
| Cattle (ii) | <16, NR | <16, NR | 84.0 (Europe/Africa/Asia) |
| Cattle (iii) | 45.0 | 4.0 | 51.0 (Europe/Africa) |
Note the relatively low percentage of variation among continental groups for goats relative to cattle. The continental groups in Goats (ii) and (iii) were chosen for comparison with published AMOVA values from cattle having the same geographic population groupings; these consist of Europe, Africa, and Asia and Europe and Africa, respectively. NR, not reported (10).
Interestingly, the degree of geographic structuring of goat mtDNA variation is far weaker than that in cattle. For example, intercontinental subdivisions account for ≈84% of the total mtDNA variation in cattle (10), compared with only ≈10% in goats (Table 2). If we exclude Asia (where a second subspecies of wild cattle might have been domesticated), the intercontinental subdivision still accounts for >50% of the mtDNA variation in cattle. The weaker genetic structure in goats probably results from more intercontinental transportation of goats than of cattle. This is consistent with the fact that goats are more portable and represent a smaller currency unit than cattle (8). Sheep and pigs, like cattle, apparently also exhibit substantial phylogenetic structuring, although the DNA data are less extensive (11, 12), and further research is needed.
It is unlikely that the limited structure among goat populations (relative to cattle) results from the very recent transport of goats between continents for three reasons. First, we sampled from remote geographic locations and not from areas where goats might have been shipped recently (e.g., large cities and coastal harbors). We also sampled only native breeds and excluded recently “improved” breeds such as the South African Boer. Second, goats are now less economically important than cattle, sheep, and pigs and thus have been transported less in recent times during which modern transport has made more feasible the long-distance shipping of livestock (especially of large-bodied cattle that are less hardy than goats). Finally, the gametes of goats have been transported less in recent times because artificial insemination and embryo transplant technology is less developed and is seldom conducted in goats compared with other livestock. These arguments suggest that if recent transport has reduced the structuring of livestock populations, it should have weakened the structuring more in cattle than goats. Nonetheless, goats have far weaker population structure than cattle.
The lack of strong phylogeographic structure (i.e., the high gene flow) in goats has intriguing implications for the history of human population movements and commerce. The weak structuring among goat populations suggests that goats have often been transported for commercial trade or during migratory and exploratory movements of humans. For example, goats were carried along on ships and occasionally released onto oceanic islands and distant continents to provide fresh sources of milk and meat (2). It is reasonable that goats have been transported more often and more successfully that other livestock because “goats are perhaps the most versatile of all ruminants in their feeding habits, a factor that has greatly affected their success as a domestic animal” and because goats “are also extremely hardy and will thrive and breed on the minimum of food and under extremes of temperature and humidity” (ref. 8, p. 75).
In summary, multiple maternal origins appear to be a general phenomenon among domestic livestock species. Diverse origins and repeated gene flow from wild stock have probably facilitated the development of the widely different and highly productive breeds we have today. Unfortunately, the mounting extinction rates among wild taxa and domestic breeds are severely diminishing the genetic diversity on which future breed improvement might depend. The finding of a divergent mtDNA lineage in Asia for all four major livestock species is consistent with a center of origin in Asia, in addition to the center in the Fertile Crescent (1–5). Compared with cattle, goats appear to have experienced far more extensive intercontinental gene flow. This is consistent with the robustness, adaptability, and relative ease of transport of goats (especially in historical times). It also suggests that goats might have played an important role in historical human colonizations, migrations, and commerce.
Supplementary Material
Acknowledgments
We thank D. Bradley, M. Bruford, P. England, I. Till-Bottraud, B. Wayne, and three anonymous reviewers for helpful comments on the manuscript. Many thanks to V. Curri and M.-P. Biju-Duval for help with laboratory analyses, and to the following, who kindly provided samples: N. Hasima, A. Virk, A. Ghaffar, O. Hanotte (ILRI), E. Bedin, P. Weinberg, R. Soriguer, S. Dunner, M. K. Sanyasi, L. O. Ngere, D. Zygoyiannis, L. O. Eik, H. Larsen, A. Amcoff, M. Gough, P. Evans, N. Azzopardi, F. Pilla, D. Matassino, V. Fet, H. Amaturado, I. Coroiu, I. Moglan, T. M. Correic, E. Zimba, S. Breznik, E. Eytorsdottir, W. Hamdine, J. Honmode, J. M. Villemot, V. I. Glazko, E. Martyniuk, M. M. Shafie, Ferme du Pic Bois, C. Courturier, M. N. Dye, R. Del Olmo, T. Faure, and especially M. Abo-Shehada, O. Ertugrul, M. Y. Zagdsuren, M. A. A. El-Barody, G. Obexer-Ruff, and G. Dolf. This work was funded by a grant from the European Commission (BIO4CT961189). L.E. was supported by Swiss National Foundation Grant 31-56755.99.
Abbreviations
- YA
years ago
- HVI
first hypervariable segment
- AMOVA
analysis of molecular variance
Footnotes
References
- 1.Pringle H. Science. 1998;282:1448. [Google Scholar]
- 2.Porter V. Goats of the World. Ipswich, U.K.: Farming Press; 1996. [Google Scholar]
- 3.Vigne J-D, Buitenhuis H. Paléorient. 1999;25:49–62. [Google Scholar]
- 4.Peters J, Helmer D, von den Driesch A, Sana-Segui M. Paléorient. 1999;25:27–47. [Google Scholar]
- 5.Zeder M A, Hesse B. Science. 2000;287:2254–2257. doi: 10.1126/science.287.5461.2254. [DOI] [PubMed] [Google Scholar]
- 6.Meadow R H. In: The Origins and Spread of Agriculture and Pastoralism in Eurasia. Harris D R, editor. Washington, DC: Smithsonian Inst.; 1996. pp. 390–412. [Google Scholar]
- 7.Shackleton D M, editor. Wild Sheep and Goats and their Relatives: Status Survey and Conservation Action Plan for Caprinae. Gland, Switzerland: IUCN; 1997. [Google Scholar]
- 8.Clutton-Brock J. Domestic Animals from Early Times. London: Heinemann and Br. Mus. of Nat. Hist.; 1981. [Google Scholar]
- 9.Loftus R T, MacHugh D E, Bradley D E, Sharp P M, Cunningham P. Proc Natl Acad Sci USA. 1994;91:2753–2761. doi: 10.1073/pnas.91.7.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradley D G, MacHugh D E, Cunningham P, Loftus R. Proc Natl Acad Sci USA. 1996;93:5131–5135. doi: 10.1073/pnas.93.10.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiendleder S, Mainz K, Plante Y, Lewalski H. J Hered. 1998;89:113–120. doi: 10.1093/jhered/89.2.113. [DOI] [PubMed] [Google Scholar]
- 12.Giuffra E, Kijas J M H, Amarger V, Carlborg O, Jeon J-T, Andersson L. Genetics. 2000;154:1785–1791. doi: 10.1093/genetics/154.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Excoffier L, Schneider S. Proc Natl Acad Sci USA. 1999;96:10597–10602. doi: 10.1073/pnas.96.19.10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson A C. Science. 1991;253:1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- 15.Wakeley J. J Mol Evol. 1993;37:613–623. doi: 10.1007/BF00182747. [DOI] [PubMed] [Google Scholar]
- 16.Yang Z, Kumar S. Mol Biol Evol. 1996;13:650–659. doi: 10.1093/oxfordjournals.molbev.a025625. [DOI] [PubMed] [Google Scholar]
- 17.Kimura M. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 18.Excoffier L, Smouse P E, Quattro J M. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savage D E, Russell D E. Mammalian Paleofaunas of the World. Reading, MA: Addision–Wesley; 1983. [Google Scholar]
- 20.Carroll R L. Vertebrate Palaeontology and Evolution. New York: Freeman; 1988. [Google Scholar]
- 21.Horwitz L K, Tchernov E, Ducos P, Becker C, von den Driesch A, Martin L, Garrard A. Paléorient. 1999;25:63–80. [Google Scholar]
- 22.Yu Y, Nie L, He Z Q, Wen J K, Jian C S, Zhang Y P. Anim Genet. 1999;30:245–250. doi: 10.1046/j.1365-2052.1999.00483.x. [DOI] [PubMed] [Google Scholar]
- 23.Pulling A, Van S. J Wildl Manage. 1945;9:82–83. [Google Scholar]
- 24.Gauthier D, Martinot J-P, Choisy J, Michallet J-C, Faure E. Le Bouquetin des Alpes Rev Ecol (Terre Vie) Suppl. 1991;6:233. [Google Scholar]
- 25.Rodgers A R, Harpending H. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 26.Schneider S, Excoffier L. Genetics. 1999;152:1079–1089. doi: 10.1093/genetics/152.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marjoram P, Donnelly P. Genetics. 1994;136:673–683. doi: 10.1093/genetics/136.2.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hongo H, Meadow R H. Proc. 4th Int. Symp. on Archaeozoology of Southwestern Asia and adjacent areas. Publication 32, Groningen, The Netherlands: Archaeological Research and Consultancy; 2000. pp. A121–A140. [Google Scholar]
- 29.Legge T. In: The Origins and Spread of Agriculture and Pastoralism in Eurasia. Harris D R, editor. Washington, DC: Smithsonian Inst.; 1996. pp. 238–262. [Google Scholar]
- 30.Vigne J-D, Carrere I, Saliege J-F, Person A, Bocherens H, Guilaine J, Briois F. Proc. 4th Int. Symp. on Archaeozoology of Southwestern Asia and adjacent areas. Publication 32, Groningen, The Netherlands: Archaeological Research and Consultancy; 2000. pp. A83–A106. [Google Scholar]
- 31.Zeder M A. Paléorient. 1999;25:11–25. [Google Scholar]
- 32.Zeder M A. J Archaeol Sci. 2001;28:61–79. [Google Scholar]
- 33.Manceau V, Boursot P, Després L, Taberlet P. Mol Phylogenet Evol. 1999;13:504–510. doi: 10.1006/mpev.1999.0688. [DOI] [PubMed] [Google Scholar]
- 34.Hassanin A, Pasquet E, Vigne J-D. J Mamm Evol. 1998;5:217–326. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.