Abstract
We describe a nonparametric framework for incorporating information from co-registered anatomical images into positron emission tomographic (PET) image reconstruction through priors based on information theoretic similarity measures. We compare and evaluate the use of mutual information (MI) and joint entropy (JE) between feature vectors extracted from the anatomical and PET images as priors in PET reconstruction. Scale-space theory provides a framework for the analysis of images at different levels of detail, and we use this approach to define feature vectors that emphasize prominent boundaries in the anatomical and functional images, and attach less importance to detail and noise that is less likely to be correlated in the two images. Through simulations that model the best case scenario of perfect agreement between the anatomical and functional images, and a more realistic situation with a real magnetic resonance image and a PET phantom that has partial volumes and a smooth variation of intensities, we evaluate the performance of MI and JE based priors in comparison to a Gaussian quadratic prior, which does not use any anatomical information. We also apply this method to clinical brain scan data using F18 Fallypride, a tracer that binds to dopamine receptors and therefore localizes mainly in the striatum. We present an efficient method of computing these priors and their derivatives based on fast Fourier transforms that reduce the complexity of their convolution-like expressions. Our results indicate that while sensitive to initialization and choice of hyperparameters, information theoretic priors can reconstruct images with higher contrast and superior quantitation than quadratic priors.
Keywords: Anatomical priors, joint entropy, mutual information, positron emission tomography
I. Introduction
THE uptake of positron emission tomographic (PET) tracers typically results in a spatial density that reflects the underlying anatomical morphology. This results in a strong correlation between the structure of coregistered anatomical and functional images for many commonly used tracers such as F18 FDG. Hence, the incorporation of anatomical information from coregistered MR/CT images into PET reconstruction algorithms can potentially improve the quality of low resolution PET images. This anatomical information is readily available from multimodality imaging equipment that is often used for acquiring data and can be incorporated into the PET reconstruction algorithm in a Bayesian framework through the use of priors.
Previous work on the use of anatomical priors can be broadly classified into: 1) methods based on anatomical boundary information, which encourage boundaries in functional images that correspond to anatomical boundaries [1]–[4], and 2) methods that use anatomical segmentation information, which encourage a smooth distribution of the tracers within each anatomical region [5]–[9]. In [10] a method that did not use explicit boundary or segmentation information was proposed, wherein the prior aimed to produce homogeneous regions in the PET image where the anatomical image had an approximately homogeneous distribution of intensities. These approaches rely on boundaries, segmentation, or the intensities themselves to model the similarity in structure between the anatomical and functional images. Mutual information (MI) between two random vectors is the Kullback-Leibler (KL) distance between their joint probability density and the product of their marginal densities. It can be interpreted as a measure of the amount of information contained by one random vector about the other and therefore can be used as a similarity metric between images collected using two different modalities [11]. In [9] a Bayesian joint mixture model was formulated such that the solution maximizes MI between class labels. A parametric method was used where each class conditional prior was assigned a Gamma distribution.
In [12] and [13] we described a nonparametric method that uses MI between feature vectors extracted from the anatomical and functional images to define a prior on the functional image. This approach did not require explicit segmentation or boundary extraction, and aimed to reconstruct images that had a distribution of intensities and/or spatial gradients that matched that of the anatomical image. MI achieves this by minimizing the joint entropy while maximizing a marginal entropy term that models the uncertainty in the PET image itself. In [14] it was shown through anecdotal examples that due to the marginal entropy term, MI tends to produce biased estimates in the case where there are differences in the anatomical and functional images, and that joint entropy (JE) is a more robust metric in these situations.
In this paper, we extend the nonparametric framework of our MI based priors to explore the use of both MI and JE as priors in PET image reconstruction. Since both these information theoretic metrics are based on global distributions of the image intensities, spatial information can be incorporated by adding features that capture the local variation in image intensities. We define features based on scale space theory, which provides a framework for the analysis of images at different levels of detail. Scale-space theory is based on generating a one parameter family of images by blurring the image with Gaussian kernels of increasing width (the scale parameter) and analyzing these blurred images to extract structural features [15]. We define the scale-space features for information theoretic (IT) priors as, the original image, the image blurred at different scales, and the Laplacians of the blurred images. These features reflect the assumption that boundaries in the two images are similar, and that the image intensities follow similar homogeneous distributions within boundaries. By analyzing the images at different scales, we aim to automatically emphasize stronger boundaries that delineate important anatomical structures, and attach less importance to internal detail and noise which is blurred out in the higher scale images.
We evaluate the performance through simulations as well as clinical data, of both MI and JE based priors in comparison with the commonly used quadratic prior (QP) [16], [17], which does not use any anatomical information. The simulations explore both the ideal case of perfect agreement between the anatomical and functional images, as well as a more realistic case of the images having some differences. Additionally, we compare reconstructions of clinical brain data obtained from a PET scan using F18 Fallypride, a tracer that binds to dopamine receptors and therefore localizes mainly in the striatum. Finally, we present an efficient method of computing the priors and their derivatives, which reduces the complexity from O(MNs) to O(M log M + Ns), where Ns is the number of feature vectors in the image and M is the number of points at which the joint density of the anatomical and functional images is estimated in order to compute the priors. This makes these priors practical to use for 3-D images where Ns can be large.
II. Methods
A. MAP Estimation Using Information Theoretic Priors
We represent the PET image by f = [f1, f2, …, fN]T, where fi represents the activity at voxel index i, and the coregistered anatomical image by a = [a1, a2, …, aN]T. Let g denote the sinogram data, which is modeled as a set of independent Poisson random variables gi, i = 1, 2, …, K. The maximum a posteriori estimate of f is given by
| (1) |
where p(f) is the prior on the image, and p(g|f) is the Poisson likelihood function given by
| (2) |
Pij represents the i, jth element of the K × N forward projection matrix P. In this paper, P not only models the geometric mapping between the source and detector but also incorporates detector sensitivity, detector blurring, and attenuation effects [18].
The prior term P(f) can be used to incorporate a priori information about the PET image. We utilize information that is available through coregistered anatomical images, by defining the prior in terms of information theoretic measures of similarity between the anatomical and functional images. Through this framework, we aim to reconstruct a PET image that maximizes the likelihood of the data, while also being maximally similar in the information theoretic sense to the anatomical image.
To define the prior, we extract feature vectors that can be expected to be correlated in the PET and anatomical images. Let the Ps feature vectors extracted from the PET and anatomical images be represented as xi and Yi respectively for i = 1, 2, …, Ns. These can be considered as independent realizations of the random vectors X and Y. Let m be the number of features in each feature vector such that X = [X1, X2, …, Xm]T. If D(X, Y) is the information theoretic similarity metric that is defined between X and Y, the prior is then defined as
| (3) |
where Z is a normalizing constant and μ is a positive hyperparameter.
Taking log of the posterior and dropping constants, the objective function L(f) is given by
| (4) |
and the MAP estimate of f is given by
| (5) |
B. Information Theoretic Priors
We use mutual information and joint entropy as measures of similarity between the anatomical and functional images. Mutual information between two continuous random variables X and Y with marginal distributions p(x) and p(y), and joint distribution is defined as
| (6) |
where the entropy H(X) is defined as
| (7) |
and the joint entropy H(X, Y) is given by
| (8) |
Mutual information can be interpreted as the reduction in uncertainty (entropy) of X by the knowledge of Y or vice versa [19]. When X and Y are independent I(X, Y) takes its minimum value of zero, and is maximized when knowledge of one random variable completely explains the other. The distribution p(x, y) that maximizes MI is sparse with localized peaks. This also corresponds to a reduction in the joint entropy H(X, Y), from (6) and (8).
We therefore define D(X, Y) in terms of MI as
| (9) |
or in terms of JE as
| (10) |
For continuous random variables, the entropy H(X) (called differential entropy) can go to –∞ unlike the Shannon entropy of a discrete random variable which has a minimum value of zero. Differential joint entropy approaches –∞ as the joint distribution approaches a delta function. In practice, the continuous integrals in (7) and (8) are approximated by numerical integration by sampling the continuous distribution at intervals of width Δx, which ensures that the value of the similarity metric is bounded.
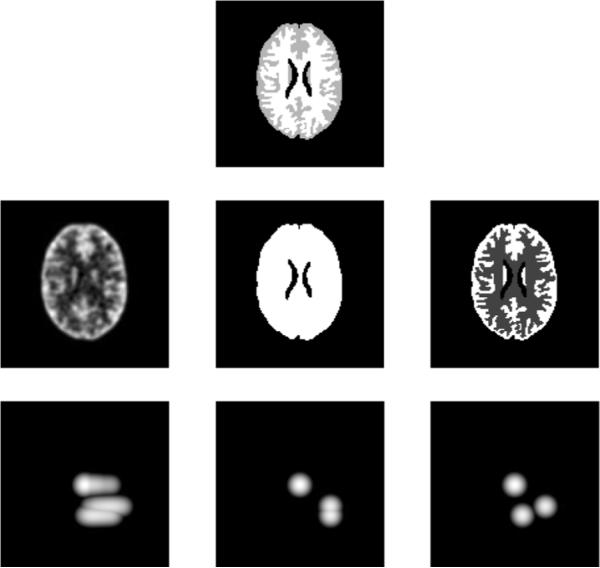
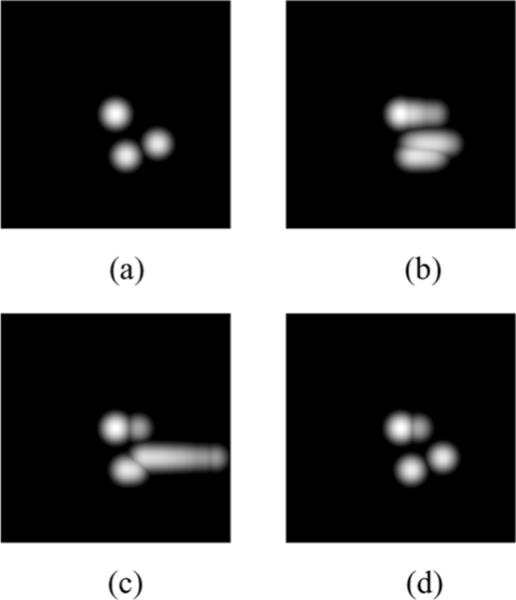
To illustrate the behavior of MI and JE as similarity metrics, we consider some example PET images and their joint pdfs (estimated from joint histograms that are blurred with Gaussian windows) with a reference image Xref, all of which are shown in Fig. 1. The horizontal axis of the pdfs shown denotes PET image intensities, and vertical axis the reference image intensities. The examples illustrate the behavior of MI and JE for 1) a blurred image Xblur, 2) an image Xhom that has the same activity in regions corresponding to two distinct anatomical regions, and 3) an image Xtrue that is identical in structure to the anatomical image. It can be seen that the joint pdf p(Xtrue, Xref) is more clustered than that of p(Xblur, Xref). The joint entropy of these images is such that H(Xblur, Xref) > H(Xtrue, Xref), and their MI is such that I(Xblur, Xref) < I(Xtrue, Xref) . Thus, by using priors that maximize MI or minimize JE, we can potentially reconstruct PET images that have higher resolution due to the structural information that is available from the anatomical image. In case of Xhom however, H(Xtrue, Xref) ≃ H(Xhom, Xref), whereas I(Xtrue, Xref) > H(Xhom, Xref). Thus the JE prior can possibly reconstruct an image that has more than one anatomical region map to a homogeneous functional region without change in its value, while MI prefers images that have the same structure as the reference image. This is due to the presence of the marginal entropy terms in MI that penalize reconstructions with fewer distinct intensity values than that in the reference image.
Fig. 1.
Log of estimated joint probability distribution functions: Top: Reference image, Middle: Example reconstructed images (L to R): Blurred image, image with same activity in two distinct anatomical regions, and image with identical structure as reference image. Bottom: Estimated joint pdf of the images in the middle row and anatomical image.
This distinction causes JE to be more robust than MI to differences between the PET and anatomical images, where the anatomical image has structures that are not present in the PET image [14]. However, this property could also lead to distinct regions in the PET image that correspond to clusters that are close to each other in the joint density to be combined into one region. It should also be noted that since the optimization is with respect to image intensities, the marginal entropy term in MI may cause the PET image intensities to have high variance, thereby increasing the entropy in the image. Both these metrics measure similarity in distributions rather than actual values of intensities, and rely on the likelihood term to converge to the correct solution.
C. Extraction of Feature Vectors
Mutual information and joint entropy between images is not directly affected by the spatial structure of the images. In other words, a random spatial reordering of corresponding pairs of voxels in the two images would produce identical measures. We introduce spatial information into the information theoretic priors by defining feature vectors that capture the local morphology of the images. The feature vectors should be chosen such that they are correlated in the anatomical and functional images, and should reflect the morphology of the images. Since the information theoretic priors defined in this paper are naturally insensitive to differences in intensity between the two images, the combination of local measures of image morphology and information theoretic measures should facilitate reconstruction of PET images whose structure is similar to that of the registered anatomical image.
Scale space theory provides a framework for the extraction of useful information from an image through a multiscale representation of the image [15]. In this approach, a family of images is generated from an image by blurring it with Gaussians of increasing width (scale parameter), thus generating images at decreasing levels of detail. Relevant features can then be extracted by analyzing this family of images rather than just the original image.
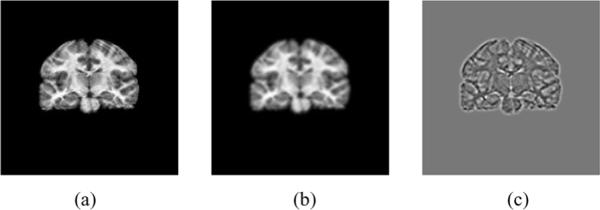
We use this approach to define the feature vectors as 1) the original image, 2) the image blurred by a Gaussian kernel of width σ1, and 3) Laplacian of the blurred image. By analyzing the image at two different scales (the original and image at scale σ1, we are giving more emphasis to those boundaries that remain in the image at the higher scale, and attach less importance to those that will be blurred out at the higher scale. A coronal slice of an MR image of the brain and its scale-space features are shown in Fig. 2. More features could be added by using different sizes of kernels, but we restrict our analysis to two scales to limit complexity. We further make the simplifying assumption of independence between features so that
| (11) |
where Xi and Yi are the random variables corresponding to the ith scale-space images. Though the scale-space images are correlated, we choose to make this independence assumption to reduce the computational cost of the prior term. Different weights on each of the scale-space features could be used to provide more emphasis to some features over others depending on the application. An investigation of the effect of weighting of the scale-space priors is left for future work.
Fig. 2.
Scale-space features of a coronal slice of an MR image of the brain. (a) Original image, (b) image blurred by a Gaussian, and (c) Laplacian of the blurred image in (b).
D. Computation of Prior Term
We use a nonparametric approach to estimate the joint pdfs using the Parzen window method [20]. Let the number of samples from which to estimate the pdf be Ns, which in our case is equal to the number of voxels N, since we are extracting one feature vector per voxel. Representing the samples by f1, f2, …, fNs the Parzen estimate of p(x) is given by
| (12) |
where ϕ(x/σ) is a windowing function of width σ. The windowing function should integrate to one, and is commonly chosen to be a Gaussian of mean zero and standard deviation σ. For estimating the joint density between X and Y corresponding to the intensities of the coregistered PET and anatomical images f and a, we use a Gaussian window function with diagonal covariance matrix . Thus, the joint density estimate is given by
| (13) |
The variance of the Parzen window is taken as a design parameter. It has been shown [20] that as Ns → ∞, the Parzen estimate converges to the true pdf in the mean squared sense. For PET reconstruction (especially for the 3-D case) Ns is large, so it can be assumed that there are sufficient number of samples to give an accurate estimate of the pdf. We therefore replace with p(x, y) in (6) or (8) to compute the prior term.
E. Computation of MAP Estimate
The objective function given in (4) can be iteratively optimized by a preconditioned conjugate gradient algorithm with an Armijo line search technique [21]. Let the estimate of the PET image at iteration k be , the gradient vector be , the preconditioner be C(k), the conjugate gragradient direction vector be, b(k), and the step size computed from the line search be α(k). The PCG update equations are given by
| (14) |
| (15) |
| (16) |
The algorithm is initialized by b(0) = C(0)d(0). We use the EM-ML preconditioner [22], which is defined as .
The kth element of the gradient vector d can be computed from (4) as
| (17) |
To compute the derivative of, D(X, Y), we first define the derivative of marginal and joint entropy terms corresponding to intensity of the original image. Replacing the integration in (7) with its numerical approximation, the derivative of marginal entropyH(X) with respect to fk is given by
| (18) |
where , and M is the number of points at which the pdf is computed. Similarly, assuming equal M for X and Y for convenience, the gradient of joint entropy is
| (19) |
The gradients corresponding to the other scale-space features can be computed easily from these equations since the Laplacian and blurring are linear operations on and . For an interpretation of these gradients through an alternative formulation, please refer to the Appendix.
We perform an approximate line search using the Armijo rule since it does not require the computation of the second derivative of L(f), which is computationally expensive for the information theoretic priors. Whenever the nonnegativity constraint is violated, we use a bent line search technique by projecting the estimate from the first line search (with negative values) onto the constraint set, and computing a second line search between the current estimate and the projected estimate. The objective function is a nonconvex function of f, so optimization of the function using gradient based techniques require a good initial estimate to converge to the correct solution.
F. Efficient Computation of Entropy and Its Gradient
The computation of the Parzen window estimate p(x) at M locations using Ns samples is O(N Ns). In addition, the gradient in (18) is also of complexity O(N Ns), since for each fk, we compute a summation of M multiplications. This can be expensive for large Ns which is the case in PET reconstruction. We take an approach similar to [23] to compute the entropy measures as well as their gradients efficiently through the use of FFTs. The Parzen window estimate in (12) can be rewritten as a convolution.
| (20) |
Here x is continuous, and the impulse train has impulses at nonequispaced locations.
Efficient fast Fourier transforms can be used to compute convolution on a uniform grid. To take advantage of these efficient implementations, we interpolate the continuous intensity values fj onto a uniform grid with equidistant discrete bin locations , where j = 1, 2, … M, with spacing . Let the image with the quantized intensity values be . The impulses in h(x) are now replaced by triangular interpolation functions thus giving
| (21) |
where
| (22) |
Thus each continuous sample fj contributes to the bin it is closest to, as well as its neighboring bins. The Parzen estimate at equispaced locations is given by
| (23) |
This convolution can now be performed using fast Fourier transforms with complexity M(M log M). We can then compute the approximate marginal entropy by replacing p(x) with in (7).
The gradient equation in (18) also has a convolution structure, and can be rewritten as
| (24) |
If we replace p(x) in the equation with , we can compute the gradient corresponding to the binned intensity values , k = 1, 2,…, M using convolution via FFTs. We represent this binned gradient as . To retrieve the gradient with respect to original intensity values, we use similar interpolation as for binning giving
| (25) |
Thus the complexity of computing the binned gradient is O(M log M) followed by interpolation which is O(Ns). Additionally, the bin locations corresponding to each fk can be stored in the pdf estimation step, and retrieved in the gradient estimation step, thus giving further computational savings. This method can be extended to the 2-D case by using bilinear interpolation instead of the triangular windowing function to compute the binned joint density and to interpolate the gradient at the binned locations to the continuous intensity values.
We compare the direct Parzen window method with the FFT based method in terms of run times and accuracy of the marginal pdf and gradient estimates. For both methods we compute the pdf at the same locations , i = 1, 2,…, M. To compute the Parzen gradient, we expressed (9) as the inner product of the vectors Lp = −Δx/Ns[(1 + log(p(x1)), 1 + log(p(x2)), …, 1 + log(p(xM))]T and Φk = [ϕ′(x1 − fk/σx), ϕ′(x2 − fk/σx), …, ϕ′(xM − fk/σx)]T. The vectorized form enables us to use efficient linear algebra libraries for evaluating the summation for each fk.
The run-times for the combined computation of pdf p(x) and the gradient ▽H(X) using the Parzen and FFT based methods are shown in Table I for different sizes of image. The reported run-times were obtained using a single processor in an AMD Opteron 870 computer with 8 dual core 1.99 GHz CPUs. The norm of the difference between the gradients of the two methods, normalized by the direct Parzen gradient was less than 1% for all the sizes of image considered. Since the main computation cost for the FFT based method is the FFT based convolutions that depend on M rather than image size, we see large speed-up factors for 3-D images compared to the Parzen method for which the complexity increases linearly with image size. It should however be noted that despite the efficient implementation, MAP with IT priors is still computationally more expensive than MAP with QP (approximately double for an image size 256 × 256 × 111 and intensity only scale-space feature) not only due to the pdf estimation and gradient computations, but also due to the Armijo line search chosen for IT priors that requires computation of the objective function at every line search iteration.
TABLE I.
Run-Timesin Seconds for Combined Computation of Pdf and Gradient for Different Image Sizes, for the Parzen and FFT Based Methods
| Image Size (No. of Bins) | Parzen | FFT-based | Speed-up |
|---|---|---|---|
| 128 × 128(M = 351) | 1.01 | 0.11 | 9.1630 |
| 128 × 128 × 111(M = 351) | 117.44 | 0.88 | 133.63 |
| 256 × 256 × 111(M = 351) | 470.64 | 3.45 | 136.41 |
III. Results
We evaluate the performance of MI and JE priors through 1) simulations that model the best case scenario of the anatomical image having identical structure as the PET image, 2) simulations of a more realistic case where there are some differences between the MR and PET regions, and 3) clinical brain imaging data.
We consider four different information theoretic priors.
-
1)
JE-Intensity: Joint entropy between the intensities of anatomical and PET images.
-
2)
MI-Intensity: Mutual information between the intensities of anatomical and PET images.
-
3)
JE-Scale: Joint entropy between scale-space feature vectors (original image, image blurred by Gaussian of width σ1, and Laplacian of blurred image) of the anatomical and PET images.
-
4)
MI-Scale: Mutual information between scale-space feature vectors of anatomical and PET images.
We compare the performance of these priors relative to each other, and to a QP, which does not use any anatomical information. QP penalizes differences in intensities within a neighborhood ηi defined for the 2-D case by eight nearest neighbors of each voxel i, and is given by [16], [17]
| (26) |
where kij is the inverse of the Euclidean distance between the centers of the voxel pair fi and fj. The weight on the prior is controlled by varying the hyperparameter μ for the information theoretic priors, and β for the QP prior.
A. Simulation Results
Our simulations are based on the F18 FDG tracer producing uniform or near-uniform uptake in the gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF) regions of a normal brain. For all the information theoretic priors considered, we computed the densities at 500 bin centers that we keep constant throughout the optimization, the range of which we set to 2.5 times the range of intensities in an initial maximum likelihood (ML) reconstruction. This assumes a reasonable range of values in the initial estimate without which the image might be thresholded at a value that is too low. We have not encountered this situation for the initializations we used. To form the scale-space features, we used a Gaussian kernel of standard deviation σ1 that will be defined separately in the later sections, and a 3 × 3 Laplacian kernel given by .
1) Anatomical and PET Images With Identical Structure
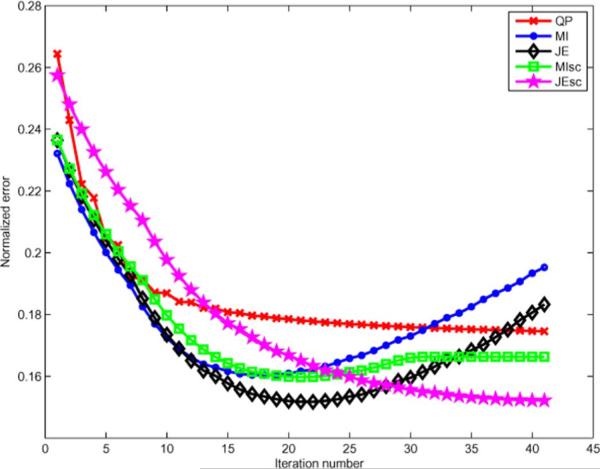
We used a 128 × 128 slice (voxel size 1.5 mm) of the Hoffman brain phantom as our functional image (activity ratios of 4:1:0 in GM, WM and CSF) and used the same image with values of 180 in GM, 255 in WM, and 0 in CSF as our anatomical image, which is shown in the top row of Fig. 1. The simulations represent a single ring PET scanner of diameter 50 cm with 504 detectors with center to center spacing of 6.2 mm leading to a sinogram size of 288 radial samples and 252 angles of view. The sinogram data had approximately 300 000 counts, corresponding to an average of 30 counts per line of response (LOR) that passes through at least one voxel of the phantom. The simulation used a simplified model with no detector blurring, randoms, or scatter. We reconstructed all images using 30 iterations of the PCG algorithm, initialized with two iterations of ordered subset expectation maximization (OSEM) using six subsets. The objective function was not observed to vary considerably after 30 iterations of MAP algorithm for any of the priors. We used a Parzen standard deviation of 15 bins, and a Gaussian kernel of σ1 = 0.2 pixels to generate the coarser scale image. The Parzen standard deviation was not found to significantly affect the reconstruction as long as it did not cause distinct peaks in the estimated pdf to be merged together. The reconstructed images using QP and the four information theoretic priors are shown in Fig. 3 along with the true image. The images shown for each prior are at values of hyperparameter that gave the least root mean squared norm of error between the true f and reconstructed image for that prior, defined by . This error is plotted as a function of iteration number in Fig. 4 for these optimized values of hyperparameter.
Fig. 3.
True and reconstructed images: Top: True PET image (left), QP reconstruction with β = 0.5 (right). Middle: MI reconstruction for μ = 7500 (left), JE reconstruction for μ = 5000 (right). Bottom: MI-scale reconstruction for μ = 2500 (left), JE-scale reconstruction for μ = 1000.
Fig. 4.
Normalized error versons iteration for best value of μ and β of the information theoretic and QP priors, respectively.
It can be seen that the images reconstructed using information theoretic priors have less overall reconstruction error, sharper boundaries, and less noise than that of QP. MI-Intensity prior produces images that follow the structure of the anatomical image, but have several high intensity pixels distributed within the GM region. Adding spatial information to the MI prior through the MI-scale prior reduces the occurence of these high intensity pixels, and gives a more stable reconstruction error versus iteration curve. The JE-Intensity prior reconstructs a near perfect image, with normalized error of about 2% with respect to the true image. This dataset illustrates the best case scenario for the JE prior, since the images are piecewise constant and the JE prior prefers piecewise homogeneous solutions. The JE-scale prior produces some smoothing along the boundaries, thus having higher error than the JE prior. However, for a more realistic case where the images are not piecewise constant, we expect the scale-space features to help the JE reconstruction by providing local spatial information.
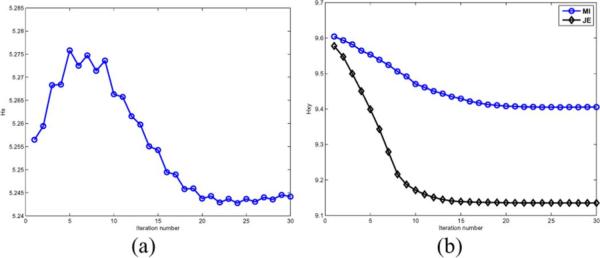
This simulation should also be the best case scenario for the MI prior, since the two images are identical except in the values of intensity, to which it is robust. However, we see that the MI priors do not reconstruct an image close to the true image. This can be explained from the joint pdfs of the anatomical image and the intensity based MI and JE reconstructions shown in Fig. 3. These pdfs are shown in Fig. 5, and the variation of the terms in the MI and JE priors as a function of iteration number are shown in Fig. 6. For the same initial joint density, MI was optimized by initially increasing the marginal entropy term with a corresponding spread in the joint pdf of the cluster that represents GM intensities, while decreasing the joint entropy by clustering these high intensity pixels together as isolated peaks in the joint histogram. This caused MI to converge to a local minimum in which the isolated peaks manifested as high intensity pixels that were distributed throughout the GM. On the other hand, minimizing the JE term alone gave a joint pdf that is more clustered. However, even the JE prior has an isolated peak in its joint pdf associated with the background/CSF of the anatomical image. This behavior of MI and JE priors was observed for a large range of hyperparameter values and is more pronounced for larger values. Introduction of the scale-space features helps the priors avoid these local minima by adding terms that penalize the formation of these isolated peaks.
Fig. 5.
Joint pdfs of the anatomical image and (a) true PET image, (b) OSEM estimate used for initialization, (c) image reconstructed using MI-intensity prior, (d) Image reconstructed using JE-Intensity prior.
Fig. 6.
Variation of marginal entropy for MI-Intensity prior and JE for MI-Intensity and JE-Intensity priors as a function of iteration. (a) Marginal entropy of MI-Intensity prior versus iteration number. (b) Joint entropy of MI-Intensity and JE-Intensity priors versus iteration number.
2) Anatomical and PET Images With Differences
To simulate a more realistic scenario, we used a 3-D MR image of the brain obtained from the IBSR dataset provided by the Center for Morphometric Analysis at Massachusetts General Hospital.1 We resized the MR image (originally of size 256×256×128 voxels of dimension 1.00 mm×1.00 mm×1.50 mm each) to 256×256×111 voxels using trilinear interpolation and performed an intensity based partial volume classification (PVC) using Brainsuite software [24]. This gives an image that has tissue fraction values that represent the amount of tissue (GM/WM/CSF) present in each voxel. Let r, w, and c denote images representing the tissue fraction, respectively of gray, white, and CSF at each voxel so that r + w + c = 1, where 1 represents a vector of ones. We then created three images with constant values zr = 4, zw = 1, and zc = 0. Uniform random noise defined on the interval (−0.1, 0.1) was added to each image and smoothed with a 2-D Gaussian kernel of size 7 × 7 and standard deviation 1.25 pixels. The final simulated image was formed as
| (27) |
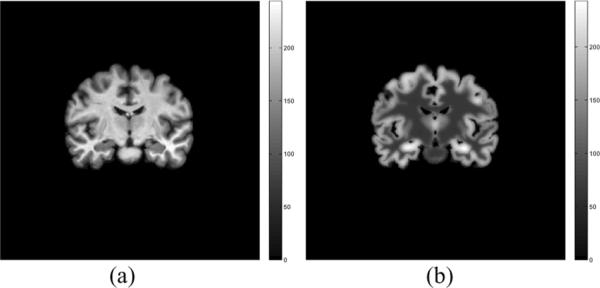
where .* denotes an element by element product. A coronal slice of MR and simulated PET image (after scaling up for appropriate number of counts) is shown in Fig. 7. The resulting phantom has a smooth intensity variation within each tissue region and a smooth transition between regions as a result of the partial fraction voxels between GM/WM/CSF regions. The standard deviation of intensities is 0.198 in WM and 0.4082 in GM due to a combination of partial volume effects and variation caused by the added smoothed noise field.
Fig. 7.
Coronal slice of an MR image of the brain, and PET image generated from partial volume classification of the MR image.
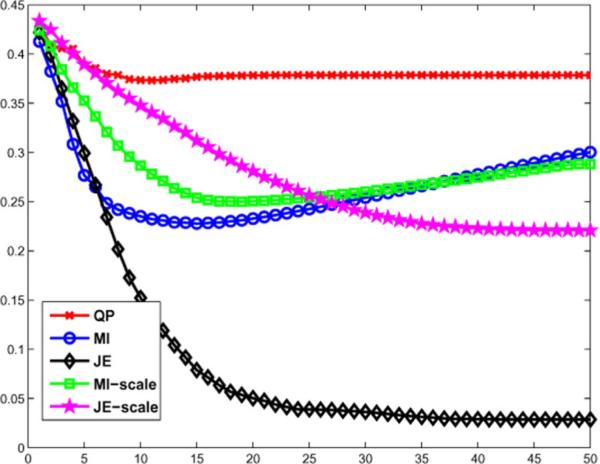
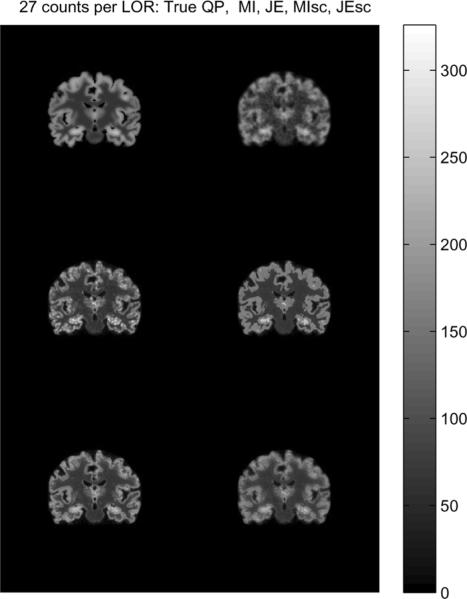
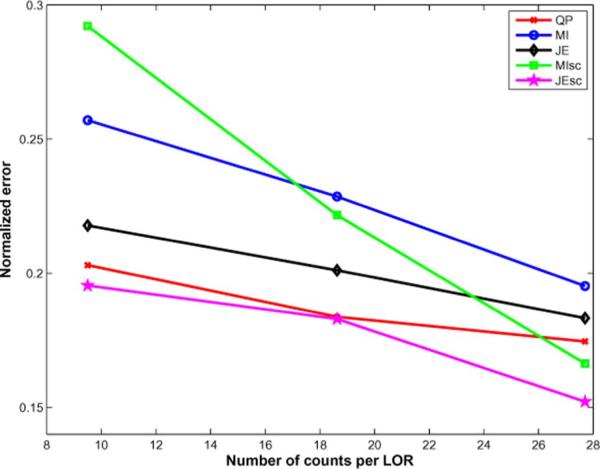
We performed 3-D simulations based on the Siemens Biograph Scanner with 672 4 mm × 4 mm detectors per ring, 55 detector rings of diameter 85.4 cm, and a total of 559 sinograms (span 11, maximum ring difference 38) of size 336 × 336. Sinograms of different average number of counts per LOR ranging from 9 to 27 were generated to study the performance of the priors as a function of counts. The forward model incorporated the geometric response and detector blurring, but no randoms and scatter. We reconstructed images using QP and information theoretic priors with an initialization of 1 iteration of expectation maximization maximum likelihood(EM-ML) algorithm, followed by 40 iterations of MAP. The joint density estimates used histograms with 500 bins with a Parzen variance of 10 bins, and the scale-space priors used a scale of σ1 = 0.5 voxels. The reconstructed images for an average of 27 counts per LOR are shown along with the true image in Fig. 8, and the normalized reconstruction error is shown as a function of iteration number in Fig. 9. The normalized error as a function of counts per LOR is shown in Fig. 10.
Fig. 8.
Coronal sections through true and reconstructed 3-D images at an average of 27 counts per LOR: Top: True PET image (left), QP reconstruction with β = 0.5 (right). Middle: MI reconstruction for μ = 25000 (left), JE reconstruction for μ = 25000, Bottom: MI-scale reconstruction for μ = 25000 (left), JE-scale reconstruction for μ = 50000.
Fig. 9.
Normalized error as a function of iteration.
Fig. 10.
Normalized error as a function of counts per LOR.
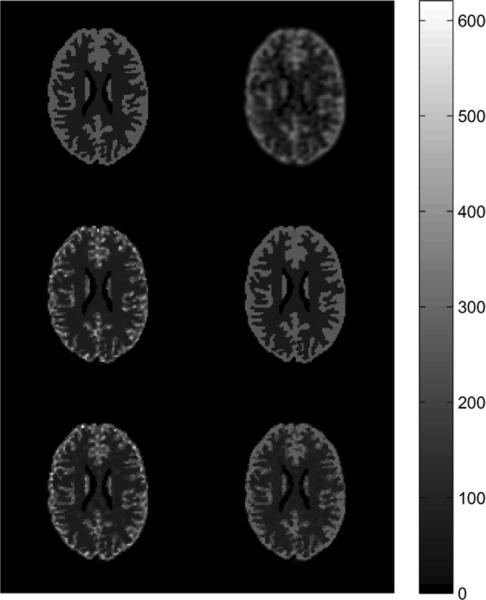
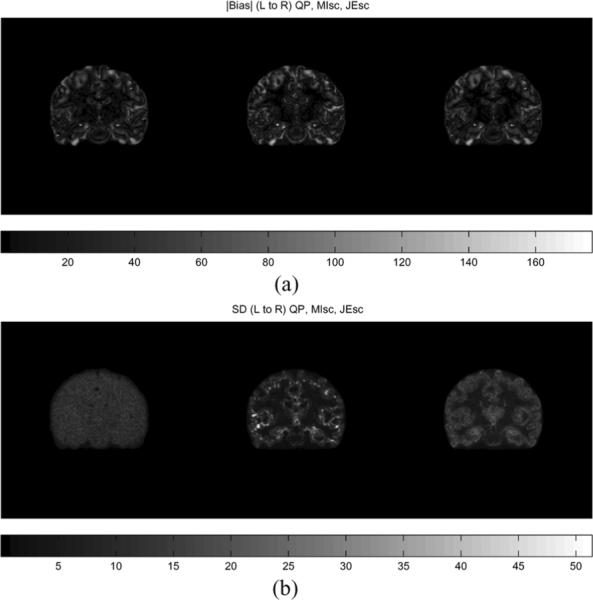
Bias and standard deviation of the QP, MI-scale, and JE-scale prior reconstructions were estimated through Monte Carlo reconstructions of 25 datasets of approximately 27 counts per LOR, initialization of five iterations of EM-ML algorithm, 30 iterations of MAP algorithm, for several different values of hyperparameter. The estimated bias and standard deviation images for the values of hyperparameter that gave the least overall normalized error for each method are shown in Fig. 11.
Fig. 11.
Coronal slice of bias and SD images: Top (L to R): QP (β = 0.5), MI-scale (μ = 50000), JE-scale (μ = 50000).
We computed the normalized bias and variance in mean uptake in regions of interest (ROIs) as
| (28) |
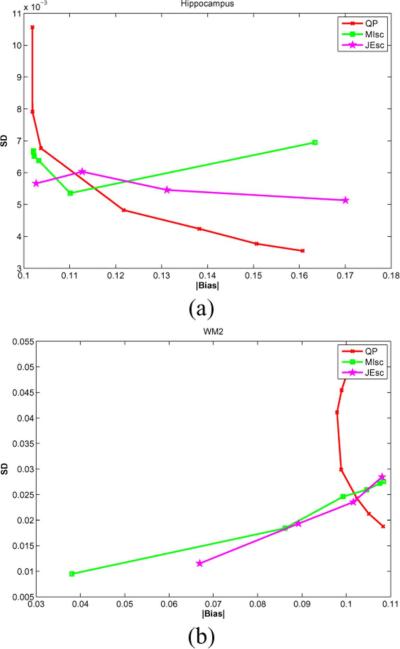
where is the mean true activity in the region of interest, is the mean activity in ROI of the estimated image from the th dataset, and Nd is the number of datasets. We drew ROIs in the hippocampus up to the edge of the region on slices where it is clearly discernible, and in regions within the WM on seven contiguous slices in the frontal lobe of the brain. The plots of bias versus standard deviation as the hyperparameter is varied are shown in Fig. 12 for these regions.
Fig. 12.
Absolute value of bias versus standard deviation curves for selected regions of interest.
The reconstructed images using the information theoretic priors show less noise (especially in the WM region) than QP, in Fig. 8. Blurring across cortical folds in the QP reconstruction is alleviated in the information theoretic prior reconstructions, showing that these priors take advantage of anatomical information to improve the resolution of the reconstruction. The MI-Intensity and JE-Intensity priors show isolated voxels with high intensity, the occurence of which are reduced in the scale-space priors. The JE-Intensity prior produces nearly piecewise homogeneous gray matter and white matter regions, consistent with the best case scenario simulations results. The MI-scale and JE-scale priors produce more realistic reconstructions that capture the variation in intensities in the true image, and have lower overall normalized reconstruction error compared to QP and MI-Intensity and JE-Intensity priors for the 27 counts per LOR data. For lower counts per LOR, while JE-scale maintains the better performance in terms of overall reconstruction error compared to QP, the performance of MI-scale worsens as the counts per LOR decreases.
The ROI curves shown in Fig. 12 indicate that the information theoretic priors do not offer a traditional trade-off between bias and variance. These priors are nonlinear functions of the image that is being estimated, and exhibit a nonmonotonic variation as the hyperparameter is changed. In WM, both the scale-space priors have decreasing bias and variance with increase in hyperparameter and lesser bias and variance than QP for higher values of hyperparameter. This is also reflected in the bias and SD images where the WM regions have lower intensity than QP. In the hippocampus, MI-scale and JE-scale priors have lower bias and variance for some values of hyperparameter, but do not consistently stay below the QP curve. This behavior was consistent in other subcortical GM regions considered (not shown), of which the hippocampus showed the best performance.
Overall, the results indicate that for this realistic phantom, the MI-scale and JE-scale priors take advantage of the anatomical information in the MR image to improve resolution and overall quantitation accuracy compared to QP. However, quantitation was improved more in the WM regions than in the GM regions considered (possibly because of the higher variance in the data for these high count regions, combined with the larger number of partial volume voxels in the GM regions), and was dependent on the choice of hyperparameter weighting the prior. For the high count dataset used for the Monte Carlo simulations described above, MI-scale and JE-scale have comparable overall performace in terms of bias and variance, but at lower counts per LOR it was seen that MI-scale has higher normalized error while JE-scale maintains a more stable behavior. This may be because for noisier datasets, the entropy term in MI tends to increase the variance in the distribution of intensities, thus causing more isolated voxels of high intensities for lower count datasets. We therefore prefer the JE-scale over the MI-scale prior because of its stability over counts per LOR.
B. Clinical Data
PET data was acquired on a Siemens Biograph True Point HD scanner from a Parkinson's disease patient who was injected with F18 Fallypride. This tracer has a high affinity to D2 receptors, which have a high density in the striatal regions of the brain. PET images using this tracer show uptake mainly in the caudate and putamen regions of the brain, and therefore this data would serve to investigate the behavior of the information theoretic priors when the PET image intensities exhibit natural variations across two closely spaced structures (putamen and caudate). Based on our simulation results, we choose the JE-scale prior to compare with the QP on this clincal dataset.
A T1 weighted MRI scan of the subject was performed on the same subject and is used as the anatomical image for reconstruction. The MR image was coregistered to a QP MAP reconstruction of the PET image using rigid mutual information based registration via the Rview software package [25]. Histogram equalization was then performed on the MR image to improve contrast. The histogram equalization improved the contrast, but did not significantly change the results. Corrections for attenuation, normalization, and scatter were done using the Siemens software, and randoms correction was based on counts within a delayed time window. Sinogram blurring was also incorporated into the system matrix. We initialized the MAP algorithm for all priors with two iterations of OSEM with 21 subsets, and ran 20 iterations of MAP. The values for bin widths and Parzen variance were chosen similar to that used for the simulations. However, for the JE-scale prior, we used a scale parameter of σ1 = 3.0 pixels, and dropped the Laplacian term. The Laplacian of these PET images is nonzero in few voxels (mainly in the striatal regions), which produces a joint density of the Laplacian images consisting of a large peak around zero, and a few small peaks corresponding to the edges. Since the JE metric can produce homogeneous solutions, it tends towards a Laplacian joint density which has one large peak around zero, producing undesirable results. This did not occur in the FDG based simulations since the PET images had large regions of activity with Laplacian images that had several nonzero voxels.
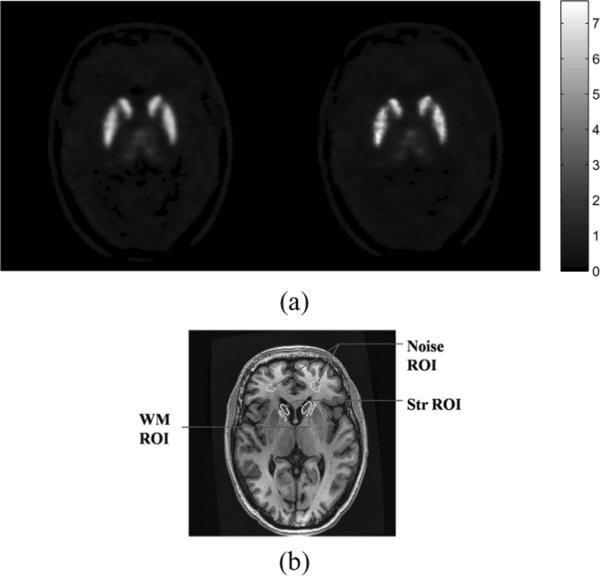
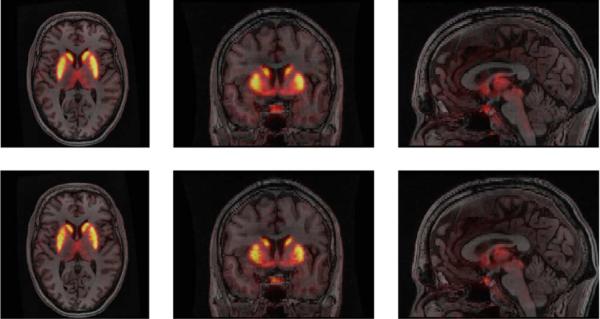
The images reconstructed using the JE-scale and QP priors are shown along with the coregistered MR image in Fig. 13. The QP and JE-scale reconstructions are overlaid on the MR image and the coronal, sagittal, and transaxial views are shown in Fig. 14. It can be seen that for QP, the activity is often spread outside the anatomical region to which it belongs, while this spreading is reduced for the JE-scale prior.
Fig. 13.
MR and reconstructed PET images. (a) Transaxial slice of reconstructed PET image using QP (left) and JE-scale (right). (b) Coregistered MR image with the ROIs for computation of CRC and noise varince outlined in yellow.
Fig. 14.
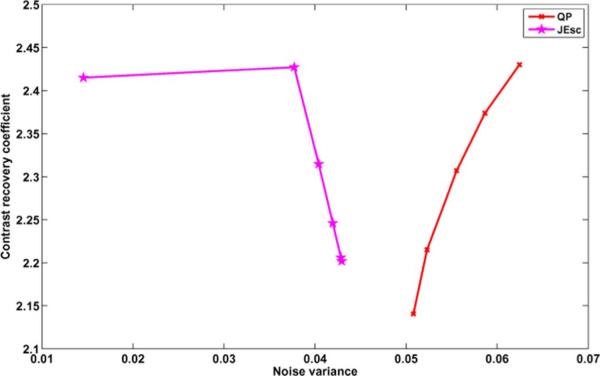
Contrast recovery coefficient versus noise variance plots as the hyperparameter is varied.
We quantify the performance of these priors in terms of their contrast recovery coefficient (CRC) as a function of noise. We define CRC between the uptake in a region of the striatum and a region in the WM close to it as
| (29) |
where is the mean activity in a region defined in the striatum, and is the mean activity in a WM region. We considered ROIs in the caudate region of the striatum, and the WM that separates the caudate from the putamen on 11 contiguous slices of the brain. We define the noise variance as the variance of intensity in regions in the brain where we do not expect significant uptake of tracer, such as the white matter in the posterior of the brain. We drew white matter ROIs on 18 contiguous slices in the brain. The ROIs used for computation of CRC and noise variance are shown on the MR image in Fig. 13. The CRC vs noise plots are shown in Fig. 15.
Fig. 15.
Overlay of PET reconstruction over coregistered MR image for QP (top) and JE-scale (bottom) priors.
These results demonstrate that the JE-scale prior can reconstruct high contrast images that take advantage of anatomical image information, in a real world scenario where the anatomical and functional images are independently acquired and then registered to each other. Additionally, the results indicate that this priors can be used even for tracers that bind nonuniformly within regions of similar intensities in the anatomical image (e.g., GM, WM, CSF for the brain), as long as the uptake is homogeneous within some clearly delineated anatomical structure that is large enough to form a prominent peak in the joint density.
The change in contrast as a function of hyperparameter illustrated in the CRC curves was seen to be a result of changes in both and for both QP and JE-scale priors. The CRC curves show that the JE-scale priors reconstructed images with higher contrast than that of QP. While QP has a decreasing contrast and noise variance as the hyperparameter is increased, the JE-scale prior has increasing contrast as μ increased. An increasing μ would give larger weight on the prior, thus causing the joint density to be more clustered, which would correspond to a higher contrast. We see that consistent to our simulation results, the scale-space priors have reduced noise variance compared to the QP based priors, without appreciable loss in contrast.
However, we note that the behavior of the information theoretic priors is unpredictable at high values of μ, where the prior dominates the likelihood term. MI tends to produce images that spuriously follow the anatomical regions, whereas JE prefers a piecewise homogeneous image. We observed that at high values of μ, the JE-scale prior tended to combine the cau-date and putamen regions into one homogeneous region. This instability makes the choice of hyperparameter especially important for these priors. We have observed that the knee of the L curve between the log likelihood and log prior terms yields reasonable values of μ, but for clinical use, these results should always be verified with the OSEM or MAP with QP results to ensure that no misleading artifacts are produced.
IV. Conclusion
We describe a scale-space based framework for incorporating anatomical information into PET image reconstruction using information theoretic priors. We validated this framework using idealized simulations that used PET and anatomical phantoms that had identical structure, realistic simulations that used a real MR brain image with a PET image that combined intensity variation within tissue type with partial volume effects, and clinical data. We presented an efficient FFT based approach for the computation of these priors and their derivatives, making it practical to use these priors for 3-D PET image reconstruction.
The reconstructed images using the information theoretic priors have visibly improved resolution compared to the quadratic prior reconstructed images and look realistic, without the mosaic-like appearance that results from anatomical priors that impose piecewise smoothness. ROI quantitation studies showed improvements in white matter, but equivocal results in the hippocampal ROI. Through simulation studies we showed that the marginal entropy term in MI tends to increase the variance in the estimate of the image. We noted that testing the information theoretic priors on piecewise constant images can give misleadingly accurate results while using only intensity as a feature, especially for JE, since it prefers piecewise constant images. The realistic simulations that we performed show that results using intensity based priors are prone to isolated hot and cold spots, and vary unpredictably with the hyperparameter. It is therefore important to incorporate spatial correlations into these priors. The scale-space based approach improved the behavior of the information theoretic priors, and gave better overall quantitation accuracy compared to QP for the optimal range of hyperparameter values while using only two additional features, the coarser scale image and its Laplacian. The clinical data results were consistent with the simulations, and show that these priors can be used even for tracers that do not have homogeneous uptake in regions corresponding to similar intensities in the anatomical image, provided the activity corresponds to a prominent anatomical region.
We expect these priors to be most useful in applications involving quantification of uptake in specific organs, where the uptake is within a region that is clearly discernible in the anatomical image. This framework of information theoretic priors can also be applied to other functional imaging modalities where the data is severely ill-conditioned and the use of anatomical information may improve the accuracy of reconstruction. Promising results were obtained for diffuse optical tomography using this framework of information theoretic anatomical priors [26].
The limitations of the current formulation of scale-space based information theoretic priors for PET reconstruction are the nonconvexity of the priors, the sensitivity of the information theoretic priors to the choice of the hyperparameter, and their tendency to converge to unpredictable solutions for high values of hyperparameter thus necessitating verification with OSEM or MAP with QP results for real data. Additionally, the quantitation results showed improvements in some regions and equivocal results in others compared to QP. In this paper we explored the properties of information theoretic measures as priors in PET reconstruction, and presented the scale-space approach as a solution to the nonspatial nature of these measures. The results presented here lend confidence to the approach as a method to incorporate anatomical information in a real world scenario, and warrant further investigation to address the limitations mentioned above.
Acknowledgment
The authors would like to thank Dr. G. Petzinger, Dr. B. Fisher, and Dr. A. Nacca for use of the Fallypride imaging data presented here.
This work was supported by the National Institutes of Health (NIH) under Grant R01 EB010197.
Appendix
Alternative Formulation to the JE Prior
Joint entropy between two random variables X and Y is defined as
| (30) |
This expectation can be approximated by the sample mean. If the intensities of image f and a are realizations of X and Y respectively, then
| (31) |
The approximate equality will be replaced by an equality symbol for convenience.
If we use Parzen windows to estimate p(fi,ai) as given in (13), then
| (32) |
The derivative of H(X,Y) with respect to fm then
| (33) |
Since we are using a symmetric Gaussian kernel with standard deviation σx for the functional image, , we get
| (34) |
If we let
and
| (35) |
then (34) can be rewritten as
| (36) |
The weights αim, and βim are both between 0 and 1, since the denominator in each of these terms is obtained by a summation of the numerator over all m or i. The weight Wim approaches 0 when fi and ai are distant from fm and am respectively, and approaches 2 when they are close. Hence, Wim selects the voxels with intensities that are within a few standard deviations of fim in both the anatomical and functional images, and updates fim such that the intensities in these voxels are clustered closer together.
The gradient in (36) can be further approximated by considering only voxels that are spatially close to the voxel m, i.e., in a neighborhood ηm, giving
| (37) |
This expression is similar to that of the quadratic prior, except that the weight Wim selects only those voxels within the neighborhood ηm that have intensities close to fm. In this case, the voxels that are spatially close to each voxel influence the gradient rather than the more global interaction described in (36).
Footnotes
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
References
- [1].Leahy R, Yan X. Incorporation of MR data for improved functional imaging with PET. Inf. Proc. Med. Imag. 1991:105–120. [Google Scholar]
- [2].Gindi G, Lee M, Rangarajan A, Zubal IG. Bayesian reconstruction of functional images using anatomical information as priors. IEEE Trans. Med. Imag. 1993 Dec.12(4):670–680. doi: 10.1109/42.251117. [DOI] [PubMed] [Google Scholar]
- [3].Ouyang X, Wong W, Johnson V, Hu X, Chen C-T. Incorporation of correlated structural images in pet image reconstruction. IEEE Trans. Med. Imag. 1994 Dec.13(4):627–640. doi: 10.1109/42.363105. [DOI] [PubMed] [Google Scholar]
- [4].Ardekani BA, Braun M, Hutton BF, Kanno I, Iida H. Minimum cross-entropy reconstruction of pet images using prior anatomical information. Phys. Med. Biol. 1996;41(11):2497–2517. doi: 10.1088/0031-9155/41/11/018. [DOI] [PubMed] [Google Scholar]
- [5].Bowsher JE, Johnson VE, Turkington TG, Jacszak RJ, C. F. Bayesian reconstruction and use of anatomical a priori information for emission tomography. IEEE Trans. Med. Imag. 1996 Oct.15(5):673–686. doi: 10.1109/42.538945. [DOI] [PubMed] [Google Scholar]
- [6].Sastry S, Carson R. Multimodality bayesian algorithm for image reconstruction in positron emission tomography: A tissue composition model. IEEE Trans. Med. Imag. 1997 Dec.16(6):750–761. doi: 10.1109/42.650872. [DOI] [PubMed] [Google Scholar]
- [7].Lipinski B, Herzog H, Kops ER, Oberschelp W, Muller-Gartner H. Expectation maximization reconstruction of positron emission tomography images using anatomical magnetic resonance information. IEEE Trans. Med. Imag. 1997 Apr.16(2):129–136. doi: 10.1109/42.563658. [DOI] [PubMed] [Google Scholar]
- [8].Baete K, Nuyts J, Paesschen WV, Suetens P, Dupont P. Anatomical-based FDG-pet reconstruction for the detection of hypo-metabolic regions in epilepsy. IEEE Trans. Med. Imag. 2004 Apr.23(4):510–519. doi: 10.1109/TMI.2004.825623. [DOI] [PubMed] [Google Scholar]
- [9].Rangarajan A, Hsiao I-T, Gindi G. A Bayesian joint mixture framework for the integration of anatomical information in functional image reconstruction. J. Math. Imag. Vis. 2000;12:199–217. [Google Scholar]
- [10].Bowsher JE, Yuan H, Hedlund LW, Turkington TG, Kurylo WC, Wheeler CT, Cofer GP, Dewhirst MW, Johnson GA. Utilizing MRI information to estimate F18-FDG distributions in rat flank tumors. Conf. Rec.: IEEE Nucl. Sci. Symp. Med. Imag. Conf. 2004 Oct.4:92–2488. [Google Scholar]
- [11].Wells W, III, Viola P, Atsumi H, Nakajima S, Nakajima S, Kikinis R. Multimodal volume registration by maximization of mutual information. Med. Image Anal. 1996;1(1):35–51. doi: 10.1016/s1361-8415(01)80004-9. [DOI] [PubMed] [Google Scholar]
- [12].Somayajula S, Asma E, Leahy RM. PET image reconstruction using anatomical information through mutual information based priors. Conf. Rec: IEEE Nucl. Sci. Symp. and Med. Imag. Conf. 2005:26–2722. [Google Scholar]
- [13].Somayajula S, Rangarajan A, Leahy R. PET image reconstruction using anatomical information through mutual information based priors: A scale space approach. Proc. 4th IEEE Int. Symp. Biomed. Imag. 2007 Apr.:165–168. [Google Scholar]
- [14].Nuyts J. The use of mutual information and joint entropy for anatomical priors in emission tomography. Conf. Rec: IEEE Nucl. Sci. Symp. Med. Imag. Conf. 2007:54–4148. [Google Scholar]
- [15].Lindeberg T. Scale-Space Theory in Computer Vision. Kluwer; Norwell, MA: 1994. [Google Scholar]
- [16].Mumcuoglu E, Leahy R, Cherry S. Bayesian reconstruction of pet images: Quantitative methodology and performance analysis. Phys. Med. Biol. 1996 Sep.41(9):1777–1807. doi: 10.1088/0031-9155/41/9/015. [DOI] [PubMed] [Google Scholar]
- [17].Qi J, Leahy RM, Cherry SR, Chatziioannou A, Farquhar TH. High resolution 3-D Bayesian image reconstruction using the micropet small-animal scanner. Phys. Med. Biol. 1998 Apr.43:1001–1013. doi: 10.1088/0031-9155/43/4/027. [DOI] [PubMed] [Google Scholar]
- [18].Leahy RM, Qi J. Statistical approaches in quantitative positron emissiontomography. Stat. Comput. 2000;10(2):147–165. [Google Scholar]
- [19].Cover T, Thomas J. Elements of Information Theory. Wiley; New York: 1991. [Google Scholar]
- [20].Duda RO, Hart PE, Stork DG. Pattern Classification. 2nd ed. Wiley; New York: 2001. [Google Scholar]
- [21].Bertsekas D. Nonlinear Programming. Athena Scientific; Nashua, NH: 1999. [Google Scholar]
- [22].Kaufman L. Implementing and accelerating the EM algorithm for positron emission tomography. IEEE Trans. Med. Imag. 1987 Mar.6(1):37–51. doi: 10.1109/TMI.1987.4307796. [DOI] [PubMed] [Google Scholar]
- [23].Shwartz S, Zibulevsky M, Schechner YY. Fast kernel entropy estimation and optimization. Signal Process., Inf. Theoretic Signal Process. 2005;85(5):1045–1058. [Google Scholar]
- [24].Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM. Magnetic resonance image tissue classification using a partial volume model. NeuroImage. 2001;13(5):856–876. doi: 10.1006/nimg.2000.0730. [DOI] [PubMed] [Google Scholar]
- [25].Studholme C, Hill DLG, Hawkes DJ. An overlap invariant entropy measure of 3-D medical image alignment. Pattern Recognit. 1999 Jan.32(1):71–86. [Google Scholar]
- [26].Panagiotou C, Somayajula S, Gibson AP, Schweiger M, Leahy RM, Arridge SR. Information theoretic regularization in diffuse optical tomography. J. Opt. Soc. Am. A. 2009;26(5):1277–1290. doi: 10.1364/josaa.26.001277. [DOI] [PubMed] [Google Scholar]