Abstract
The NAD+-dependent deacetylase SIRT1 is an evolutionarily conserved metabolic sensor of the Sirtuin family that mediates homeostatic responses to certain physiological stresses such as nutrient restriction. Previous reports have implicated fluctuations in intracellular NAD+ concentrations as the principal regulator of SIRT1 activity. However, here we have identified a cAMP-induced phosphorylation of a highly conserved serine (S434) located in the SIRT1 catalytic domain that rapidly enhanced intrinsic deacetylase activity independently of changes in NAD+ levels. Attenuation of SIRT1 expression or the use of a non-phosphorylatable SIRT1 mutant prevented cAMP-mediated stimulation of fatty acid oxidation and gene expression linked to this pathway. Overexpression of SIRT1 in mice significantly potentiated the increases in fatty acid oxidation and energy expenditure caused by either pharmacological β-adrenergic agonism or cold exposure. These studies support a mechanism of Sirtuin enzymatic control through the cAMP/PKA pathway with important implications for stress responses and maintenance of energy homeostasis.
Keywords: Sirtuin activity, PGC-1α, adrenergic signaling, fatty acid oxidation and energy expenditure
Introduction
Sirtuins are a group of phylogenetically conserved NAD+-dependent deacetylases (Imai et al., 2000) that play a role in chromatin remodeling, transcriptional regulation, and control of biological processes such as energy homeostasis (Picard et al., 2004; Rodgers et al., 2005; Lagouge et al., 2006; Gerhart-Hines et al., 2007; Schwer and Verdin, 2008), DNA damage repair (Luo et al., 2001; Yuan et al., 2007), neuronal protection (Donmez et al., 2010), circadian patterning (Asher et al., 2008; Nakahata et al., 2008), and longevity (Kaeberlein et al., 1999; Viswanathan et al., 2005; Viswanathan and Guarente, 2011). Precisely how the catalytic activity of Sirtuin proteins is regulated is unclear, though several studies have demonstrated that blanket changes in NAD+ substrate levels are sufficient to alter activity of the mammalian Sir2 homologue, SIRT1, under different conditions (Revollo et al., 2004; Fulco et al., 2008; Cantó et al., 2009). Nevertheless, several of the models proposed by these reports require the induction of genes involved in NAD+ biosynthesis and, consequently, are less suited to facilitate prompt regulatory responses. There have also been previously identified post-translational modifications on SIRT1 that have been purported to increase deacetylase activity on the Sirtuin target, p53 (Sasaki et al., 2008; Kang et al., 2009; Guo et al., 2010). However, these modifications occur outside the catalytic domain and the molecular mechanisms regarding SIRT1 activation, dependency on NAD+ changes, and physiological relevance, remain unknown.
Evidence from both cellular and in vivo studies suggests that a primary function of SIRT1 is to provide protection against a multitude of stresses (Donmez and Guarente, 2010; Herranz and Serrano, 2010; Vinciguerra et al., 2010). The adrenergic signaling cascade is also commonly associated with stress responses and relies upon transcriptional and post-translational regulatory events to mediate metabolic responses. During conditions of cold stress, adrenergic stimulation increases fatty acid consumption and net energy expenditure in brown adipose tissue (BAT) and skeletal muscle through upregulation of mitochondrial oxidative transcriptional programs (Kumar et al., 2008; Nedergaard and Cannon, 2010). Importantly, this cAMP/protein kinase A (PKA)-dependent induction of fatty acid utilization and energy expenditure in peripheral tissues is facilitated in part by peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1α) (Finck and Kelly, 2006; Uldry et al., 2006; Miura et al., 2007). We and others have previously shown PGC-1α to be deacetylated and subsequently activated by SIRT1 (Rodgers et al., 2005; Lagouge et al., 2006; Gerhart-Hines et al., 2007). However, current SIRT1 regulatory models rely on wholesale changes in NAD+ which take place over a time span of hours and are unable to provide the rapid control of Sirtuin function necessary for more quickly occurring bioenergetic responses. Hence, we suspected that SIRT1 deacetylase activity could be directly modulated during metabolic and environmental stresses by the cAMP/PKA pathway to induce fatty acid oxidation. This would provide a more immediate and dynamic form of SIRT1 regulation under conditions in which changes in NAD+ levels are nominal or absent.
Here we show that stimulation of the cAMP/PKA signaling cascade resulted in a phosphorylation of the highly conserved residue, S434, in the SIRT1 catalytic domain. This modification significantly enhanced intrinsic deacetylase activity and led to increased expression of lipid metabolism genes and elevated fatty acid utilization independently of changes in NAD+ concentration. Finally, adrenergic-mediated induction of fatty acid oxidation and energy expenditure was significantly potentiated in transgenic mice overexpressing SIRT1 compared to wildtype littermates.
Results
Activation of the cAMP signaling pathway induces rapid deacetylation of SIRT1 substrates
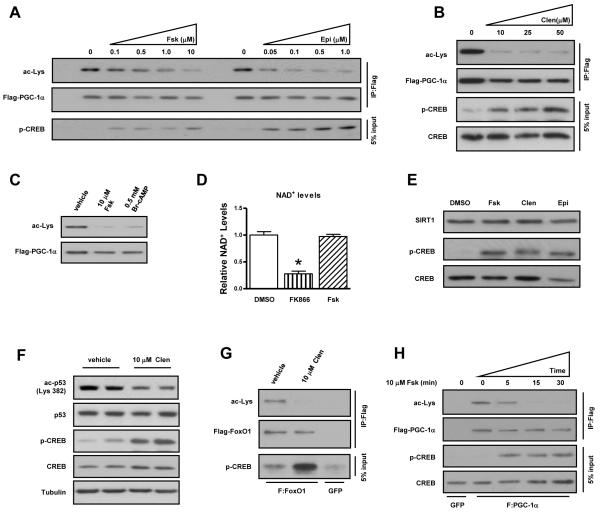
We first tested whether activation of the cAMP/PKA pathway affected the acetylation status of the SIRT1 substrate, PGC-1α. Treatment with an adenylyl cyclase activator (forskolin), catecholamine (epinephrine), non-hydrolyzable cAMP analog (8-BrcAMP), or β2-adrenergic receptor agonist (clenbuterol) resulted in the complete deacetylation of PGC-1α independently of changes in NAD+ or SIRT1 levels (Figure 1AE). Forskolin had no effect on NAD+ concentrations whereas FK866, an inhibitor of the NAD+ biosynthetic enzyme NAMPT, produced a significant decrease. Adrenergic stimulation also resulted in comparable deacetylation of endogenous p53 and ectopically expressed FoxO1, two additional SIRT1 substrates (Figure 1F-G) (Luo et al., 2001; Vaziri et al., 2001; Motta et al., 2004; Nemoto et al., 2004). Strikingly, PGC-1α deacetylation occurred within 5-15 minutes following forskolin treatment and was temporally correlated with the induction of PKA activity as evidenced by cAMP response element binding protein (CREB) phosphorylation (Figure 1H). This dynamic regulation was in stark contrast to AMP-activated protein kinase (AMPK)-mediated control of SIRT1, which has previously been shown to increase intracellular NAD+ levels and, in turn, cause SIRT1-dependent PGC-1α deacetylation after a period of several hours (Fulco et al., 2008; Cantó et al., 2009; Iwabu et al., 2010). Consistent with these earlier studies, activation of AMPK by AICAR did not affect PGC-1α acetylation within the 15 minute time frame in which forskolin acts (Figure S1). Moreover, the effect of forskolin was not blocked by the AMPK inhibitor, compound-C (Ara-C). These data collectively demonstrate that acute activation of cAMP signaling produces rapid deacetylation of established SIRT1 targets and, importantly, occurs independently of perturbations in intracellular NAD+ levels and AMPK signaling cascades.
Figure 1. Activation of the cAMP signaling pathway induces rapid deacetylation of SIRT1 substrates.
PGC-1α is deacetylated following acute stimulation of the cAMP/PKA signaling cascade. Transfected, FLAG-tagged PGC-1α was immunoprecipitated from U2OS cells treated for 30 minutes with, A, forskolin (Fsk) or epinephrine (Epi), B, clenbuterol (clen), and C, Br-cAMP; acetylation status was assessed using a pan-acetyl-lysine antibody (Cell Signaling). D, Intracellular NAD+ and, E, SIRT1 protein levels are not affected by short-term acute PKA activation. NAD+ concentrations were assessed from U2OS cells treated with DMSO (n=4), FK866 (n=4), or Fsk (n=4). Cells were treated with FK866 overnight or with forskolin for 1 h and then harvested in NAD+ extraction buffer. NAD+ levels were measured according to manufacturer’s instructions (BioAssay Systems). F, Endogenous p53 and, G, overexpressed FOXO1 are deacetylated following activation of β-adrenergic pathways. H, Forskolin-mediated PGC-1α deacetylation is rapid. Data are presented as means ± S.D.
cAMP-mediated PGC-1α deacetylation/activation is SIRT1 and PKA-dependent
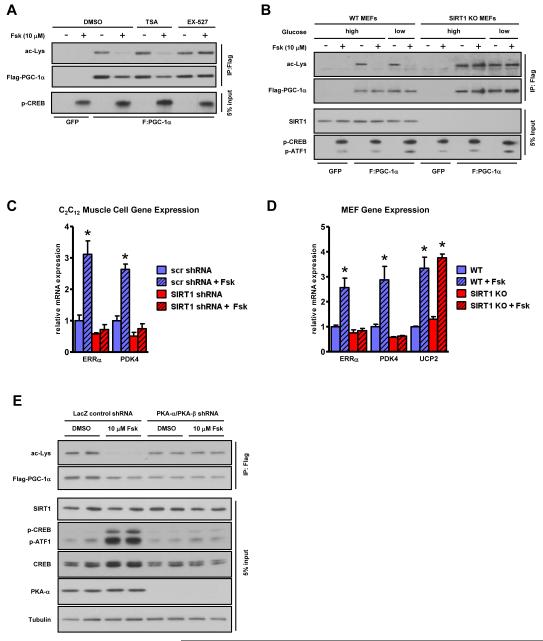
The SIRT1 dependence of cAMP-induced PGC-1α deacetylation was tested using the SIRT1 inhibitor, EX-527, which completely blocked the effect of forskolin, whereas the class I/II HDAC inhibitor, trichostatin A, did not (Figure 2A). In addition, forskolin was able to induce PGC-1α deacetylation in WT MEFs but not in fibroblasts isolated from SIRT1 knockout mice (SIRT1 KO) (Figure 2B). Moreover, a PGC-1α mutant (R13), in which 13 lysines known to be targeted by SIRT1 were mutated to arginines (Rodgers et al., 2005), was no longer sensitive to cAMP-induced deacetylation (Figure S2A). Further underscoring the SIRT1 dependency of cAMP/PKA-mediated control of PGC-1α targets, shRNA knockdown and genetic ablation of SIRT1 eliminated the forskolin induction of fatty acid oxidative regulatory genes such as estrogen related-receptor alpha (ERRα) and pyruvate dehydrogenase kinase 4 (PDK4) (Figure 2C-D, S2B-C). Interestingly, uncoupling protein 2 (UCP2) was equally induced in both wildtype and SIRT1 KO MEF lines in response to cAMP agonism indicating that only a subset of forskolin-sensitive PGC-1α gene targets relies on SIRT1 in these cells. Neither pharmacological inhibition of SIRT1 nor shRNA knockdown produced significant effects on intracellular NAD+ concentrations (Figure S2D). In order to assess the PKA dependence of cAMP-mediated PGC-1α deacetylation, PKA-α and PKA-β catalytic isoforms were depleted using specific shRNAs (Figure S2E). This knockdown ablated the forskolin-triggered deacetylation of PGC-1α (Figure 2E). Taken together, these results indicate that PKA and SIRT1 are required components of the signaling pathway by which cAMP alters PGC-1α deacetylation and its co-transcriptional activity on endogenous target genes independently of changes in NAD+.
Figure 2. cAMP-mediated PGC-1α deacetylation/activation is SIRT1 and PKA-dependent.
A, Pharmacological inhibition and, B, genetic ablation of SIRT1 abrogates cAMP-mediated PGC-1α deacetylation. MEFs were subject to cAMP agonism under conditions of high (25 mM) and low (2.5 mM) glucose. C, SIRT1 knockdown by shRNA in skeletal muscle cells or, D, genetic ablation in fibroblasts abolishes the forskolin-mediated increase in fatty acid oxidative genes. PGC-1α was adenovirally expressed in differentiated C2C12 myotubes in addition to either scrambled (scr) control or SIRT1 shRNA for 72 hours to ensure knockdown. Skeletal muscle cells and MEFs were treated with Fsk for 6 h before RNA isolation and mRNA quantification; statistical significance was assessed using one-way ANOVA with a Tukey post-test, *, p<0.01, DMSO vs Fsk. E, cAMP-mediated PGC-1α deacetylation is PKA dependent. PGC-1α was immunoprecipitated from DMSO or Fsk-treated U2OS cell lines expressing either control LacZ shRNA or shRNAs against both PKA-α and β catalytic subunits (Figure S2D). Data are presented as means ± S.D.
PKA activation results in phosphorylation of SIRT1 on serine 434
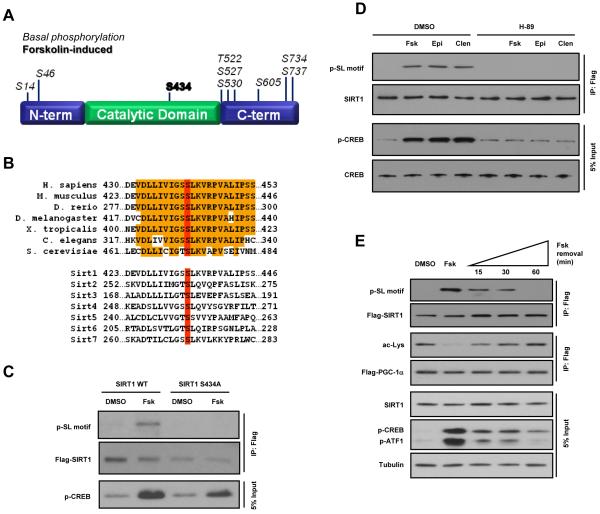
The SIRT1 and PKA dependence of cAMP-induced PGC-1α deacetylation and the rapid rate at which it occurred suggested that SIRT1 might be a target of the cAMP/PKA signaling pathway. SIRT1 does not contain any perfect canonical PKA consensus sites (RRXS/T) but does possess two residues, S161 and S605, in RXS motifs. However, mutation of these two sites to alanine did not prevent cAMP-mediated PGC-1α deacetylation, indicating that other residues were likely responsible for the activating effects of the cAMP/PKA pathway on SIRT1 (data not shown). Using mass spectrometry, we mapped differentially phosphorylated residues on immunopurified FLAG-tagged SIRT1 from DMSO-treated or forskolin-treated cells (Figure 3A, S3A). Among the phospho-residues identified, S434 was the only site found to be phosphorylated exclusively following forskolin treatment. This residue is located in the NAD+ binding pocket of the catalytic domain and is phylogenetically conserved across all known Sir2 orthologs, including the seven mammalian Sirtuin paralogs (Figure 3B). The primary sequence flanking S434 (IGSSLKVRP) lacks any elements of a PKA motif and recombinant PKA weakly phosphorylated SIRT1 S434 in in vitro kinase assays (Figure S3B), suggesting that other kinases, downstream of PKA, might be responsible for the direct phosphorylation of SIRT1 in vivo. We validated the mass spectrometric identification of S434 using a polyclonal antibody that specifically recognizes phosphorylated serine-leucine (SL) motifs (Cell Signaling, F7152). Mutation of S434 to alanine abolished the forskolin-induced SIRT1 phosphorylation (Figure 3C). Moreover, phosphorylation of SIRT1 triggered by forskolin, epinephrine, and clenbuterol was completely blocked by the PKA inhibitor, H-89 (Figure 3D). Removal of forskolin resulted in rapid attenuation of SIRT1 and CREB phosphorylation and re-establishment of PGC-1α acetylation in parallel experiments (Figure 3E). These data indicate that activation of cAMP/PKA signaling results in phosphorylation of the highly conserved SIRT1 S434 residue situated in the deacetylase catalytic core.
Figure 3. PKA activation results in phosphorylation of SIRT1 on serine 434.
A, Schematic depicting phosphorylation sites detected using mass spectrometric analysis of SIRT1 protein immunoprecipitated from U2OS cells incubated with Fsk or DMSO. Murine SIRT1 residue, S434 (bold), was found to be differentially phosphorylated following stimulation of the cAMP signaling pathway. B, Phylogenetic alignment shows complete conservation of SIRT1 S434 (red) among orthologs as well as other mammalian Sirtuin family members. C, Mutation of S434 abolishes forskolin-induced SIRT1 phosphorylation. D, Phosphorylation of SIRT1 S434 induced by forskolin, epinephrine, or clenbuterol is blocked by the PKA inhibitor, H-89. E, Removal of forskolin attenuates cAMP signaling and SIRT1 phosphorylation and re-establishes PGC-1α acetylation. Transfected, FLAG-tagged PGC-1α was immunoprecipitated from U2OS cells treated with Fsk for 30 min, washed with PBS, and then incubated in Fsk-free medium for the indicated time points.
The cAMP/PKA pathway increases intrinsic SIRT1 enzymatic activity through serine 434 phosphorylation
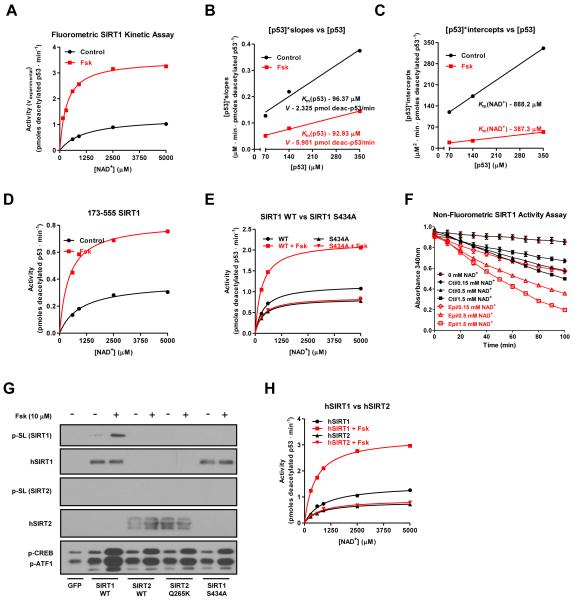
To mechanistically determine how cAMP/PKA-induced S434 phosphorylation was modulating SIRT1 activity independently of changing NAD+ concentrations, we performed in vitro deacetylation assays using a fluorescently-labeled, acetylated p53 peptide (ENZO). Consistent with data obtained from cells, the deacetylase activity of ectopically expressed SIRT1 protein immunopurified from nuclear extracts was significantly higher following forskolin treatment; EX-527 inhibitor was used to demonstrate the SIRT1-dependence of the observed deacetylation activity in the eluates (Figure 4A, S4A-C). By using a range of NAD+ concentrations at fixed amounts of p53 substrate, kinetic parameters for enzyme velocity (V), Km for NAD+, and Km for acetylated p53 were calculated (Figure 4B-C, S4D). Based on this analysis, forskolin caused the V of SIRT1 to increase by ~250% and the Km for NAD+ to decrease by ~55%. There was, however, no significant change in the Km for acetylated p53. Furthermore, forskolin did not change SIRT1 sensitivity to nicotinamide-mediated inhibition (Figure S4E). The cAMP-mediated induction of SIRT1 deacetylase activity was still evident in a truncated SIRT1 mutant containing only the catalytic domain (residues 173-555), thus indicating that the highly conserved enzymatic core is sufficient to facilitate the forskolin effect (Figure 4D, S4F). These results are consistent with the mass spectrometry analysis that identified S434, a residue located in the deacetylase domain, to be differentially phosphorylated in response to forskolin treatment. Therefore, we assessed whether non-phosphorylatable S434 mutants affected basal and/or forskolin-induced SIRT1 activity. As shown in Figure 4E and S4G-J, SIRT1 WT, the S433A mutant, and the non-phosphorylatable S434A or S434C mutants exhibited similar Km’s for NAD+ in the absence of PKA activation. Unlike the WT and S433A mutant, however, the S434 phosphorylation mutants did not display an increase in deacetylase activity following forskolin treatment. The experiments using the fluorescently labeled p53 peptide were performed in order to quantify relative changes in the kinetic parameters of intrinsic enzymatic activity and were further validated with a non-fluorometric SIRT1 deacetylation assay (Smith et al., 2009) using a H3K14 peptide (Figure 4F, S4K).
Figure 4. The cAMP/PKA pathway increases intrinsic SIRT1 enzymatic activity through serine 434 phosphorylation.
A, Forskolin treatment increases SIRT1 activity at multiple concentrations of acetylated substrate and three different NAD+ concentrations (140 μM acetyl-p53 peptide results are shown; data for 70 and 350 μM peptide are found in Figure S4B). Transfected FLAG-tagged SIRT1 was immunoprecipitated from DMSO or Fsk-treated U2OS cells and its activity was quantified using a fluorophore-conjugated acetylated p53 peptide; error bars are present but may be smaller than data point symbols. B-C, Forskolin treatment decreases the SIRT1 Km for NAD+ and increases the reaction rate, V, while leaving the Km for p53 unaffected. Kinetic data from Figure 4A were first plotted reciprocally as [NAD+]/vexperimental vs [NAD+] (Figure S4D) and the slopes and y-intercepts from that graph were used to make secondary plots from which the reaction velocity, V, and the Km’s for both p53 and NAD+ were calculated. V was converted from arbitrary fluorescence units to pmoles of deacetylated p53/min using a deacetylated standard (ENZO). D, The SIRT1 catalytic domain (amino acids 173-555) is sufficient to mediate the effects of forskolin on deacetylase activity. E, S434 mutation abolishes the effect of cAMP on SIRT1 in vitro activity. F, Activation of PKA signaling by epinephrine increases SIRT1 activity in a non-fluorometric based in vitro assay using a H3K14 peptide. G-H, Forskolin induces the phosphorylation and activation of human SIRT1 but not SIRT2 in vitro. Data are presented as means ± S.D.
Notably, forskolin-induced phosphorylation and activation was specific to SIRT1 and was not seen in another reported p53 deacetylase, SIRT2 (Figure 4G-H, S4L). The cAMP insensitivity of SIRT2 may be due, in part, to poor conservation of the primary sequence immediately flanking the serine in SIRT2 that is analogous to SIRT1 S434 or to different cognate binding partners. Notably, mutation of the glutamine residue (Q265) in the +2 position of SIRT2 S263 to a lysine, which more closely mimics the SIRT1 primary sequence, did not result in phosphorylation following forskolin treatment. Taken together, these in vitro results indicate that cAMP/PKA activation directly controls SIRT1 catalytic function through phosphorylation of a single residue, S434, located in the core deacetylase domain. This modification accounts for the rapid increase in enzymatic efficiency independently of discernible fluctuations in intracellular NAD+.
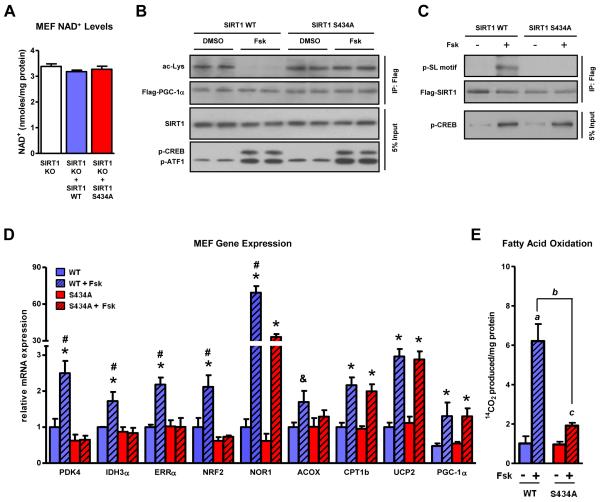
cAMP-mediated increases in fatty acid oxidation and corresponding transcriptional programs are largely dependent on SIRT1 serine 434 phosphorylation
In order to evaluate the physiological relevance of SIRT1 S434 phosphorylation in the context of the cAMP signaling, we established retroviral cell lines over-expressing either SIRT1 WT or the S434A mutant in SIRT1 KO MEFs to eliminate any confounding effects from endogenous SIRT1 protein. Intracellular NAD+ levels were similar between the two reconstituted lines (Figure 5A). Forskolin-triggered CREB phosphorylation in both cell lines, however, PGC-1α was only deacetylated in cells expressing SIRT1 WT but not SIRT1 S434A (Figure 5B). Moreover, the antibody targeting phosphorylated SL motifs recognized a cAMP-triggered modification only on WT SIRT1 and not the non-phosphorylatable mutant (Figure 5C). Most importantly, the forskolin-mediated upregulation of oxidative genes was completely ablated and the forskolin induction of fatty acid oxidation was significantly attenuated in the SIRT1 S434A expressing MEFs (Figure 5D-E). Consistent with Figure 2D, a subset of forskolin-sensitive PGC-1αtargets (UCP2 and CPT1b) and even PGC-1α itself were not dependent on SIRT1 S434 phosphorylation for cAMP responsiveness. These data provide a plausible mechanism for how the cAMP/PKA pathway sensitizes and stimulates SIRT1 enzymatic activity in the absence of NAD+ fluctuations to increase the expression of lipid metabolism genes and induce appreciable changes in fatty acid oxidation.
Figure 5. cAMP-mediated increases in fatty acid oxidation and corresponding transcriptional programs are largely dependent on SIRT1 serine 434 phosphorylation.
A, Intracellular NAD+ levels are similar in SIRT1 KO MEFs and MEF retroviral cell lines overexpressing SIRT1 wildtype and the S434A mutant. Mutation of S434 to a non-phosphorylatable residue in cells ablates the forskolin-triggered, B, deacetylation of PGC-1α, C, phosphorylation of SIRT1, D, induction of fatty acid oxidation genes, and significantly blunts, E, utilization of oleic acid. Retroviral cell lines expressing either WT or S434A SIRT1 were established using SIRT1−/− fibroblasts. MEFs were treated with Fsk for 6 h before RNA isolation and mRNA quantification; statistical significance was assessed using one-way ANOVA with a Tukey post-test, *, p<0.05, DMSO vs Fsk; *, p<0.01, DMSO vs Fsk; #, p<0.01, WT/Fsk vs S434A/Fsk. Fatty acid oxidation was quantified by measuring 14CO2 production and statistical significance was assessed using one-way ANOVA with a Tukey post-test; a, p<0.01, WT/DMSO vs WT/Fsk; b, p<0.01, WT/Fsk vs S434A/Fsk; c, p<0.01, S434A/DMSO vs S434A/Fsk. Data are presented as means ± S.D.
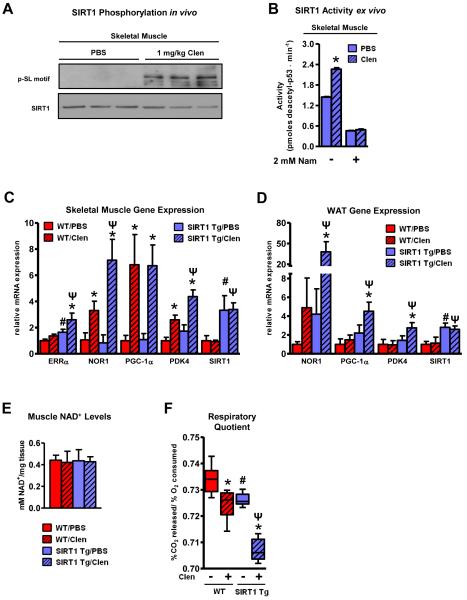
Adrenergic signaling induces SIRT1 phosphorylation/activation which promotes lipid oxidation and energy expenditure in vivo
We next investigated whether activation of the cAMP pathway exerted similar regulatory control on SIRT1 enzymatic activity and function in whole animals using both pharmacological and physiological approaches. The former was addressed by administering the β2-adrenergic receptor agonist, clenbuterol, to mimic the catecholamine response animals undergo following various metabolic or thermogenic challenges. One of the actions of clenbuterol, as well as endogenous β-agonists such as epinephrine, is the elevation of fatty acid oxidation in peripheral tissues, particularly skeletal muscle. This phenomenon is at least partially dependent upon the increased expression of PGC-1α target genes (Miura et al., 2007; Pearen et al., 2008). We have previously shown that SIRT1 is capable of mediating this lipid-utilizing response by deacetylating PGC-1α and inducing the expression of targets such as ERRα and PDK4 (Gerhart-Hines et al., 2007). In order to determine what role SIRT1 might play under conditions of increased adrenergic signaling, we analyzed SIRT1 activity and phosphorylation status as well as the expression of PGC-1α target genes in transgenic mice moderately overexpressing SIRT1 (SIRT1 Tg; (Banks et al., 2008)) following a single, acute administration of clenbuterol. Consistent with the results obtained from cellular cAMP agonism, SIRT1 immunoprecipitated from clenbuterol-treated mice was highly phosphorylated at S434 and more active compared to SIRT1 from saline control animals (Figure 6A-B). At the transcriptional level, a subset of clenbuterol-regulated genes associated primarily with driving fatty acid oxidation (NOR1, ERRα and PDK4) (Huss et al., 2004; Maxwell et al., 2005; Wende et al., 2005; Hummasti and Tontonoz, 2008) was further increased in SIRT1 Tg mice (Figure 6C). This striking pattern of SIRT1 influence on clenbuterol-induced gene expression was also observed in the expression profile from white adipose tissue (WAT) following adrenergic stimulation (Figure 6D). Importantly, neither clenbuterol nor SIRT1 transgeneity affected NAD+ levels (Figure 6E). The whole body physiological significance of the increased expression of these fatty acid metabolism and energy expenditure genes in mice was then determined using Comprehensive Lab Animal Monitoring System (CLAMS). Within 6 hours of clenbuterol administration, the respiratory quotient (RQ) of SIRT1 Tg mice was significantly decreased relative to WT mice indicating elevated oxidation of fatty acids (Figure 6F). This SIRT1-mediated enhancement in lipid utilization was associated with a corresponding elevation in total energy expenditure or heat dissipation (Figure S5A-B). These pharmacological data suggest that the elevation in fatty acid oxidation rates and acceleration of energy expenditure caused by β-adrenergic signaling in vivo are mediated in part by an increase in SIRT1 activity in the absence of changes in intracellular NAD+ levels.
Figure 6. Adrenergic signaling induces SIRT1 phosphorylation/activation which promotes lipid oxidation and energy expenditure in vivo.
A, SIRT1 is phosphorylated in response to cAMP signaling in vivo. B, SIRT1 catalytic function is enhanced by adrenergic stimulation in vivo. Activity of FLAG-tagged SIRT1 immunoprecipitated from pooled gastrocnemius/soleus samples from Tg mice treated with PBS (n=6) or Clen (n=6) was assessed using the SIRT1 fluorometric activity assay as described in Figure 4 and measured in duplicate; *, p<0.01, PBS vs Clen. Ectopic expression of SIRT1 enhances the clenbuterol-mediated induction of fatty acid oxidative genes in vivo in both, C, skeletal muscle and, D, white adipose tissue (WAT). RNA was isolated from the gastrocnemius and epididymal fat pads of fasted WT (n=5) and SIRT1 transgenic (Tg) (n=6) mice intraperitoneally injected with either saline (PBS) or clenbuterol (Clen-1 mg/kg) overnight for 14 h; statistical significance was assessed using one-way ANOVA with a Tukey post-test, *, p<0.01, PBS vs Clen treatment; #, p<0.01, WT/PBS vs Tg/PBS; Ψ, P<0.01, WT/Clen vs Tg/Clen. Gene expression results are representative of two separate experiments. E, Skeletal muscle NAD+ levels are not affected by clenbuterol administration or SIRT1 overexpression. NAD+ concentrations were assessed from gastrocnemius tissue from treated WT (n=4) and SIRT1 Tg mice (n=4). Muscle samples were homogenized in NAD+ extraction buffer and NAD+ was measured according to the manufacturer’s instructions (BioAssay Systems). F, SIRT1 transgenic mice exhibit lower respiratory quotients (RQ) than WT mice in response to clenbuterol. Metabolic parameters of WT (n=8) and SIRT1 Tg (n=8) mice were measured using CLAMS analysis over the course of a 14 h fast and an initial refeeding (RF). Statistical significance of RQ measurements was determined by one-way ANOVA with a Tukey post-test; *, p<0.01, PBS vs Clen treatment; #, p<0.01, WT/PBS vs Tg/PBS; Ψ, P<0.01, WT/Clen vs Tg/Clen. Data are presented as means ± S.D.
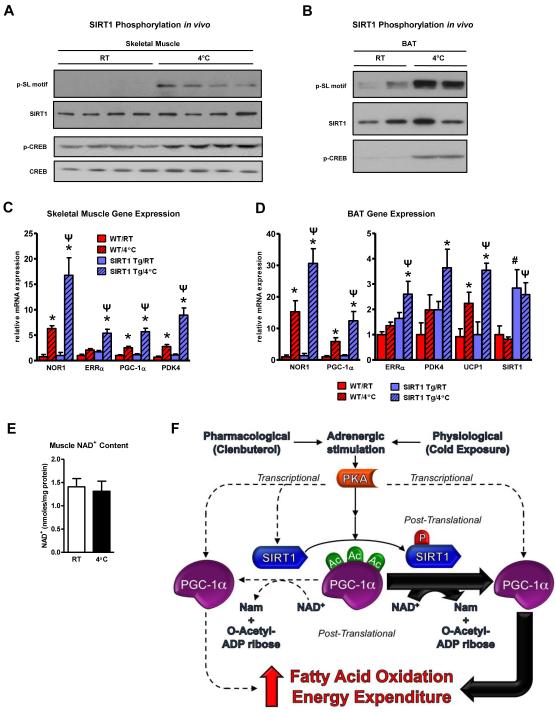
Cold challenge induces SIRT1 phosphorylation/activity which potentiates the thermogenic transcriptional programming in vivo
Given the enhanced fatty acid oxidation and energy expenditure observed in the SIRT1 Tg mice following pharmacological adrenergic agonism, we decided to investigate whether SIRT1 played a role in the physiologically-relevant, cAMP-mediated stress pathway of adaptive thermogenesis. This fundamental response is predominantly orchestrated by adrenergic signaling to ensure maintenance of body temperature and is characterized by increased lipid oxidation and energy expenditure (Puigserver et al., 1998; Nedergaard and Cannon, 2010). Wildtype and SIRT1 Tg mice were maintained at 25°C or cold-challenged at 4°C at which point SIRT1 phosphorylation and gene expression linked to fatty acid oxidation and energy expenditure were assessed. Similar to clenbuterol administration, SIRT1 was highly phosphorylated in skeletal muscle and BAT following cold exposure (Figure 7A-B) and the cold-induction of fatty acid oxidation genes was greatly enhanced in SIRT1 Tg mice compared to wildtype mice whereas NAD+ concentrations remained unchanged (Figure 7C-E). Collectively, the results from clenbuterol administration and cold exposure are consistent with a model whereby activation of the PKA pathway in mice, either pharmacologically or physiologically, causes SIRT1 phosphorylation and a rapid increase in SIRT1 activity independently of changes in NAD+ levels (Figure 7F). This heightened Sirtuin enzymatic capacity then leads to PGC-1α deacetylation and upregulation of transcriptional networks associated with fatty acid oxidation and energy expenditure. In addition to the more immediate post-translational control, PGC-1α transcription is also induced following cAMP agonism thereby facilitating a more prolonged response to homeostatic perturbations (Puigserver et al., 1998).
Figure 7. Cold challenge induces SIRT1 phosphorylation/activity which potentiates the thermogenic transcriptional programming in vivo.
A, SIRT1 is phosphorylated in skeletal muscle and, B, BAT following cold exposure. C, Ectopic expression of SIRT1 enhances the cold-induction of fatty acid oxidation and energy expenditure genes in skeletal muscle and, D, brown adipose tissue (BAT). RNA was isolated from WT (n=6) and SIRT1 Tg (n=6) mice that were either maintained at room temperature (RT, 25°C) or exposed to cold (4°C) for 6 h; statistical significance was assessed using one-way ANOVA with a Tukey post-test, *, p<0.01, RT vs 4°C treatment; #, p<0.01, WT/RT vs Tg/RT; Ψ, P<0.01, WT/4°C vs Tg/4°C. E, Skeletal muscle NAD+ levels are not affected by cold exposure. F, Stimulation of the cAMP/PKA pathway results in phosphorylation of SIRT1 which enhances the efficiency of NAD+ co-substrate utilization and increases overall catalytic capacity. Adrenergic agonism also induces transcription of both SIRT1 and PGC-1α. Data are presented as means ± S.D.
Discussion
Our findings support a regulatory mechanism through which the cAMP/PKA pathway significantly increases the intrinsic catalytic function of SIRT1 through S434 phosphorylation, resulting in deacetylation of key metabolic target, PGC-1α, and increased fatty acid oxidation and thermogenic function in the absence of discernible fluctuations in NAD+. Unlike earlier studies that have postulated that changes in the Sirtuin co-substrate NAD+ were the central regulatory link between energetic state and SIRT1 function, our work demonstrates that nutrient and hormonal signals can converge to directly modulate SIRT1 deacetylase activity. This enables SIRT1 to more rapidly and dynamically manage homeostatic responses to metabolic and environmental stresses. Integrating our results with previously reported mechanisms of SIRT1 control creates a more comprehensive regulatory model of Sirtuin function. Adrenergic-mediated phosphorylation of SIRT1 triggers an immediate increase in catalytic function and this heightened activity can then be sustained through AMPK-dependent elevation of NAD+ levels in the face of persistent nutrient or energy demand. Although our data strongly indicate that PKA is required for cAMP-induced SIRT1 phosphorylation and PGC-1α deacetylation (Figures 2E; 3D), whether it is the kinase that directly phosphorylates S434 is not yet clear. Recombinant PKA weakly phosphorylates SIRT1 in vitro and the residues flanking S434 do not constitute a canonical PKA consensus motif. The possibility remains, therefore, that other kinases downstream of PKA are phosphorylating SIRT1 in response to increased cAMP levels in vivo—a possibility that we are actively pursuing. Most recently, SIRT1 has been shown to also undergo transcriptional regulation by the cAMP/PKA/CREB axis following prolonged nutrient deprivation (Noriega et al., 2011). This secondary layer of Sirtuin control could further enhance long-term responses to metabolic stresses.
While it remains unclear how S434 phosphorylation coordinates a lower Km for NAD+ and a higher V, structural data from some Sir2 orthologs suggests that this residue hydrogen bonds with a highly conserved arginine (R266 in SIRT1), forming a clasp over the diphosphate backbone of NAD+ (Figure S6A-B) (Hoff et al., 2006). Therefore, modification of S434 might affect this interaction and promote a more efficient, “active” coordination of NAD+. However, without crystallographic evidence from a structure containing phosphorylated S434, no definitive remarks can be made regarding molecular mechanisms. The complete conservation of this serine among Sir2 orthologues suggests that phosphorylation of this residue may serve a more general regulatory role in modulation of Sirtuin catalytic function. Variation among the different Sir2 orthologs in the primary amino acid sequence that flanks this serine as well as differential protein interactions may allow for targeted specificity by different kinase pathways and biological stimuli. This method of control could provide an added degree of selectivity that is not possible by blanket changes in the general Sirtuin co-substrate, NAD+. For example, SIRT1 and SIRT2 are similarly localized in the nucleus and share p53 as a substrate, yet only SIRT1 deacetylates p53 in response to cAMP agonism (Figure 4H).
The cAMP pathway and SIRT1 have both been independently shown to increase fatty acid oxidation, at least in part, through the activation of the transcriptional coactivator, PGC-1α(Gerhart-Hines et al., 2007; Miura et al., 2007; Kumar et al., 2008). Here, we provide a mechanistic connection between PKA signaling and SIRT1 that directly affects nutrient selection/utilization and total energy expenditure under both pharmacological adrenergic activation and the well-established, physiological adaptive thermogenic response. The adaptation to cold exposure mediated by adrenergic stimulation is associated with rapid increases in expression of genes encoding enzymes linked to fatty acid oxidation and energy expenditure in brown fat and skeletal muscle (Nedergaard and Cannon, 2010). Our studies show that SIRT1 is rapidly phosphorylated and activated in these two target tissues during cold exposure through the PKA signaling and contributes to the expression of oxidative metabolic genes. It is not clear at this point whether these transcriptional effects are mediated solely by PGC-1α or if other SIRT1 substrates are involved in this environmental adaptation.
While our results advance the basic understanding of how the nutrient “sensor,” SIRT1, coordinates metabolic responses to environmental and dietary stresses, the identification of additional components of the adaptive thermogenic pathway possesses added clinical implications. This fundamental homeostatic response has recently garnered a considerable amount of attention as a potential therapeutic avenue for the treatment of metabolic dysfunction (Nedergaard and Cannon, 2010; Tseng et al., 2010). This emerging interest stems from the prevailing notion that increases in thermogenic pathways, specifically utilization of lipids, are thought to be an important mechanism of body weight regulation and protection against the onset and pathological progression of diet-induced obesity (Nedergaard and Cannon, 2010). Therefore, a promising, non-invasive approach to combat obesity would be to pharmacologically exploit these catabolic, fat-burning pathways in skeletal muscle and BAT. To this end, studies examining the selective targeting and stimulation of fatty acid oxidation and energy expenditure through PKA and SIRT1 might provide opportunities for clinical intervention to ameliorate the detrimental effects of metabolic syndrome.
Methods
Animal Studies
All animal studies were performed with an approved protocol from the Beth Israel Deaconess Medical Center Institutional Animal Care and Use Committee. Mice were housed on a 12:12-h light-dark cycle (lights on at 6 AM). Gene expression analysis was carried out on 14-20 week old male SIRT1 Tg and WT mice that were intraperitoneally injected with 1 mg/kg clenbuterol (Sigma) or sterile PBS (GIBCO) and fasted for 14 hrs. Comprehensive laboratory animal monitoring system (CLAMS) (Columbus instruments) experiments were performed on SIRT1 Tg mice and WT littermates to assess respiratory quotient (RQ), energy expenditure, and physical activity. Prior to clenbuterol or saline administration, mice were acclimated for 48 h in individual metabolic cages without bedding. CLAMS data are presented as rolling averages (± 3 30 min time points) ± S.D. Cold exposure experiments were performed in climate controlled units set to 25 and 4°C. All the WT and SIRT1 Tg mice used in the experiment were placed in individual cages with access to food and water and allowed to acclimate to the 25°C unit for 24 hours before a select number of mice were moved to 4°C for the indicated period of time.
Cell Culture
C2C12, U2OS, and MEF cell lines were cultured in 4.5 g/L glucose DMEM (GIBCO) supplemented with 10% FBS (Hyclone) and 1% penicillin/streptomycin (GIBCO) in 5% CO2. C2C12 myoblasts were differentiated by switching the medium to DMEM containing 2% horse serum (GIBCO) upon reaching ~75% confluency. Transfections were carried out on MEF cell lines using Lipofectamine 2000 (Invitrogen) and on U2OS using Polyfect (Invitrogen) according to manufacturer’s instructions. Cellular assays were performed by replacing growth media with serum-free DMEM at which point inhibitors - EX-527 (Tocris), trichostatin A (Wako), Compound C (Calbiochem), and H-89 (Calbiochem) - were added 1 h prior to administration of agonists - forskolin (Sigma), clenbuterol (Sigma), epinephrine (Sigma), Br-cAMP (Sigma), and AICAR (Sigma). Cells were then incubated for an additional 30 minutes before being harvested in nonidet P-40 lysis buffer (1% nonidet P-40, 20 mM HEPES pH 7.9, 125 mM NaCl, 10 mM NaF, 10 mM glycerol-2-phosphate, and protease inhibitors (Roche)); overnight immunoprecipitation was carried out using FLAG-M2 agarose beads (Sigma). FK866 (Cayman) was added overnight prior to measuring NAD+ levels according to manufacturer’s instructions (BioAssay Systems). Nuclear fractions for Sirtuin activity assays and phosphorylation mapping were obtained by first lysing cells in a hypotonic lysis solution (10 mM HEPES pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5% nonidet P-40, 10 mM NaF, 10 mM glycerol-2-phosphate, and protease inhibitors), centrifuging to pellet nuclei, and then washing once with hypotonic lysis buffer before resuspending in nonidet P-40 lysis buffer. Nuclear contents were liberated by sonication.
Fluorometric SIRT1 Activity Assay
FLAG-tagged SIRT1 was immunoprecipitated from nuclear fractions from forskolin-treated U2OS cells or from whole-cell extracts from saline/clenbuterol injected C57BL/6J mice. Buffer exchange was performed on eluted protein from nonidet P-40 lysis buffer to fluorometric activity assay buffer (50 mM Tris-Cl pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1 mg/ml BSA) and incubated with various concentrations of NAD+ and fluorescently-labeled acetylated p53 peptide for 45 or 60 min at 30°C according to the manufacturer’s instructions (ENZO). The reaction was terminated by addition of a developer solution containing 2 mM nicotinamide to inhibit SIRT1 and protease to digest deacetylated p53 peptide. SIRT1 activity was assessed by measuring the fluorescent emission at 460 nm, following excitation at 360 nm.
Non-fluorometric SIRT1 Activity Assay
FLAG-tagged SIRT1 was immunoprecipitated from whole-cell extracts from epinephrine-treated U2OS cells. Buffer exchange was performed on eluted protein from nonidet P-40 lysis buffer to non-fluorometric activity assay buffer (20 mM K3PO4, 150 mM NaCl, 1 mM DTT, and 5% glycerol) and assays were carried out as previously described (Smith et al., 2009). Briefly, eluted SIRT1 enzyme was incubated with 500 μM acetylated H3K14 peptide (Tufts Core Facility), 0.2 mM NADPH, 1 mM DTT, 3.3 mM α-ketoglutarate, 1.5 μM recombinant Nicotinamidase (MBP-PncA), and ~1.5 U glutamate dehydrogenase from Proteus over a range of NAD+ concentrations (150, 500, and 1500 μM) at 30°C. SIRT1 activity was assessed by measuring the disappearance of NAPDH from the coupled reaction system at a 380 nm absorbance every 10 min for 600 minutes.
Fatty Acid Oxidation
Oxidation assays were performed as previously described (Gerhart-Hines et al., 2007). Briefly, MEFs were treated with Fsk and 0.3 mM unlabeled fatty acid in low glucose DMEM (~5 mM glucose, GIBCO) for 6 h before incubation with 14C-oleic acid (Perkin Elmer) for an additional 3 h. 14CO2 produced by oxidation of the radiolabeled fatty acid was liberated from the media by perchloric acid, captured in phenylethylamine-soaked Whatman paper, and quantified on a scintillation counter.
Mass Spectrometry
In-gel protein digests were resuspended in 8 μL 1% formic acid, and 4 μL were analyzed by microcapillary liquid chromatography electrospray ionization tandem mass spectrometry (LC-MS/MS). Analyses were done on a LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific, Germany) equipped with a Thermo Fisher Scientific nanospray source, an Agilent 1100 Series binary HPLC pump, and a Famos autosampler. Peptides were separated on a 100 μm × 16 cm fused silica microcapillary column with an in-house made needle tip. The column was packed with MagicC18AQ C18 reversed-phase resin (particle size, 5 μm; pore size, 200 Å; Michrom Bioresources). Separation was achieved through applying a 30 min gradient from 0 to 28 % acetonitrile in 0.125 % formic acid. The mass spectrometer was operated in a data dependent mode with a full MS scan acquired with the Orbitrap, followed by up to 10 LTQ MS/MS spectra on the most abundant ions detected in the MS scan. Mass spectrometer settings were: full MS (AGC, 1×106; resolution, 6×104; m/z range, 375-1800; maximum ion time, 1000 ms); MS/MS (AGC, 5×103; maximum ion time, 120 ms; minimum signal threshold, 4×103; isolation width, 2 Da; dynamic exclusion time setting, 30 sec).
All MS/MS spectra were then exported as individual DTA files and searched using the Sequest algorithm. These spectra were then searched against a database containing sequence of mouse SIRT1 in both forward and reversed orientations. The following parameters were selected to identify SIRT1: 50 ppm precursor mass tolerance, 0.8 Da product ion mass tolerance, fully tryptic digestion, and up to two missed cleavages. Variable modifications were as follows: oxidation of methionine (+15.994915), phosphorylation of serines, threonines, and tyrosines (+79.966330).
The AScore algorithm was used to quantify the confidence with which each phosphorylation modification could be assigned to a particular residue in each peptide (Beausoleil et al., 2006). Peptides with AScores above 13 were considered to be localized to a particular residue (p < 0.05). For sites that could not be confidently localized, the algorithm provided the range of possible residues on which the modification could be localized, together with scores for each possible site.
RNA
Total RNA was isolated by Trizol (Invitrogen) extraction and 2 μg of total RNA was used for cDNA synthesis. Relative mRNA levels were determined using qPCR on a BioRad iQ5. Primer sequences are available upon request.
in vitro PKA Kinase Assay
FLAG-tagged SIRT1 was immunoprecipitated from nuclear fractions and incubated in PKA assay buffer (40 mM Tris, pH 7.5, 20 mM MgOAc, and 1 mM DTT) with 200 μM ATP, 2 μCi [32P-γ] ATP, and 100 ng recombinant PKA-α catalytic subunit (Millipore) according to manufacturer’s instructions.
Statistics
Data are presented as means ± S.D. Statistical analysis was performed using one-way ANOVA for mouse gene expression studies and repeated measures ANOVA for CLAMS analysis on GraphPad Prism software. Comparisons among specific treatment groups were done using post-tests as indicated in the respective figure legends.
Supplementary Material
Acknowledgements
We thank members of the Puigserver lab for important discussions on the project. We particularly thank Dr. Joseph Rodgers and Dr. Timothy Kelly for critical reading of the manuscript and Dr. Wei Gu for providing the SIRT1 transgenic mice. We also thank Dr. Fred Alt and Dr. Raul Mostoslavsky for providing the WT and SIRT1 KO MEFs and Dr. John Denu for providing reagents necessary for the coupled deacetylase assay. The authors were supported in part by fellowships from the American Heart Association (ZGH), NRSA Kirschstein from the National Institutes of Health (JED), and Swiss National Science Foundation (SMB). The research was supported from funds from the Dana-Farber Cancer Institute and grants from the American Diabetes Association, Department of Defense and NIH/NIDDK, RO1 069966, (PP).
ZGH and PP conceived the project. ZGH designed and performed cellular deacetylation and phosphorylation experiments, phosphorylation-mapping, in vitro kinase and Sirtuin activity assays, mouse experiments, and wrote the manuscript. JED designed and performed cellular deacetylation and phosphorylation experiments, phosphorylation-mapping, in vitro Sirtuin activity assays, mouse experiments, and wrote the manuscript. SMB and ASB designed and performed mouse experiments. MPJ and SPG performed and analyzed mass spectrometry experiments. JL and HC performed cellular phosphorylation experiments. PP designed experiments and wrote the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing financial interests:
The authors declare no competing financial interests.
References
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Banks AS, Kon N, Knight C, Matsumoto M, Gutiérrez-Juárez R, Rossetti L, Gu W, Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beausoleil SA, Villén J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G, Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 2010;9:285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- Donmez G, Wang D, Cohen DE, Guarente L. SIRT1 suppresses beta-amyloid production by activating the alpha-secretase gene ADAM10. Cell. 2010;142:320–332. doi: 10.1016/j.cell.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev. Cell. 2008;14:661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim S-H, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Williams JG, Schug TT, Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J. Biol. Chem. 2010;285:13223–13232. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Serrano M. SIRT1: recent lessons from mouse models. Nat. Rev. Cancer. 2010;10:819–823. doi: 10.1038/nrc2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoff KG, Avalos JL, Sens K, Wolberger C. Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide. Structure. 2006;14:1231–1240. doi: 10.1016/j.str.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Hummasti S, Tontonoz P. Adopting new orphans into the family of metabolic regulators. Mol. Endocrinol. 2008;22:1743–1753. doi: 10.1210/me.2007-0566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss JM, Torra IP, Staels B, Giguère V, Kelly DP. Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle. Mol. Cell. Biol. 2004;24:9079–9091. doi: 10.1128/MCB.24.20.9079-9091.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Iwabu M, Yamauchi T, Okada-Iwabu M, Sato K, Nakagawa T, Funata M, Yamaguchi M, Namiki S, Nakayama R, Tabata M, et al. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature. 2010;464:1313–1319. doi: 10.1038/nature08991. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Jung J-W, Kim MK, Chung JH. CK2 is the regulator of SIRT1 substrate-binding affinity, deacetylase activity and cellular response to DNA-damage. PLoS ONE. 2009;4:e6611. doi: 10.1371/journal.pone.0006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Liu D, Wang H, Robidoux J, Collins S. Orphan nuclear receptor NOR-1 enhances 3′,5′-cyclic adenosine 5′-monophosphate-dependent uncoupling protein-1 gene transcription. Mol. Endocrinol. 2008;22:1057–1064. doi: 10.1210/me.2007-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Maxwell MA, Cleasby ME, Harding A, Stark A, Cooney GJ, Muscat GEO. Nur77 regulates lipolysis in skeletal muscle cells. Evidence for cross-talk between the beta-adrenergic and an orphan nuclear hormone receptor pathway. J. Biol. Chem. 2005;280:12573–12584. doi: 10.1074/jbc.M409580200. [DOI] [PubMed] [Google Scholar]
- Miura S, Kawanaka K, Kai Y, Tamura M, Goto M, Shiuchi T, Minokoshi Y, Ezaki O. An increase in murine skeletal muscle peroxisome proliferator-activated receptor-gamma coactivator-1alpha (PGC-1alpha) mRNA in response to exercise is mediated by beta-adrenergic receptor activation. Endocrinology. 2007;148:3441–3448. doi: 10.1210/en.2006-1646. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedergaard J, Cannon B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell Metab. 2010;11:268–272. doi: 10.1016/j.cmet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Noriega LG, Feige JN, Canto C, Yamamoto H, Yu J, Herman MA, Mataki C, Kahn BB, Auwerx J. CREB and ChREBP oppositely regulate SIRT1 expression in response to energy availability. EMBO Rep. 2011;12:1069–1076. doi: 10.1038/embor.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearen MA, Myers SA, Raichur S, Ryall JG, Lynch GS, Muscat GEO. The orphan nuclear receptor, NOR-1, a target of beta-adrenergic signaling, regulates gene expression that controls oxidative metabolism in skeletal muscle. Endocrinology. 2008;149:2853–2865. doi: 10.1210/en.2007-1202. [DOI] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S.-ichiro. The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J. Biol. Chem. 2004;279:50754–50763. doi: 10.1074/jbc.M408388200. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Sasaki T, Maier B, Koclega KD, Chruszcz M, Gluba W, Stukenberg PT, Minor W, Scrable H. Phosphorylation regulates SIRT1 function. PLoS ONE. 2008;3:e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Smith BC, Hallows WC, Denu JM. A continuous microplate assay for sirtuins and nicotinamide-producing enzymes. Anal. Biochem. 2009;394:101–109. doi: 10.1016/j.ab.2009.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng Y-H, Cypess AM, Kahn CR. Cellular bioenergetics as a target for obesity therapy. Nat Rev Drug Discov. 2010;9:465–482. doi: 10.1038/nrd3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Vinciguerra M, Fulco M, Ladurner A, Sartorelli V, Rosenthal N. SirT1 in muscle physiology and disease: lessons from mouse models. Dis Model Mech. 2010;3:298–303. doi: 10.1242/dmm.004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477:E1–E2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- Viswanathan M, Kim SK, Berdichevsky A, Guarente L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev. Cell. 2005;9:605–615. doi: 10.1016/j.devcel.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Wende AR, Huss JM, Schaeffer PJ, Giguère V, Kelly DP. PGC-1alpha coactivates PDK4 gene expression via the orphan nuclear receptor ERRalpha: a mechanism for transcriptional control of muscle glucose metabolism. Mol. Cell. Biol. 2005;25:10684–10694. doi: 10.1128/MCB.25.24.10684-10694.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol. Cell. 2007;27:149–162. doi: 10.1016/j.molcel.2007.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.