Abstract
Wnt proteins bind to Frizzled family members on the surface of the cell to regulate developmental processes (e.g., cell fate and morphogenesis).
Members of the Wnt family are secreted ligands that regulate numerous developmental pathways (Cadigan and Peifer 2009; Van Amerongen and Nusse 2009; and see Neel and Muthuswarmy 2012; Perrimon and Shilo 2012). Wnt binds to members of the Frizzled family, activating a canonical signaling pathway that targets members of the LEF/TCF transcription factor family (Fig. 1). These control gene expression programs that regulate cell fate and morphogenesis (Van Amerongen and Nusse 2009). Wnt also activates so-called non-canonical pathways (Fig. 2), which regulate planar cell polarity by stimulating cytoskeletal reorganization and can also lead to calcium mobilization (Veeman et al. 2003).
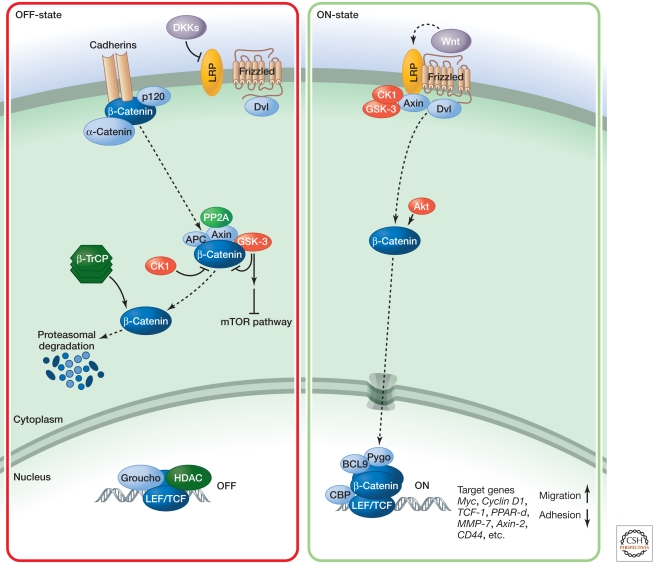
Figure 1.
Canonical Wnt signaling.
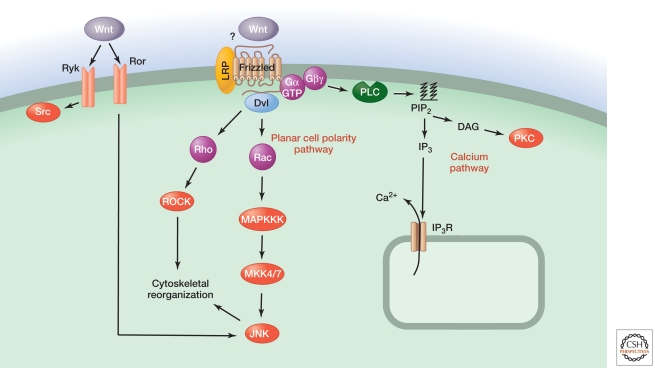
Figure 2.
Non-canonical Wnt signaling pathways.
The canonical Wnt signaling pathway is regulated at many levels, including by extensive negative control steps. In cells not exposed to Wnt signals, the major signaling components including the receptors and the β-catenin protein are kept in an off state (Fig. 1, left). Active Wnt signaling rearranges these complexes (Fig. 1, right). In the inactive state, β-catenin levels are kept low through interactions with the protein kinases GSK-3b and CK1, the adenomatous polyposis coli tumor suppressor protein (APC), and the scaffolding protein axin. β-Catenin is degraded, after phosphorylation by GSK-3 and CK1 through the ubiquitin pathway, which involves interactions with β-TrCP. β-Catenin is also regulated by adhesion complexes containing cadherins and α-catenin. At the level of receptors, the negative regulator DKK can bind to the LRP receptor and inhibit Wnt signaling.
During signaling (right), Wnt proteins interact with Frizzled receptors; the transmembrane protein LRP is also required for Wnt signaling. When Wnt proteins bind, the receptors presumably rearrange, leading to the activation of β-catenin. The cytoplasmic tail of LRP binds to axin in a Wnt- and phosphorylation-dependent manner. Phosphorylation of the tail of LRP is regulated by CK1, and Dishevelled (Dvl) and Frizzled also have roles in this process. In a current model, Wnt signaling initially leads to formation of a complex involving Dvl, axin, and GSK3. The DIX domain in axin is similar to the NH2 terminus in Dvl and promotes interactions between Dvl and axin. As a consequence, GSK does not phosphorylate β-catenin anymore, releasing it from the axin complex and allowing it to accumulate. The stabilized β-catenin then enters the nucleus to interact with TCF/LEF transcription factors. Note that GSK3 also participates in other pathways, such as the mTor and Akt pathway (see Hemmings 2012; Laplante and Sabatini 2012).
In the nucleus, in the absence of the Wnt signal, TCF/LEF acts as a repressor of Wnt target genes, in a complex with Groucho. β-Catenin can convert TCF/LEF into a transcriptional activator of the same genes that are repressed by TCF/LEF alone. Three other key players in this complex are BCL9, Pygopos, and CBP. There are many target genes for the canonical Wnt pathway. Most of these genes are cell type specific, with the possible exception of axin 2, which acts as a negative feedback regulator (Grigoryan 2008).
In non-canonical Wnt signaling, Wnt stimulates the planar cell polarity pathway by activating the small GTPases Rho and Rac. These induce cytoskeletal rearrangements that lead to the development of lateral asymmetry in epithelial sheets and other structures. Wnt can also provoke release of calcium from intracellular stores, probably via heterotrimeric G-proteins. A less-well-understood mechanism involves activation of the Ror and Ryk tyrosine kinase receptors, which control the activities of the JNK and Src kinases, respectively (van Amerongen et al. 2008).
Acknowledgments
Figure 1 adapted, with permission, from Cell Signaling Technology (http://www.cellsignal.com.)
Footnotes
Editors: Lewis Cantley, Tony Hunter, Richard Sever, and Jeremy W. Thorner
Additional Perspectives on Signal Transduction available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Cadigan KM, Peifer M 2009. Wnt signaling from development to disease: Insights from model systems. Cold Spring Harb Perspect Biol 1: a002881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan T, Wend P, Klaus A, Birchmeier W 2008. Deciphering the function of canonical Wnt signals in development and disease: Conditional loss- and gain-of-function mutations of β-catenin in mice. Genes Dev 22: 2308–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hemmings B 2012. P13K/AKT pathway. Cold Spring Harb Perspect Biol 10.1101/cshperspect.011189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lapalante M, Sabatini D 2012. mTOR signaling. Cold Spring Harb Perspect Biol 10.1101/cshperspect.011593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Neel B, Muthuswarmy S 2012. Signaling in development 2—cell differentiation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.005884 [DOI] [Google Scholar]
- *.Perrimon N, Shilo B 2012. Signaling in development 1—cell fate and patterning. Cold Spring Harb Perspect Biol 10.1101/cshperspect.005975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Amerongen R, Nusse R 2009. Towards an integrated view of Wnt signaling in development. Development 136: 3205–3214 [DOI] [PubMed] [Google Scholar]
- van Amerongen R, Mikels A, Nusse R 2008. Alternative Wnt signaling is initiated by distinct receptors. Sci Signal 1: re9. [DOI] [PubMed] [Google Scholar]
- Veeman MT, Axelrod JD, Moon RT 2003. A second canon. Functions and mechanisms of β-catenin-independent Wnt signaling. Dev Cell 5: 367–377 [DOI] [PubMed] [Google Scholar]