Abstract
INH: We report two new approaches, using click-chemistry and disulfide bond bridges, to surface-immobilize nucleic acids for single-molecule fluorescence experiments using covalent bonds and self-assembled monolayers. Both approaches are specific and yield comparable results to the avidin-biotin linkage, but offer new surface chemical properties that might be advantageous to prevent non-specific binding of biopolymers to the surface and to expand the range of fluorescent probes that can be employed in single-molecule studies.
Keywords: Single-Molecule Fluorescence, Surface Immobilization, Click Chemistry, Thiol Chemistry, Self-Assembled Monolayers
Single-molecule fluorescence spectroscopy has become a widely used approach to study the mechanisms of folding and catalysis of proteins and nucleic acids.[1, 2] Typically, molecules are surface-immobilized to extend observation times to the minute timescale or longer.[3] An avidin-biotin linkage is commonly used for this purpose because of its stability and high affinity constant.[4] Poly(ethylene glycol) is also typically used to study protein-nucleic acid interactions because it minimizes non-specific binding to the surface.[5] Here, we present two new approaches to immobilize nucleic acids using covalent bonds (Figure 1). We have used a commercially available 18-nucleotide long double stranded DNA oligo with a random sequence modified with a 5’-Cy3 and a 3’-NH2 for single molecule imaging and immobilization. However, these protocols can also be applied to RNA. All microscope slides were cleaned and surface activated prior to being functionalized as described (Experimental Section).
Figure 1.

New fluorophore-labeled DNA immobilization approaches for single molecule imaging using click (A) and thiol (B) chemistries. Top: Single-molecule fluorescence set up with total internal reflection excitation at 532 nm. Inset: Cy3-labeled DNA molecules immobilized on the slide surface using covalent bonds (blue). A) A triazole linkage is generated from a surface azide (yellow) and a 3’-alkyne modified DNA oligo. The approach can be reversed (A, bottom). B) A disulfide bond between the surface and the 3’-DNA thiols immobilizes the DNA onto the quartz slide.
The first approach consists of applying the Hüisgen cycloaddition reaction between an alkyne and an azide group, an example of Sharpless’ “click” chemistry reaction (Figure 1A).[6, 7] The resulting triazole linkage can immobilize a 3’-modified DNA to a functionalized microscope slide surface (Scheme 1A-B). This linkage is stable under physiological conditions, and the reaction can take place with or without a CuI catalyst.[7, 8] Cleaned and activated slide surfaces were functionalized with a terminal azide in a two-step reaction by first forming a self-assembled monolayer (SAM) of 11-bromoundecyltrichlorosilane (BUTS) followed by substitution of the Br for N3 in a saturated solution of NaN3 (Figure S1 in the Supplementary Information). The 3’-NH2 on the DNA was substituted for an alkyne group by reacting with N-hydroxysuccimide activated-pentynoic acid (Figure S2 in the Supplementary Information). The alkyne-modified DNA (10 pM) can be readily immobilized on the N3-functionalized surface by incubating in standard buffer (50 mM Tris-HCl, pH 7.5 and 5 mM MgCl2) for 10 minutes at room temperature (Figure S3 in the Supplementary Information) in the presence or absence of catalyst (1 mM CuSO4 in saturated tris-(benzyltriazolylmethyl)amine (TBTA)).[9] All immobilizations were monitored using a home-built, prism-based total internal reflection fluorescence microscope (Figure 1).[3] A characteristic image is shown in Figure 2A (left). The surface-immobilized fluorophore-labelled DNAs appear as bright yellow dots on the non-fluorescent (black) background. We confirmed the presence of isolated single-molecules by single-step photobleaching of Cy3 (Figure 2B). Occasionally, two or more molecules may be immobilized under the same diffraction limited spot. In this case, the fluorophore of each DNA will photobleach sequentially. We can readily determine the number of molecules under a given spot by simply counting the number of photobleaching steps observed in the fluorescence time trace (Figure 2B), where each photobleaching step corresponds to one immobilized molecule.[10] Under our conditions, the rate of photobleaching is kphotobleach= 0.06 ± 0.02 s-1. We then calculated the surface density of immobilized molecules for each experiment by dividing the total number of molecules by the area of the probe surface. An average surface density of 0.06 molecules/μm2 was obtained (Table 1), which is comparable to immobilization with the avidin-biotin linkage.[11] Out of 326 molecules analyzed, 89% exhibited single-step photobleaching, while 11% photobleached in two or more steps (Table 1), indicating that a small fraction of molecules were immobilized in the same diffraction limited spot. In the presence of the catalyst, the surface density increased to 0.09 molecules/μm2 under the same conditions, and the fraction of spots photobleaching in a single-step did not change (Figure 2A, middle and Table 1).
Scheme 1.
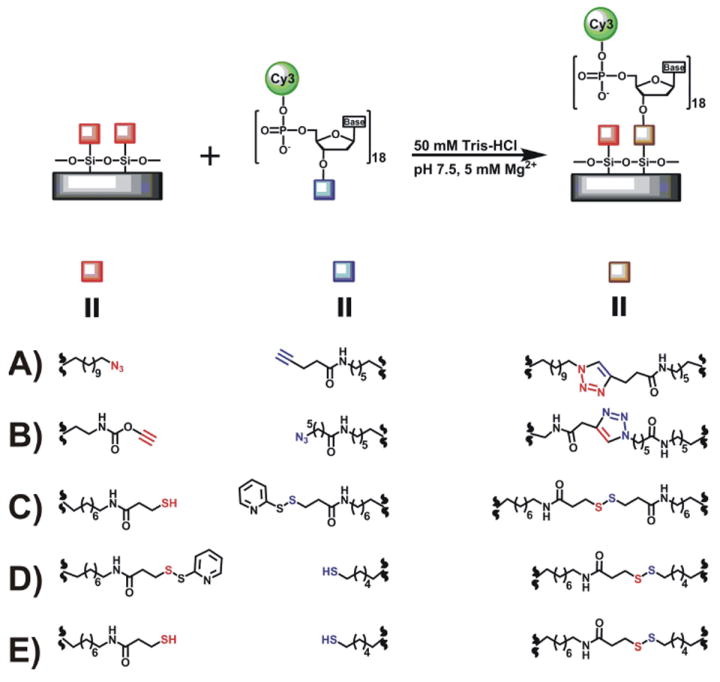
Covalent linkage formation in the click-chemistry (A-B) and the disulfide-bridge (C-E) immobilization approaches. Surface modified group is shown in red and DNA modified terminal in blue.
Figure 2.

Characteristic single-molecule fluorescence images (A, C and E) and fluorescence time trajectories (B, D and F) of covalently immobilized 5’-Cy3 DNA molecules. The fluorescent DNA molecules appear as yellow spots. A) Click chemistry approach with the azide surface and the 3’-alkyne modified DNA in the absence (left) and presence (center) of catalyst. In the absence of azide groups (-N3, right), the average surface density decreases by 40-fold. C) Reverse click chemistry approach with the alkyne surface and the 3’-azide modified DNA in the absence (left) and presence (center) of catalyst. In the absence of surface alkyne groups (-CCH, right), the average surface density decreases by 20-fold. E) Thiol chemistry approach with the thiol surface and 3’-thiol modified DNA (left), pyridyl surface and 3’-thiol modified DNA (middle), and thiol surface and 3’-pyridyl modified DNA (right). B, D, and F) 89% of molecules photobleached in a single step.
Table 1.
Frequencies of the number of molecules per trace found in the single molecule experiments of each immobilization approach.
| Modifications
|
N[a] | Density[b] | Frequencies[c] |
||||
|---|---|---|---|---|---|---|---|
| Surface | Oligo | P(1) | P(2) | P(3) | P(4) | ||
| N3 | C≡CH | 327 | 0.06 ± 0.02 | 0.89 | 0.10 | <0.01 | <0.01 |
| N3 (catalyzed) | C≡CH | 214 | 0.09 ± 0.02 | 0.89 | 0.09 | 0.02 | - |
| C≡CH | N3 | 240 | 0.04 ± 0.01 | 0.89 | 0.10 | 0.01 | - |
| C≡CH (catalyzed) | N3 | 197 | 0.04 ± 0.01 | 0.74 | 0.22 | 0.04 | 0.01 |
| SH | SH | 318 | 0.03 ± 0.01 | 0.89 | 0.10 | 0.01 | - |
| PyrSS | SH | 102 | 0.06 ± 0.01 | 0.88 | 0.11 | 0.01 | - |
| SH | PyrSS | 71 | 0.04 ± 0.01 | 0.96 | 0.04 | - | - |
N indicates the number of molecules analyzed.
molecules/μm2.
P(n) indicates the fraction of fluorescent spots that photobleach in n steps. The number of steps (n) indicates the number of molecules under the same diffraction limited spot. Frequency error bars are ~1%.
To test for the specificity of this immobilization approach we functionalized the surface with BUTS but did not substitute Br for N3, which prevents the formation of the triazole linkage. After incubation with the alkyne-modified DNA, the surface density decreased 20-fold on average (Figure 2A, right), showing that ≤5% of the alkyne-modified DNA binds to the surface non-specifically, similar to the avidin-biotin linkage.[12]
This approach can be easily inverted by functionalizing the surface with the alkyne and modifying the DNA with the azide (Scheme 1B). One advantage of the inverse approach is that the alkyne-functionalized surface can be prepared in a single step (Figure S4 in the Supplementary Information). The azide-modified DNA is prepared by incubating the 3’-NH2 DNA with 6-azidohexanoic acid succinimidyl ester (Figure S5 in the Supplementary Information). After incubation at room temperature of 10 pM azide-DNA on the alkyne surface in standard buffer for 10 minutes (Figure S8 in the Supplementary Information), the inverse “click” approach yields an average surface density of 0.04 molecules/μm2 (Figure 2C, left, and Table 1), and out of 240 molecules analyzed 89% photobleached in a single step (Figure 2d). Addition of catalyst did not significantly affect the average surface density, but increased the fraction of spots that photobleach in multiple steps to 27% (Figure 2C, center and Table 1). This result indicates that the catalyst may promote multiple immobilizations in nearby locations. Incubating with non-azide modified 3’-NH2 DNA decreases the surface density 40-fold on average (Figure 2B, right), showing that ≤2.5% of the DNA binds to the alkyne-functionalized surface non-specifically.
The second approach exploits the efficient coupling between two thiol groups to immobilize nucleic acids by forming disulfide bridges (Figure 1B and Scheme 1C-E). A commercially available 3’-thiol-modified DNA was used, while the microscope slide was functionalized with a thiol in a three-step process (Figure S7 in the Supplementary Information). First, cleaned and surface-activated slides were treated with 3-aminopropyltriethoxysilane, followed by N-succinimidyl-3-(2-pyridylthio) propionate (SPDP), and finally with tris-(2-carboxyethyl) phosphyne (TCEP). Then, 10 pM thiol-modified DNA was surface-immobilized by incubating it in standard buffer for 10 minutes (Figure S8 in the Supplementary Information). An average surface density of 0.03 molecules/μm2 was obtained (Figure 2e, left), and out of 318 molecules analyzed, 89% photobleached in a single step (Table 1). In the absence of surface thiols, the surface density decreased by more than 100-fold (data not shown), demonstrating the specificity of this immobilization approach.
Interestingly, the thiol-modified DNA oligo could also be immobilized on the SPDP-functionalized surface, thus eliminating the need for the TCEP treatment step (Figures S7 and S9 in the Supplementary Information). The resulting single-molecule image (Figure 2E, middle), shows that the surface density is two-fold higher than with the thiol, while the fraction of fluorescent spots photobleaching in a single step remains unchanged (Table 1).
This approach can be also reversed by linking SPDP to the 3’-NH2-modified DNA (Figure S10 in the Supplementary Information) and incubating it on the thiol-functionalized surface (Figures S11 in the Supplementary Information). The resulting average surface density is 0.04 molecules/μm2 (Figure 2E, right), and only 4% of 71 molecules analyzed showed two-step photobleaching (Table 1).
In summary, we have shown two new approaches to surface-immobilize DNA or RNA oligos onto microscope slides for single-molecule fluorescence spectroscopy. This is the first example where click and thiol chemistries have been used for this purpose. Both approaches result in efficient and specific surface immobilization of DNA and low fluorescence background, comparable to the avidin-biotin linkage. However, these new approaches offer different chemical properties of the surface, which may be useful for the surface-immobilization of biopolymers. For example, the covalent nature of the linking bond is expected to be more stable than the typical avidin-biotin linkage, however the thiol approach can only be used under non-reducing conditions. The removal of the avidin protein opens up the possibility to employ UV probes that would otherwise be masked by the tryptophan fluorescence from avidin. Both the azide and alkyne SAMs are expected to make the slide surface highly hydrophobic, and thus help to prevent non-specific interactions with the glass surface. Both the click chemistry and the thiol approaches have been previously shown to be compatible with downstream processing.[13, 14] We found that the “click” chemistry approach with the alkyne-functionalized surface was the least time consuming and required the least materials. Both of these covalent bond-based approaches may also prove useful for single-molecule force spectroscopy studies when other polymer types must be immobilized.
Experimental Section
DNA sample
Commercially available 18 nucleotide-long DNA oligos (Sigma-Genosys) with a random sequence, and a 5’ Cy3 dye and a 3’ functional amine (5’-Cy3-TGC TAC CAG GCG AAG ATA-NH2-3’, and the unmodified complementary strand) were purified by poly-acrylamide gel electrophoresis followed by HPLC, as described.[15] The oligo concentration was determined by its absorption at 260 nm (ε260= 186800 L/mole•cm). These two DNA strands were heated at 90° C for 45 sec and annealed for 10 minutes in standard buffer (50 mM Tris-HCl, pH 7.5 and 5 mM MgCl2).
Microscope slide cleaning and activation procedure
The slides were rubbed with Alconox/ddH2O (10%), sonicated in preheated (80°C) Alconox (10%) for 10 minutes and rinsed with ddH2O to remove all the soap. The slides were soaked into a basic piranha solution (NH4OH:30% H2O2, 3:1) and heated (60°C) for 25 minutes. The quartz substrates were rinsed with ddH2O and passed over a flame to remove any organic residual. The slides were soaked into a NaOH solution (10%) at 80°C for 10 minutes and incubated at room temperature in HCl (1%) for 2 minutes. The slides were heated until boiling in ddH2O and immediately sonicated for 10 minutes, rinsed with EtOH and dried under nitrogen. The substrates were stored under N2 until used for the surface modification reactions.
Azide slide surface modification[16]
Clean and surface activated slides were incubated in a stirring solution of 11-bromoundecyltrichlorosilane (Gelest) in toluene (0.1% v/v) for 45 minutes. The substrates were rinse and sonicated in fresh toluene for 5 minutes to remove any non-specific silanation layer. The slides were rinsed with toluene, acetone, and ethanol, and sonicated in ethanol for 5 minutes. After sonication, bromide modified slides were rinsed with ethanol, dried, and stored under nitrogen. The surface Br were displaced by N3 by incubating the slides in a saturated solution of NaN3 in DMF for ~48 hours at room temperature. The slides were rinsed with ddH2O, acetone, and ethanol and sonicated in ethanol for 5 minutes. The azide modified slides were rinsed with ethanol and dried and stored under nitrogen until used.
N-hydroxysuccimide activated-pentynoic acid (3)
Compound 3 was synthesized according to published procedures.[14] Pentynoic acid (0.2 g, 2.0 mmol) (1, Acros Organics) was dissolved in dry CH2Cl2 (60 mL) under Argon atmosphere. N-hydroxysuccimide (0.23 g, 2.00 mmol) (2, Acros Organics) and 1,3-dicyclohexylcarbodiimide (DCC) (0.46 g, 2.2 mmol) of were added to the resulting solution. The reaction was stirred at room temperature under Argon for 2.5 hours. The reaction mixture was filtered and washed with saturated aqueous solution of NaHCO3 and saturated solution of NaCl. The organic layer was separated, washed with CH2Cl2, dried with anhydrous Na2SO4, filtered and concentrated to dryness until all solvent was evaporated and a white solid was obtained. This solid was purified using chromatography on silica (20 g) and eluted with CH2Cl2:ethyl acetate (40:60). Main product 3 (Figure S2a) was collected and concentrated to dryness until all solvent was evaporated and a white solid (0.17 g, 42% yield) was obtained. Characterization of product 3 was in agreement with previously reported results;[14] m.p. 71°C; 1H NMR (500 MHz, CDCl3) δ 2.05 (t, 1H), 2.62 (dt, 2H), 2.84 (s, 4H), 2.88 (t, 2H); 13C NMR (125 MHz, CDCl3): δ 14.3 (CH2), 25.8 (CH2), 30.5 (CH2), 70.3 (C), 81.1 (CH), 167,2 (C), 169.1 (C); ESI-MS: m/z found [MH]+ 196.23.
3’-alkyne modified oligo (CC02)
A reaction mixture of 3 (0.374 μmol, 19 μL, 19.7 mM in DMF), amino modified oligo (CC01) (10.7 nmol) and sodium phosphate (Na-Pi) (75 μL, 0.1 M pH 7.9) was incubated at room temperature for 1 hour. The mixture was diluted with Na-Pi (200 μL, 0.1 M pH 7.9) and extracted with CHCl3 (200 μL). After extraction, the aqueous layer was desalted and HPLC purified. Compound CC02 (Figure S2b in the Supplementary Information) was characterized by MALDI-TOF: m/z found [MH]+ 6298.442.
Alkyne slide surface modification
Clean slides were incubated in a solution of o-(propargyloxy)-N-(triethoxysilylpropyl) urethane (Gelest) (60 mL, 0.1% v/v) in ethanol (95%) for 45 minutes. The substrates were rinse and sonicated in ethanol for 5 minutes to remove any non-specific silanation layer. The slides were rinsed with ddH2O, then with ethanol and dried and kept under nitrogen atmosphere until they were used in the immobilization reaction.
6-Azidohexanoic acid (5)
Compound 5 was synthesized as described.[17] 6-Bromohexanoic acid (4, Acros Organics) (1.5 g, 7.7 mmol) was dissolved by stirring in DMF (5 mL). To this solution was added sodium azide (1.0 g, 15.4 mmol), and the mixture was heated at 85°C in an oil bath for 3 hours. The mixture was diluted in CH2Cl2 (20 mL) and washed with HCl (10 mL, 0.1 N). The organic layer was extracted, washed with HCl (0.1 N) and dried over anhydrous Na2SO4. The solution was filtered and concentrated until all the solvent was evaporated under vacuum to give 5 (Figure S5a in the Supplementary Information), a yellow oil-like solution. Rf = 0.6 (ethyl acetate:CH2Cl2, 3:7); ESI-MS m/z found [M]- 156.28.
6-Azidohexanoic acid succinimidyl ester (6)
Compound 6 was synthesized according to published procedures.[17] To a stirred solution mixture of CHCl3:DMF (1 mL, 9:1), 5 (590 mg, 3.76 mmol) and N-hydroxysuccinimide (Acros Organics) (432 mg, 3.76 mmol) was added 1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC, Acros Organics) (720 mg, 3.76 mmol). The reaction mixture was stirred at room temperature overnight. The reaction mixture was diluted with CH2Cl2 (20 mL) and washed with HCl (10 mL, 0.1 N). The organic layer was extracted, rinsed with HCl (0.1 N), washed with aqueous NaHCO3 (5%) and with concentrated NaCl. The organic layer was dried over anhydrous Na2SO4, filtered and concentrated until all solvent evaporated under vacuum. The obtained yellowish solution was purified using chromatography on silica (20 g) and eluted with a ethyl acetate:CH2Cl2 (3:7). Main product 6 (Figure S5a in the Supplementary Information) was collected and concentrated to dryness until all solvent was evaporated and an oil solution was obtained (700 mg, 73% yield). Characterization of product 6 was in agreement with previously reported results;[17] Rf = 0.77 (ethyl acetate:CH2Cl2 3:7); 1H NMR (500 MHz, CDCl3) δ 1.54-1.50 (m, 2H), 1.67-1.62 (m, 2H), 1.82-1.77 (q, 2H), 2.66-2.63 (q, 2H), 2.85 (br s, 4H), 3.32-3.30 (t, 2H); ESI-MS: m/z found [MH]+ 255.3.
3’-azide modified oligo (CC03)
A reaction mixture of of 6 (0.380 μmol, 19 μL, 20 mM in DMF), amino modified oligo (CC01) (10.7 nmol) and Na-Pi (75 μL, 0.1 M pH 7.9) was incubated at room temperature for 1 hour. The mixture was diluted with Na-Pi (200 μL, 0.1 M pH 7.9) and extracted with CHCl3 (200 μL) After extraction, the aqueous layer was desalted and HPLC purified. Compound CC03 (Figure S5b in the Supplementary Information) was characterized by MALDI-TOF: m/z found [MH]+ 6358.157.
Pyridyl disulfide slide surface modification via SPDP (PyrSS-slide)
Clean slides were incubated in a solution of 3-aminopropyltriethoxysilane (Gelest) (60 mL, 0.1% v/v) in ethanol (95%) for 45 minutes. The substrates were rinse and sonicated in ethanol for 5 minutes to remove any non-specific silanation layer. The slides were rinsed with ddH2O, then with ethanol and dried and kept under nitrogen atmosphere until they were used in the immobilization reaction. Placed amino silanated slides in a humid chamber with SPDP (70 μL, 20 mM in DMF/PBS) spotted in the single molecule experiment probe area on the surface of the slide and incubated overnight. Rinsed slides with DMF, with ethanol and dried and stored under nitrogen atmosphere until the immobilization reaction.
Thiol slide surface modification
To produce the SH terminals, use the pyridyl disulfide-modified slide (as described above), and incubate the sample chamber with TCEP (100 μL, 20 mM) for 20 minutes. Flush the excess TCEP and pyridine-2-thione groups by washing the chamber with standard buffer (100 μL).
Thiol slide surface modification via silanation
Clean slides were incubated in a solution of 3-mercaptopropyltriethoxysilane (Gelest) (60 mL, 0.1% v/v) in ethanol (95%) for 45 minutes. The substrates were rinse and sonicated in ethanol for 5 minutes to remove any non-specific silanation layer. The slides were rinsed with ddH2O, then with ethanol and dried and kept under nitrogen atmosphere until they were used in the immobilization reaction.
3’-pyridyl disulfide modified oligo (PyrSS)
A reaction mixture of SPDP (0.380 μmol, 19 μL, 20 mM in DMF), CC01 (10.7 nmol), and Na-Pi (75 μL, 0.1 M pH 7.9) was incubated at room temperature for 1 hour. The mixture was diluted with Na-Pi (200 μL 0.1 M pH 7.9) and extracted with CHCl3 (200 μL). After extraction the aqueous layer was desalted. The main product PyrSS was used without further purification.
Single-molecule experiments
After the slide surface modification, the sample probe volume chamber was assembled as described.[3] Modified oligo (10 pM) was incubated inside the chamber in standard buffer (50 mM Tris-HCl pH 7.5, 5 mM Mg2+) for 10 minutes. When the catalyst (CuI/TBTA) was used in the click chemistry reaction, the buffer was supplemented with 1 mM CuSO4 in saturated TBTA. Non-immobilized oligos were flushed from the chamber with standard buffer. All single molecule experiments were performed using a home-built, prism-based, total internal reflection fluorescence microscope as described.[3] Fluorescence from single, surface-immobilized molecules was imaged onto a back illuminated, electron-multiplied CCD camera (iXon+, Andor Technology, South Windsor, CT) with 33 ms time resolution.
Supplementary Material
Acknowledgments
This research was supported by NSF Postdoctoral Fellowship (DBI-0805651) to E.A.A., and NSF Career (MCB-0747285) and NIH (R01-GM085116) to D. R. Special thanks to Santosh Mahto and Christine Chow for help in the synthesis of 3 and 6.
References
- 1.Alemán EA, Lamichhane R, Rueda D. Curr Opin Chem Biol. 2008;12:647. doi: 10.1016/j.cbpa.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Schuler B, Eaton WA. Curr Opin Struct Biol. 2008;18:16. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao R, Rueda D. Methods. 2009 doi: 10.1016/j.ymeth.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 4.Ha T. Methods. 2001;25:78. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 5.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Nature. 2002;419:638. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 6.Kolb HC, Finn MG, Sharpless KB. Angew Chem Int Ed Engl. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 7.Rozkiewicz DI, Janczewski D, Verboom W, Ravoo BJ, Reinhoudt DN. Angew Chem Int Ed. 2006;45:5292. doi: 10.1002/anie.200601090. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Seo TS, Ju J. Tetrahedron Lett. 2004;45:3143. [Google Scholar]
- 9.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Org Lett. 2004;6:2853. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 10.Shu D, Zhang H, Jin J, Guo P. EMBO J. 2007;26:527. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roy R, Hohng S, Ha T. Nat Methods. 2008;5:507. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. Science. 2000;288:2048. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 13.Chu BC, Orgel LE. Nucleic Acids Res. 1988;16:3671. doi: 10.1093/nar/16.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humenik M, Huang Y, Wang Y, Sprinzl M. ChemBioChem. 2007;8:1103. doi: 10.1002/cbic.200700070. [DOI] [PubMed] [Google Scholar]
- 15.Rueda D, Walter NG. Methods Mol Biol. 2006;335:289. doi: 10.1385/1-59745-069-3:289. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Cai J, Rauscher H, Behm RJ, Goedel WA. Chem Eur J. 2005;11:3968. doi: 10.1002/chem.200400896. [DOI] [PubMed] [Google Scholar]
- 17.Grandjean C, Boutonnier A, Guerreiro C, Fournier JM, Mulard LA. J Org Chem. 2005;70:7123. doi: 10.1021/jo0505472. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


