Abstract
Structures of the bacterial ribosome have provided a framework for understanding universal mechanisms of protein synthesis. However, the eukaryotic ribosome is much larger than it is in bacteria, and its activity is fundamentally different in many key ways. Recent cryo-electron microscopy reconstructions and X-ray crystal structures of eukaryotic ribosomes and ribosomal subunits now provide an unprecedented opportunity to explore mechanisms of eukaryotic translation and its regulation in atomic detail. This review describes the X-ray crystal structures of the Tetrahymena thermophila 40S and 60S subunits and the Saccharomyces cerevisiae 80S ribosome, as well as cryo-electron microscopy reconstructions of translating yeast and plant 80S ribosomes. Mechanistic questions about translation in eukaryotes that will require additional structural insights to be resolved are also presented.
New cryo-electron microscopy and X-ray crystal structures of the ribosome in different steps of the translation cycle have provided mechanistic insights into eukaryotic protein synthesis.
All ribosomes are composed of two subunits, both of which are built from RNA and protein (Figs. 1 and 2). Bacterial ribosomes, for example of Escherichia coli, contain a small subunit (SSU) composed of one 16S ribosomal RNA (rRNA) and 21 ribosomal proteins (r-proteins) (Figs. 1A and 1B) and a large subunit (LSU) containing 5S and 23S rRNAs and 33 r-proteins (Fig. 2A). Crystal structures of prokaryotic ribosomal particles, namely, the Thermus thermophilus SSU (Schluenzen et al. 2000; Wimberly et al. 2000), Haloarcula marismortui and Deinococcus radiodurans LSU (Ban et al. 2000; Harms et al. 2001), and E. coli and T. thermophilus 70S ribosomes (Yusupov et al. 2001; Schuwirth et al. 2005; Selmer et al. 2006), reveal the complex architecture that derives from the network of interactions connecting the individual r-proteins with each other and with the rRNAs (Brodersen et al. 2002; Klein et al. 2004). The 16S rRNA can be divided into four domains, which together with the r-proteins constitute the structural landmarks of the SSU (Wimberly et al. 2000) (Fig. 1A): The 5′ and 3′ minor (h44) domains with proteins S4, S5, S12, S16, S17, and S20 constitute the body (and spur or foot) of the SSU; the 3′ major domain forms the head, which is protein rich, containing S2, S3, S7, S9, S10, S13, S14, and S19; whereas the central domain makes up the platform by interacting with proteins S1, S6, S8, S11, S15, and S18 (Fig. 1B). The rRNA of the LSU can be divided into seven domains (including the 5S rRNA as domain VII), which—in contrast to the SSU—are intricately interwoven with the r-proteins as well as each other (Ban et al. 2000; Brodersen et al. 2002) (Fig. 2A). Structural landmarks on the LSU include the central protuberance (CP) and the flexible L1 and L7/L12 stalks (Fig. 2A).
Figure 1.
The bacterial and eukaryotic small ribosomal subunit. (A,B) Interface (upper) and solvent (lower) views of the bacterial 30S subunit (Jenner et al. 2010a). (A) 16S rRNA domains and associated r-proteins colored distinctly: b, body (blue); h, head (red); pt, platform (green); and h44, helix 44 (yellow). (B) 16S rRNA colored gray and r-proteins colored distinctly and labeled. (C–E) Interface and solvent views of the eukaryotic 40S subunit (Rabl et al. 2011), with (C) eukaryotic-specific r-proteins (red) and rRNA (pink) shown relative to conserved rRNA (gray) and r-proteins (blue), and with (D,E) 18S rRNA colored gray and r-proteins colored distinctly and labeled.
Figure 2.
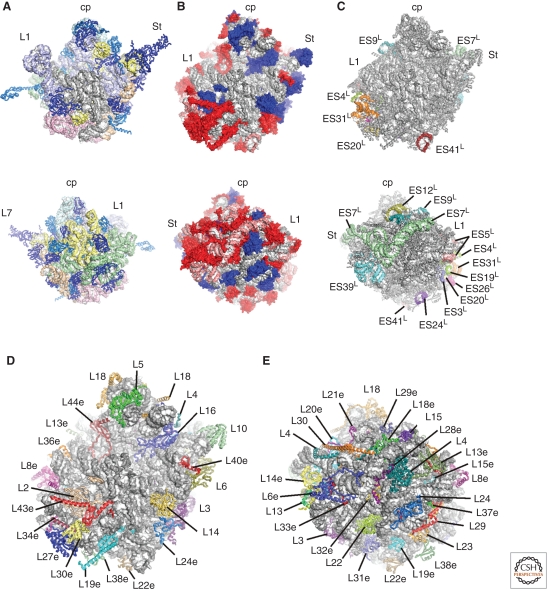
The bacterial and eukaryotic large ribosomal subunit. (A) Interface (upper) and solvent (lower) views of the bacterial 50S subunit (Jenner et al. 2010b), with 23S rRNA domains and bacterial-specific (light blue) and conserved (blue) r-proteins colored distinctly: cp, central protuberance; L1, L1 stalk; and St, L7/L12 stalk (or P-stalk in archeaa/eukaryotes). (B–E) Interface and solvent views of the eukaryotic 60S subunit (Klinge et al. 2011), with (B) eukaryotic-specific r-proteins (red) and rRNA (pink) shown relative to conserved rRNA (gray) and r-proteins (blue), (C) eukaryotic-specific expansion segments (ES) colored distinctly, and (D,E) 28S rRNA colored gray and r-proteins colored distinctly and labeled.
In contrast to their bacterial counterparts, eukaryotic ribosomes are much larger and more complex, containing additional rRNA in the form of so-called expansion segments (ES) as well as many additional r-proteins and r-protein extensions (Figs. 1C–E and 2C–E). Compared with the ∼4500 nucleotides of rRNA and 54 r-proteins of the bacterial 70S ribosome, eukaryotic 80S ribosomes contain >5500 nucleotides of rRNA (SSU, 18S rRNA; LSU, 5S, 5.8S, and 25S rRNA) and 80 (79 in yeast) r-proteins. The first structural models for the eukaryotic (yeast) ribosome were built using 15-Å cryo–electon microscopy (cryo-EM) maps fitted with structures of the bacterial SSU (Wimberly et al. 2000) and archaeal LSU (Ban et al. 2000), thus identifying the location of a total of 46 eukaryotic r-proteins with bacterial and/or archaeal homologs as well as many ES (Spahn et al. 2001a). Subsequent cryo-EM reconstructions led to the localization of additional eukaryotic r-proteins, RACK1 (Sengupta et al. 2004) and S19e (Taylor et al. 2009) on the SSU and L30e (Halic et al. 2005) on the LSU, as well as more complete models of the rRNA derived from cryo-EM maps of canine and fungal 80S ribosomes at ∼9 Å (Chandramouli et al. 2008; Taylor et al. 2009). Recent cryo-EM reconstructions of plant and yeast 80S translating ribosomes at 5.5–6.1 Å enabled the correct placement of an additional six and 10 r-proteins on the SSU and LSU, respectively, as well as the tracing of many eukaryotic-specific r-protein extensions (Armache et al. 2010a,b). The full assignment of the r-proteins in the yeast and fungal 80S ribosomes, however, only became possible with the improved resolution (3.0–3.9 Å) resulting from the crystal structures of the SSU and LSU from Tetrahymena thermophila (Klinge et al. 2011; Rabl et al. 2011) and the Saccharomyces cerevisiae 80S ribosome (Figs. 1D,E and 2D,E) (Ben-Shem et al. 2011).
RIBOSOMAL RNA OF THE EUKARYOTIC RIBOSOME
In terms of rRNA, the major differences between bacterial and eukaryotic ribosomes is the presence in eukaryotes of five expansion segments (ES3S, ES6S, ES7S, ES9S, and ES12S, following the nomenclature of Gerbi [1996]) and five variable regions (VRs) (h6, h16, h17, h33, and h41) on the SSU, as well as 16 expansion segments (ES3L, ES4L, ES5L, ES7L, ES9L, ES10L, ES12L, ES15L, ES19L, ES20L, ES24L, ES26L, ES27L, ES31L, ES39L, and ES41L) and two VRs (H16–18 and H38) on the LSU (Figs. 1C and 2C) (Cannone et al. 2002). On the LSU most ES are located on the back and sides of the particle, leaving the subunit interface and exit tunnel regions essentially unaffected (Taylor et al. 2009; Armache et al. 2010a; Ben-Shem et al. 2010; Klinge et al. 2011). The largest concentration of additional rRNA (∼40%) on the yeast LSU is positioned behind the P stalk and is formed by ES7L (∼200 nucleotides) and ES39L (∼150 nucleotides), with a second patch (∼150 nucleotides) located behind the L1 stalk formed by the clustering of ES19L, ES20L, ES26L, and ES31L (Figs. 2C and 3A). In addition, the highly flexible ES27L (150 nucleotides), which was not observed in the crystal structures (Ben-Shem et al. 2011; Klinge et al. 2011), adopts two distinct conformations in cryo-EM reconstructions of yeast ribosomes (Beckmann et al. 2001; Armache et al. 2010a). On the yeast SSU the majority (∼75%) of the additional rRNA comprises ES3S (∼100 nucleotides) and ES6S (∼200 nucleotides), which interact and cluster together to form the left foot of the particle (Figs. 1C and 3B) (Armache et al. 2010a; Ben-Shem et al. 2011; Rabl et al. 2011).
Figure 3.
Structural and functional aspects of the eukaryotic ribosome. Interweaving of rRNA and r-proteins on the (A) LSU near ES7L and ES39L (Klinge et al. 2011), and (B) SSU near ES3 and ES6 (Rabl et al. 2011). Extension of r-proteins at the tRNA-binding sites on the (C) SSU (Armache et al. 2010b; Rabl et al. 2011), LSU of the (D) bacterial (Jenner et al. 2010b), and (E) eukaryotic (Armache et al. 2010b) peptidyltransferase centers. R-proteins located at the mRNA (F) exit, and (G) entry sites (Klinge et al. 2011).
Comparison of rRNA sequences of diverse organisms, ranging from bacteria to mammals, reveals that the major differences in ES are restricted to four sites on the LSU, namely, ES7L, ES15L, ES27L, and ES39L. These ES are significantly longer (∼850, ∼180, ∼700, and ∼220 nucleotides) in human 80S ribosomes than in yeast (∼200, ∼20, ∼150, and ∼120 nucleotides, respectively) (Cannone et al. 2002). Moreover, cryo-EM reconstructions of mammalian ribosomes (Dube et al. 1998; Morgan et al. 2000; Spahn et al. 2004b; Chandramouli et al. 2008; Budkevich et al. 2011) reveal little to no density for the longer ES in mammalian ribosomes, indicating that they are highly mobile elements. In Tetrahymena, deletion of ES27L is lethal (Sweeney et al. 1994), suggesting a functionally important role for this ES. Despite the high variability in length of ES27L, ranging from ∼150 nucleotides in yeast to ∼700 nucleotides in mammals (Cannone et al. 2002), deletion of ES27L can be complemented with a corresponding ES27L from other species (Sweeney et al. 1994). ES27L has been suggested to play a role in coordinating the access of nonribosomal proteins to the tunnel exit (Beckmann et al. 2001), but this remains to be shown. The role of other ES remains unclear. Their presence in eukaryotic ribosomes may reflect the increased complexity of translation regulation in eukaryotic cells, as evident for assembly, translation initiation, and development, as well as the phenomenon of localized translation (Sonenberg and Hinnebusch 2009; Freed et al. 2010; Wang et al. 2010).
RIBOSOMAL PROTEINS OF THE EUKARYOTIC RIBOSOME
The yeast 80S ribosome contains 79 r-proteins (SSU, 33; LSU, 46), 35 of which (SSU, 15; LSU, 20) have bacterial/archaeal homologs, whereas 32 (SSU, 12; LSU, 20) have only archaeal homologs (Lecompte et al. 2002). Thus, 12 (SSU, 6; LSU, 6) r-proteins of the yeast 80S are specific for eukaryotes. Cytoplasmic 80S ribosomes of Tetrahymena and higher eukaryotes, such as humans, contain an additional LSU r-protein, L28e, and thus have 13 eukaryotic-specific r-proteins and 80 (SSU, 33; LSU, 47) in total. Together with the ES, the additional r-proteins/r-protein extensions form an intricate layer of additional RNA–protein mass that locates predominantly to the solvent surfaces of the ribosome (Figs. 1C and 2B). More than half of the conserved r-proteins contain extensions, which in some cases, such as S5, L4, L7, and L30, establish long-distance interactions far (50–100 Å) from the globular core of the protein. Interaction of eukaryotic-specific extensions with conserved core proteins using interprotein shared β-sheets has been noted, for example, between L14e and L6 (Ben-Shem et al. 2011) as well as L21e and L30 (Klinge et al. 2011).
The eukaryotic LSU contains ∼1 MDa of additional protein: 200 kDa of eukaryotic-specific domains or extensions and 800 kDa of r-proteins that are absent in bacteria. Most of this additional protein mass is located in a ring around the back and sides of the LSU, where it interacts with ES (Fig. 2B). Two large concentrations of additional RNA–protein mass exemplify the intertwined and coevolving nature of the ribosome (Yokoyama and Suzuki 2008). One cluster on the LSU comprises ES7L, ES39L, five eukaryotic r-proteins (L6e, L14e, L28e, L32e, and L33e), as well as eukaryotic-specific extensions of conserved r-proteins (L4, L13, and L30) (Fig. 3A). In this cluster yeast ES7L comprises three helices, ES7La–c, whereas wheat germ (plant) ES7L has five helices, ES7La–e, including a three-way junction extending from ES7Lc (Armache et al. 2010b). Curiously, the extension of L6e is longer in wheat germ as compared with yeast and appears to wrap around ES7L and insert through the three-way junction of ES7La–c (Armache et al. 2010b). ES7La is stabilized by L28e in wheat germ and Tetrahymena, whereas this helix is more flexible in baker’s yeast lacking L28e. Stabilization of ES by eukaryotic r-proteins is also evident for ES27L, with the two different yeast conformations being stabilized by interaction with either L38e or L27e (Armache et al. 2010b). The second major ES cluster comprises ES19L, ES20L, ES26L, and ES31L, which are intimately associated with eukaryotic-specific r-proteins L27e, L30e, L34e, L43e, and the carboxy-terminal extension of L8e (Fig. 2C–E) (Ben-Shem et al. 2011). A single-stranded loop region of ES31L provides an interaction platform for many of these r-proteins, notably the carboxy-terminal helix of L34e. Similarly, ES39L also has many single-stranded loop regions that provide interaction sites for r-proteins, such as L20e and L14e.
The protein-to-RNA ratio of bacterial SSU is ∼1:2, whereas the dramatic increase in r-protein mass for the eukaryotic SSU results in an almost 1:1 ratio. The SSU structures reveal that most of the additional eukaryotic-specific r-proteins and extensions cover the back of the SSU particle, forming a web of interactions with each other as well as with conserved r-proteins and rRNA (Fig. 1C–E) (Ben-Shem et al. 2011; Rabl et al. 2011). The beak of the eukaryotic SSU has acquired three r-proteins, S10e, S12e, and S31e, which appear to compensate for the reduced h33 compared with the bacterial SSU rRNA (Rabl et al. 2011). R-proteins are also seen to interact with the expansion segments ES3S and ES6S, via r-proteins S4e, S6e, S7e, and S8e (Fig. 3B). S6e has a long carboxy-terminal helix that stretches from the left to right foot, and that is phosphorylated in most eukaryotes (Meyuhas 2008). Based on the peripheral position of S6e, any regulation of translation via S6e phosphorylation is likely to be via indirect recruitment of specific regulatory factors (Rabl et al. 2011). The mRNA exit site on the eukaryotic SSU also differs from the bacterial one because of the presence of S26e and S28e surrounding the 3′ end of the 18S rRNA (Fig. 3F) (Armache et al. 2010a; Rabl et al. 2011). S26e overlaps the binding position of the E. coli r-protein S21p (Schuwirth et al. 2005), whereas S28e has a similar fold to the bacterial RNA-binding domain of r-protein S1p (Rabl et al. 2011). Such differences may reflect the distinct elements found in the 5′ untranslated regions of eukaryotic mRNAs, as well as the divergence in the translation initiation phase from bacteria (Sonenberg and Hinnebusch 2009). Indeed, eIF3, which is absent in bacteria, interacts with this general region of the SSU (Bommer et al. 1991; Srivastava et al. 1992; Siridechadilok et al. 2005), as do internal ribosome entry site (IRES) elements present in the 5′ untranslated region of viral mRNAs (Spahn et al. 2001b; Schuler et al. 2006; Muhs et al. 2011). S30e replaces part of S4 at the mRNA entry site of the eukaryotic SSU and has conserved lysine residues that extend into the mRNA channel (Fig. 3G), suggesting that S30e, together with S3, plays a role in unwinding mRNA secondary structure (Rabl et al. 2011). S3 has a long carboxy-terminal extension that spans across S17e and interacts with RACK1 (Fig. 3G) (Rabl et al. 2011). RACK1 is a scaffold protein that binds to several signaling proteins, therefore connecting signaling transduction pathways with translation (Nilsson et al. 2004). Thus, in addition to stabilization of rRNA ES architecture of the ribosome, eukaryotic-specific r-proteins and extensions appear to be important for binding of eukaryotic-specific regulatory factors, particularly factors that interact with the SSU to regulate translation initiation of specific mRNAs.
THE tRNA-BINDING SITES ON THE EUKARYOTIC RIBOSOME
The binding sites for the aminoacyl-transfer RNA (tRNA) (A site), peptidyl-tRNA (P site), and deacylated tRNA (exit or E site) on the bacterial ribosome are composed predominantly of rRNA (Yusupov et al. 2001; Selmer et al. 2006). This rRNA is conserved in archaeal and eukaryotic ribosomes, suggesting that the basic mechanism by which the ribosome distinguishes the cognate tRNA from the near- or noncognate tRNAs at the A site during decoding (Ogle and Ramakrishnan 2005; Schmeing et al. 2011) is also likely to be conserved. Nevertheless, many r-proteins encroach on the tRNA-binding sites and appear to play important roles in decoding, accommodation, and stabilization of tRNAs (Fig. 3C) (Yusupov et al. 2001; Selmer et al. 2006; Jenner et al. 2010b). These r-proteins may be responsible for the slightly different positioning of tRNAs on the eukaryotic ribosome compared with the bacterial ribosome (Budkevich et al. 2011). On the SSU a conserved loop of S12 participates in monitoring of the second and third positions of the mRNA–tRNA codon–anticodon duplex (Ogle and Ramakrishnan 2005). Additionally, the carboxy-terminal extensions of r-proteins S19 and S9/S13 stretch from globular domains located on the head of the SSU to interact with anticodon stem-loop (ASL) regions of A- and P-tRNA, respectively, whereas S7, and to a lesser extent S11, interacts with the ASL of E-tRNA (Fig. 3C) (Yusupov et al. 2001; Selmer et al. 2006; Jenner et al. 2010b). Although these tRNA interactions are likely to be maintained in eukaryotic 80S ribosomes, additional interactions are probable on the SSU because of the presence of extensions of four eukaryotic r-proteins that approach the tRNA-binding sites, namely, the amino-terminal extensions of S30e and S31e that reach into the A site; S25e, which is positioned between the P and E sites; and S1e at the E site (Fig. 3C) (Armache et al. 2010b; Ben-Shem et al. 2011; Rabl et al. 2011). S31e is expressed with an amino-terminal ubiquitin fusion, suggesting that the lethality from lack of cleavage (Lacombe et al. 2009) arises because of the inability of tRNA and/or initiation factors to bind to the SSU (Rabl et al. 2011).
Additional stabilization of tRNA binding is observed via interaction between LSU r-proteins with the elbow regions of tRNAs, namely, the A- and P-tRNA, through contact with conserved r-proteins L16 and L5, respectively, as well as the E-tRNA with the L1 stalk (Yusupov et al. 2001; Selmer et al. 2006; Jenner et al. 2010b). The carboxyl terminus of the bacterial-specific r-protein L25p also interacts with the elbow region of A-tRNA (Jenner et al. 2010b). This r-protein is absent in archaeal and eukaryotic ribosomes. At the peptidyltransferase center (PTC) of the LSU, the CCA ends of the A- and P-tRNAs are stabilized through interaction with the conserved A- and P-loops of the 23S rRNA, thus positioning the α-amino group of the A-tRNA for nucleophilic attack on the carbonyl carbon of the peptidyl-tRNA (Leung et al. 2011). The high sequence and structural conservation of the PTC and of the tRNA substrates suggests that the insights into the mechanism of peptide bond formation gained from studying archaeal and bacterial ribosomes (Simonovic and Steitz 2009) are transferable to eukaryotic ribosomes. Nevertheless, the varying specificity for binding of antibiotics to the PTC of bacterial versus eukaryotic LSU indicates that subtle differences do in fact exist (Wilson 2011). In addition to differences in the conformation of rRNA nucleotides, one of the major differences between the bacterial and eukaryotic PTC is related to r-proteins. Eukaryotic L16 contains a highly conserved loop that reaches into the PTC and contacts the CCA end of the P-tRNA (Fig. 3D) (Armache et al. 2010b; Bhushan et al. 2010b). This loop is absent in bacteria, and instead the space is occupied by the amino-terminal extension of bacterial-specific r-protein L27p (Fig. 3E) (Voorhees et al. 2009). The binding site of the CCA end of the E-tRNA on the eukaryotic LSU resembles the archaeal, rather than the bacterial, context. Whereas bacterial-specific r-protein L28p contributes to the E site of the bacterial LSU (Selmer et al. 2006), the archaeal and eukaryotic r-protein L44e contains an internal loop region (Fig. 2D) through which the CCA end of the E-tRNA inserts (Schmeing et al. 2003). Moreover, the carboxyl terminus of L44e is longer in eukaryotes, such as yeast, than in archaea, providing the potential for additional interactions with the P- and/or E-tRNA. Nevertheless, the E site restricts binding of only deacylated tRNAs via a direct interaction between the 2′OH of A76 and the base of C2394 (E. coli 23S rRNA numbering) (Schmeing et al. 2003; Selmer et al. 2006). The base equivalent to C2394 is conserved across all kingdoms (Cannone et al. 2002), suggesting a universal mechanism of deacylated-tRNA discrimination at the E site on the LSU.
BINDING SITES OF INITIATION FACTORS ON THE RIBOSOME
In bacteria, translation initiation is driven in large part by base pairing between the mRNA just 5′ of the start codon and the 3′ end of 16S rRNA—the Shine–Dalgarno interaction—which defines the ribosome binding site (Geissmann et al. 2009; Simonetti et al. 2009). Three proteins contribute to bacterial initiation, termed initiation factors 1, 2, and 3 (IF1, IF2, and IF3), and help to load initiator tRNA into the small-subunit P site at the correct start codon (Simonetti et al. 2009). In eukaryotes, translation initiation generally requires a scanning mechanism that starts at the 5′-7-methyl-guanosine (5′-m7G) cap and proceeds to the appropriate AUG start codon, often the first AUG codon encountered by the initiation machinery (Jackson et al. 2010). To accomplish scanning, a whole suite of eukaryotic translation initiation factors (eIFs) is involved, with names from eIF1 through eIF6, as described in more detail by Lorsch et al. (2012). Only two of the three bacterial proteins, IF1 and IF2, are conserved in eukaryotes, as counterparts of eIF1A and eIF5B, respectively (Benelli and Londei 2009). However, eIF1A and eIF5B have augmented or divergent roles to play in eukaryotic translation initiation (Jackson et al. 2010). IF3 is not conserved in eukaryotes, but seems to have a functional counterpart in eIF1 (Lomakin et al. 2003, 2006). Similar to what is observed for r-proteins in eukaryotes, eIF1 and eIF1A have extensions or “tails” that are important for their function (Olsen et al. 2003; Fekete et al. 2005, 2007; Cheung et al. 2007; Reibarkh et al. 2008; Saini et al. 2010). Most of the interactions between the 40S subunit and eukaryotic translation initiation factors are only known from genetic, biochemical, and low-resolution cryo-EM reconstructions and models of partial initiation complexes (Lomakin et al. 2003; Valasek et al. 2003; Fraser et al. 2004, 2007; Unbehaun et al. 2004; Siridechadilok et al. 2005; Passmore et al. 2007; Szamecz et al. 2008; Shin et al. 2009; Yu et al. 2009; Chiu et al. 2010; Kouba et al. 2011). With the determination of the recent X-ray crystal structures of the T. thermophila 40S and 60S subunits, in complexes with eIF1 and eIF6, respectively (Klinge et al. 2011; Rabl et al. 2011), our understanding of the structural basis for translation initiation in eukaryotes has increased greatly, but still lags behind our structural knowledge of bacterial translation initiation (Simonetti et al. 2009).
Initiation factor eIF1 promotes binding of initiator tRNA, in the form of a ternary complex of eIF2–GTP–Met–tRNAiMet, to preinitiation complexes of the SSU. It also serves to prevent initiation at non–start codons, likely by promoting an “open” state of the SSU (Jackson et al. 2010; Hinnebusch 2011). Consistent with this model, a cryo-EM reconstruction of the yeast 40S subunit in complex with eIF1 and eIF1A revealed that these two proteins induce an opening of the mRNA- and tRNA-binding groove in the 40S subunit that may contribute to scanning and correct start codon selection (Passmore et al. 2007). Release of eIF1 when the start codon is recognized is proposed to result in the closing of this groove, thereby locking the mRNA and initiator tRNA in place (Nanda et al. 2009). In the structure of the 40S subunit, eIF1 is bound adjacent to the SSU P site, in such a way that it would prevent full docking of the initiator tRNA ASL in the P-site cleft (Fig. 4A). Notably, the position of eIF1 is more compatible with tRNA docked in a hybrid configuration seen in the bacterial ribosome, in which the tRNA is bound in the SSU P site and LSU E site (P/E-tRNA) (Fig. 4B) (Dunkle et al. 2011). As part of start codon selection, dissociation of eIF1 may allow initiator tRNA to adopt an intermediate P/I orientation, observed in bacterial initiation complexes with IF2 (Allen et al. 2005; Julian et al. 2011), or the P/P configuration, in which it could access the LSU P site upon subunit association (Jackson et al. 2010).
Figure 4.
Positioning of eIF1 near the SSU P site. (A) Steric clash between eIF1 and P-site tRNA in the canonical P/P configuration. Structure of the 40S subunit–eIF1 complex superimposed with the unrotated state of the ribosome in Dunkle et al. (2011). (B) Binding of eIF1 is more compatible with tRNA in the P/E configuration. Structure of the 40S subunit–eIF1 complex superimposed with the rotated state of the ribosome in Dunkle et al. (2011). Nucleotides in 18S rRNA that would contribute to contacts with the LSU in bridge B2a are colored red.
The binding site for eIF1 would also block the premature binding of the 60S subunit, because it is situated right where a critical contact (“bridge” B2a) forms between the two ribosomal subunits (Fig. 4) (Rabl et al. 2011). Part of eIF1 also extends into the mRNA-binding groove, adjacent to where the P-site codon would be situated. From biochemical and genetic experiments, the amino-terminal tail of eIF1 plays an important role in recruiting the eIF2–GTP–Met–tRNAiMet ternary complex to preinitiation complexes (Cheung et al. 2007). However, the structure of the eIF1–40S complex provides only the first structural hints into how the ternary complex is recruited and how start codons are selected. Future structures with more of the translation initiation factors, as well as with initiator tRNA, will be needed to unravel the molecular basis for start codon selection.
The role in initiation of translation initiation factor eIF6 is not as clearly defined. It has been proposed to be an antiassociation factor that prevents premature association of the two ribosomal subunits, and it also acts in late stages of pre-60S assembly (Brina et al. 2011). In the recent X-ray crystal structure of the 60S subunit (Klinge et al. 2011), and as previously observed (Gartmann et al. 2010), eIF6 binds to the GTPase center, the region of the LSU where GTPases such as those responsible for mRNA decoding (eukaryotic elongation factor 1 [eEF1]) and mRNA and tRNA translocation (eEF2) interact with the ribosome. The location of eIF6 would sterically prevent SSU interactions with the LSU, helping to explain its antiassociative activity. Its position near the GTPase center is also highly suggestive of how it might be released in a GTPase-dependent manner during LSU assembly (Senger et al. 2001; Menne et al. 2007; Finch et al. 2011), and also how it might be used to regulate the availability of 60S subunits as a means to control cell growth and proliferation (Gandin et al. 2008).
THE RIBOSOMAL TUNNEL OF EUKARYOTIC RIBOSOMES
As the nascent polypeptide chain (NC) is being synthesized, it passes through a tunnel within the LSU and emerges at the solvent side, where protein folding occurs. Cryo-EM reconstructions and X-ray crystallography structures of bacterial, archaeal, and eukaryotic cytoplasmic ribosomes have revealed the universality of the dimensions of the ribosomal tunnel (Frank et al. 1995; Beckmann et al. 1997; Ban et al. 2000; Ben-Shem et al. 2011; Klinge et al. 2011). The ribosomal tunnel is ∼80 Å long, 10–20 Å wide, and predominantly composed of core rRNA (Nissen et al. 2000), consistent with an overall electronegative potential (Lu et al. 2007). The extensions of the r-proteins L4 and L22 contribute to formation of the tunnel wall, forming a so-called constriction where the tunnel narrows (Nissen et al. 2000). Near the tunnel exit the ribosomal protein L39e is present in eukaryotic and archaeal ribosomes (Nissen et al. 2000), whereas a bacterial-specific extension of L23 occupies an overlapping position in bacteria (Harms et al. 2001).
For many years the ribosomal tunnel was thought of only as a passive conduit for the NC. However, growing evidence indicates that the tunnel plays a more active role in regulating the rate of translation, in providing an environment for early protein folding events, and in recruiting translation factors to the tunnel exit site (Wilson and Beckmann 2011). At the simplest level, long stretches of positively charged residues, such as arginine or lysine, in an NC can reduce or halt translation, most likely through interaction with the negatively charged rRNA in the tunnel (Lu and Deutsch 2008). More specific regulatory systems also exist in bacteria and eukaryotes, in which stalling during translation of upstream open reading frames (uORFs of the cytomegalovirus [CMV] gp48 and arginine attenuator peptide [AAP] CPA1 genes) or leader peptides (TnaC, SecM) leads to modulation of expression of downstream genes (Tenson and Ehrenberg 2002). Interestingly, the translational stalling events depend critically on the sequence of the NC and the interaction of the NC with the ribosomal tunnel. Cryo-EM reconstructions of bacterial TnaC- and SecM-stalled 70S ribosomes (Seidelt et al. 2009; Bhushan et al. 2011) and eukaryotic CMV- and AAP-stalled 80S ribosomes (Bhushan et al. 2010b) reveal the distinct pathways and conformations of the NCs in the tunnel as well as the interactions between the NCs and tunnel wall components. Compared with bacteria, eukaryotic r-protein L4 has an insertion that establishes additional contacts with the CMV- and AAP-NCs (Bhushan et al. 2010b), whereas the bacterial stalling sequences interact predominantly with L22 (Seidelt et al. 2009; Bhushan et al. 2011). The dimensions of the ribosomal tunnel preclude the folding of domains as large as an IgG domain (∼17 kDa) (Voss et al. 2006), whereas α-helix formation has been demonstrated biochemically (Deutsch 2003; Woolhead et al. 2004) and visualized structurally within distinct regions of the tunnel (Bhushan et al. 2010a). Folding of NCs within the tunnel may have implications for not only protein folding, but also downstream events, such as recruitment of chaperones or targeting machinery (Bornemann et al. 2008; Berndt et al. 2009; Pool 2009).
INTERACTIONS BETWEEN THE RIBOSOMAL SUBUNITS
During translation the ribosome undergoes global conformational rearrangements that are required for mRNA decoding, mRNA and tRNA translocation, termination, and ribosome recycling. These changes involve intersubunit rotation, as well as swiveling of the head domain of the SSU (Fig. 5A). The interactions between the ribosomal subunits, or “bridges,” change with each of these rearrangements, and are therefore dynamic in composition. The intersubunit bridges were originally mapped in bacteria by modeling high-resolution SSU and LSU structures into cryo-EM reconstructions and low-resolution X-ray crystal structures (Gabashvili et al. 2000; Yusupov et al. 2001; Valle et al. 2003), and in more recent high-resolution structures of the intact bacterial ribosome (Schuwirth et al. 2005; Dunkle et al. 2011). The bridges in eukaryotic ribosomes have been mapped using similar approaches. The high-resolution structures of the yeast 80S ribosome now provide an atomic-resolution view of the bridges for rotated states of the ribosome (Ben-Shem et al. 2011), and cryo-EM reconstructions of translating ribosomes at ∼5- to 6-Å resolution reveal the intersubunit bridges in the unrotated state of the ribosome (Armache et al. 2010a,b).
Figure 5.
Intersubunit rotation required for translation. (A) Key conformational rearrangements in the ribosome. Rotation of the SSU body, head domain, and opening of the mRNA- and tRNA-binding groove during mRNA and tRNA translocation (asterisk) are indicated by arrows. Closing of the SSU body toward the LSU during mRNA decoding is also indicated by an arrow. Dynamic regions of the LSU (L1 arm, P proteins, and GTPase center) are labeled. (B) Bridges eB12 and eB13 in the yeast ribosome at the periphery of the subunits. LSU proteins contributing to the bridges are marked. The view is indicated to the left. (C) Bridge eB14 in the yeast ribosome, near the pivot point of intersubunit rotation. LSU protein L41e and 18S rRNA helices in the SSU contributing to the bridge (gold) are indicated.
Whereas the bacterial ribosome preferentially adopts the unrotated state of the two subunits, the eukaryotic ribosome seems to adopt rotated states more readily (Spahn et al. 2004a; Chandramouli et al. 2008; Ben-Shem et al. 2011; Budkevich et al. 2011). A possible reason for this difference in behavior is the fact that the interaction surface between the two ribosomal subunits has nearly doubled in eukaryotes compared with bacteria, primarily because of the appearance of numerous additional bridges at the periphery of the subunit interface. These new bridges are composed mainly of protein–protein and protein–rRNA contacts, some of the more notable involving long extensions from the LSU to contact the body and platform of the SSU, bridges eB12 and eB13 (Fig. 5B) (Ben-Shem et al. 2011). One striking exception to this general trend is one new bridge right at the center of the subunit interface, near the pivot point of intersubunit rotation (Ben-Shem et al. 2011). This bridge, termed eB14, is composed of a single short α-helical peptide, designated L41e, that is nearly entirely buried in a pocket composed of 18S rRNA in the SSU. Remarkably, this pocket is highly conserved in eukaryotes and in bacteria (Fig. 5C) (Schluenzen et al. 2000; Wimberly et al. 2000; Cannone et al. 2002; Ben-Shem et al. 2011), but no corresponding peptide in bacteria has been identified. The importance of this peptide in eukaryotic ribosome function remains unknown.
MECHANISMS OF mRNA DECODING, TRANSLOCATION, TERMINATION, AND RIBOSOME RECYCLING
Remarkably for processes that are functionally conserved in all domains of life, the mechanisms used by eukaryotes for mRNA decoding, mRNA and tRNA translocation, translation termination, and ribosome recycling differ in significant ways from those in bacteria (Triana-Alonso et al. 1995; Andersen et al. 2000; Gaucher et al. 2002; Jorgensen et al. 2003; Alkalaeva et al. 2006; Khoshnevis et al. 2010; Pisarev et al. 2010). The recent breakthroughs in the structural biology of the eukaryotic ribosome provide a structural framework to unravel these differences. The large number of approximately nanometer or subnanometer cryo-EM reconstructions of eukaryotic ribosomes in different functional states (Halic et al. 2004, 2005, 2006a,b; Spahn et al. 2004a; Gao et al. 2005; Andersen et al. 2006; Schuler et al. 2006; Taylor et al. 2007, 2009; Chandramouli et al. 2008; Sengupta et al. 2008; Becker et al. 2009, 2011, 2012; Armache et al. 2010a,b; Bhushan et al. 2010a,b; Gartmann et al. 2010; Budkevich et al. 2011) now can be interpreted using high-resolution structures of the ribosome (Jarasch et al. 2011) in combination with X-ray crystal structures of the individual factors (Noble and Song 2008; Chen et al. 2010).
Although there are many differences in the translation elongation and termination factors between bacteria and eukaryotes, these factors seem to exploit common features of the ribosome conserved in all domains of life. One notable example is the mechanism for GTPase activation in mRNA decoding, in which the sarcin–ricin loop was shown to reorganize the catalytic center in bacterial EF-Tu (eukaryotic ortholog of eEF1A) during mRNA decoding (Voorhees et al. 2010). A second example is the convergent evolution of a motif in release factors that is responsible for stimulating the hydrolysis of completed proteins from peptidyl-tRNA during termination. Bacterial and eukaryotic release factors (RF1 and RF2 in bacteria, eRF1 in eukaryotes) are composed of entirely different protein topologies (Song et al. 2000; Vestergaard et al. 2001; Shin et al. 2004). Furthermore, eukaryotic RF1 requires the GTPase eRF3 and ATPase ABCE1 to stimulate termination and ribosome recycling (Khoshnevis et al. 2010; Pisarev et al. 2010; Becker et al. 2012), whereas bacterial termination and ribosome recycling use different factors (Zavialov et al. 2001; Savelsbergh et al. 2009). Strikingly, given these differences, the key residues in RFs that insert into the PTC to promote peptidyl-tRNA hydrolysis, a GGQ motif, are universally conserved. A second example occurs with the GTPases involved in elongation. Bacteria rely on the GTPases EF-Tu and EF-G, whereas eukaryotes use the GTPases eEF1A and eEF2. Eukaryotic eEF2 cannot function on the bacterial ribosome, unless the bacterial L10 and L12 proteins in the LSU are replaced by the eukaryotic acidic proteins P0 and P1/P2 (Uchiumi et al. 1999, 2002). Notably, this protein-swapping experiment also illustrates how the underlying rRNA functions are probably universal.
CONCLUSIONS
The last few years have witnessed a surge of new structures of the bacterial and eukaryotic ribosome in different steps of the translation cycle. The recent X-ray crystal structures of the T. thermophila 40S and 60S ribosomal subunits and yeast 80S ribosome now provide an unprecedented framework for interpreting the many cryo-EM reconstructions of the eukaryotic ribosome and biochemical insights into the eukaryotic translation mechanism. In a few years, it is not hard to imagine that many of the steps in eukaryotic translation will be understood in atomic detail based on new cryo-EM and X-ray crystal structures of the eukaryotic ribosome.
ACKNOWLEDGMENTS
This work is supported by the EMBO Young Investigator program (to D.N.W.) and by the National Institutes of Health grant R56-AI095687 (to J.H.D.C).
Footnotes
Editors: John W.B. Hershey, Nahum Sonenberg, and Michael B. Mathews
Additional Perspectives on Protein Synthesis and Translational Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136 [DOI] [PubMed] [Google Scholar]
- Allen GS, Zavialov A, Gursky R, Ehrenberg M, Frank J 2005. The cryo-EM structure of a translation initiation complex from Escherichia coli. Cell 121: 703–712 [DOI] [PubMed] [Google Scholar]
- Andersen GR, Pedersen L, Valente L, Chatterjee I, Kinzy TG, Kjeldgaard M, Nyborg J 2000. Structural basis for nucleotide exchange and competition with tRNA in the yeast elongation factor complex eEF1A:eEF1Bα. Mol Cell 6: 1261–1266 [DOI] [PubMed] [Google Scholar]
- Andersen CB, Becker T, Blau M, Anand M, Halic M, Balar B, Mielke T, Boesen T, Pedersen JS, Spahn CM, et al. 2006. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature 443: 663–668 [DOI] [PubMed] [Google Scholar]
- Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. 2010a. Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-Å resolution. Proc Natl Acad Sci 107: 19748–19753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. 2010b. Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map of the 80S eukaryotic ribosome. Proc Natl Acad Sci 107: 19754–19759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920 [DOI] [PubMed] [Google Scholar]
- Becker T, Bhushan S, Jarasch A, Armache JP, Funes S, Jossinet F, Gumbart J, Mielke T, Berninghausen O, Schulten K, et al. 2009. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science 326: 1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R 2011. Structure of the no-go mRNA decay complex Dom34–Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol 18: 715–720 [DOI] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, et al. 2012. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature 482: 501–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R, Bubeck D, Grassucci R, Penczek P, Verschoor A, Blobel G, Frank J 1997. Alignment of conduits for the nascent polypeptide chain in the ribosome—Sec61 complex. Science 278: 2123–2126 [DOI] [PubMed] [Google Scholar]
- Beckmann R, Spahn CM, Eswar N, Helmers J, Penczek PA, Sali A, Frank J, Blobel G 2001. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107: 361–372 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Jenner L, Yusupova G, Yusupov M 2010. Crystal structure of the eukaryotic ribosome. Science 330: 1203–1209 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529 [DOI] [PubMed] [Google Scholar]
- Benelli D, Londei P 2009. Begin at the beginning: Evolution of translational initiation. Res Microbiol 160: 493–501 [DOI] [PubMed] [Google Scholar]
- Berndt U, Oellerer S, Zhang Y, Johnson AE, Rospert S 2009. A signal-anchor sequence stimulates signal recognition particle binding to ribosomes from inside the exit tunnel. Proc Natl Acad Sci 106: 1398–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Gartmann M, Halic M, Armache JP, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R 2010a. α-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol 17: 313–317 [DOI] [PubMed] [Google Scholar]
- Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, Berninghausen O, Sattler M, Wilson DN, Beckmann R 2010b. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol Cell 40: 138–146 [DOI] [PubMed] [Google Scholar]
- Bhushan S, Hoffmann T, Seidelt B, Frauenfeld J, Mielke T, Berninghausen O, Wilson DN, Beckmann R 2011. SecM-stalled ribosomes adopt an altered geometry at the peptidyl transferase center. PLoS Biol 9: e1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer UA, Lutsch G, Stahl J, Bielka H 1991. Eukaryotic initiation factors eIF-2 and eIF-3: Interactions, structure and localization in ribosomal initiation complexes. Biochimie 73: 1007–1019 [DOI] [PubMed] [Google Scholar]
- Bornemann T, Jockel J, Rodnina MV, Wintermeyer W 2008. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat Struct Mol Biol 15: 494–499 [DOI] [PubMed] [Google Scholar]
- Brina D, Grosso S, Miluzio A, Biffo S 2011. Translational control by 80S formation and 60S availability: The central role of eIF6, a rate limiting factor in cell cycle progression and tumorigenesis. Cell Cycle 10: 3441–3446 [DOI] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM Jr, Carter AP, Wimberly BT, Ramakrishnan V 2002. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: Structure of the proteins and their interactions with 16 S RNA. J Mol Biol 316: 725–768 [DOI] [PubMed] [Google Scholar]
- Budkevich T, Giesebrecht J, Altman RB, Munro JB, Mielke T, Nierhaus KH, Blanchard SC, Spahn CM 2011. Structure and dynamics of the mammalian ribosomal pretranslocation complex. Mol Cell 44: 214–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. 2002. The Comparative RNA Web (CRW) Site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli P, Topf M, Menetret JF, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW 2008. Structure of the mammalian 80S ribosome at 8.7 Å resolution. Structure 16: 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Muhlrad D, Hauryliuk V, Cheng Z, Lim MK, Shyp V, Parker R, Song H 2010. Structure of the Dom34–Hbs1 complex and implications for no-go decay. Nat Struct Mol Biol 17: 1233–1240 [DOI] [PubMed] [Google Scholar]
- Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG 2007. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev 21: 1217–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WL, Wagner S, Herrmannova A, Burela L, Zhang F, Saini AK, Valasek L, Hinnebusch AG 2010. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol Cell Biol 30: 4415–4434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch C 2003. The birth of a channel. Neuron 40: 265–276 [DOI] [PubMed] [Google Scholar]
- Dube P, Wieske M, Stark H, Schatz M, Stahl J, Zemlin F, Lutsch G, van Heel M 1998. The 80S rat liver ribosome at 25 Å resolution by electron cryomicroscopy and angular reconstitution. Structure 6: 389–399 [DOI] [PubMed] [Google Scholar]
- Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH 2011. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332: 981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete CA, Applefield DJ, Blakely SA, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG 2005. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. EMBO J 24: 3588–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete CA, Mitchell SF, Cherkasova VA, Applefield D, Algire MA, Maag D, Saini AK, Lorsch JR, Hinnebusch AG 2007. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J 26: 1602–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch AJ, Hilcenko C, Basse N, Drynan LF, Goyenechea B, Menne TF, Gonzalez Fernandez A, Simpson P, D’Santos CS, Arends MJ, et al. 2011. Uncoupling of GTP hydrolysis from eIF6 release on the ribosome causes Shwachman–Diamond syndrome. Genes Dev 25: 917–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Zhu J, Penczek P, Li Y, Srivastava S, Verschoor A, Radermacher M, Grassucci R, Lata RK, Agrawal RK 1995. A model of protein synthesis based on cryo–electron microscopy of the E. coli ribosome. Nature 376: 441–444 [DOI] [PubMed] [Google Scholar]
- Fraser CS, Lee JY, Mayeur GL, Bushell M, Doudna JA, Hershey JW 2004. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J Biol Chem 279: 8946–8956 [DOI] [PubMed] [Google Scholar]
- Fraser CS, Berry KE, Hershey JW, Doudna JA 2007. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell 26: 811–819 [DOI] [PubMed] [Google Scholar]
- Freed EF, Bleichert F, Dutca LM, Baserga SJ 2010. When ribosomes go bad: Diseases of ribosome biogenesis. Mol Biosyst 6: 481–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabashvili IS, Agrawal RK, Spahn CM, Grassucci RA, Svergun DI, Frank J, Penczek P 2000. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell 100: 537–549 [DOI] [PubMed] [Google Scholar]
- Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC, Biffo S 2008. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature 455: 684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Ayub MJ, Levin MJ, Frank J 2005. The structure of the 80S ribosome from Trypanosoma cruzi reveals unique rRNA components. Proc Natl Acad Sci 102: 10206–10211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartmann M, Blau M, Armache JP, Mielke T, Topf M, Beckmann R 2010. Mechanism of eIF6-mediated inhibition of ribosomal subunit joining. J Biol Chem 285: 14848–14851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher EA, Das UK, Miyamoto MM, Benner SA 2002. The crystal structure of eEF1A refines the functional predictions of an evolutionary analysis of rate changes among elongation factors. Mol Biol Evol 19: 569–573 [DOI] [PubMed] [Google Scholar]
- Geissmann T, Marzi S, Romby P 2009. The role of mRNA structure in translational control in bacteria. RNA Biol 6: 153–160 [DOI] [PubMed] [Google Scholar]
- Gerbi SA 1996. Expansion segments: Regions of variable size that interrupt the universal core secondary structure of ribosomal RNA. In Ribosomal RNA—Structure, evolution, processing, and function in protein synthesis (ed. Zimmermann RA, Dahlberg AE), pp. 71–87 CRC Press, Boca Raton, FL [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427: 808–814 [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Frank J, Spahn CM, Beckmann R 2005. Localization and dynamic behavior of ribosomal protein L30e. Nat Struct Mol Biol 12: 467–468 [DOI] [PubMed] [Google Scholar]
- Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R 2006a. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 444: 507–511 [DOI] [PubMed] [Google Scholar]
- Halic M, Gartmann M, Schlenker O, Mielke T, Pool MR, Sinning I, Beckmann R 2006b. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science 312: 745–747 [DOI] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG 2011. Molecular mechanism of scanning and start codon selection in eukaryotes. Microbiol Mol Biol Rev 75: 434–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarasch A, Dziuk P, Becker T, Armache JP, Hauser A, Wilson DN, Beckmann R 2011. The DARC site: A database of aligned ribosomal complexes. Nucleic Acids Res 40: D495–D500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenner L, Demeshkina N, Yusupova G, Yusupov M 2010a. Structural rearrangements of the ribosome at the tRNA proofreading step. Nat Struct Mol Biol 17: 1072–1078 [DOI] [PubMed] [Google Scholar]
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M 2010b. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol 17: 555–560 [DOI] [PubMed] [Google Scholar]
- Jorgensen R, Ortiz PA, Carr-Schmid A, Nissen P, Kinzy TG, Andersen GR 2003. Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase. Nat Struct Biol 10: 379–385 [DOI] [PubMed] [Google Scholar]
- Julian P, Milon P, Agirrezabala X, Lasso G, Gil D, Rodnina MV, Valle M 2011. The cryo-EM structure of a complete 30S translation initiation complex from Escherichia coli. PLoS Biol 9: e1001095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnevis S, Gross T, Rotte C, Baierlein C, Ficner R, Krebber H 2010. The iron-sulphur protein RNase L inhibitor functions in translation termination. EMBO Rep 11: 214–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA 2004. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol 340: 141–177 [DOI] [PubMed] [Google Scholar]
- Klinge S, Voigts-Hoffmann F, Leibundgut M, Arpagaus S, Ban N 2011. Crystal structure of the eukaryotic 60S ribosomal subunit in complex with initiation factor 6. Science 334: 941–948 [DOI] [PubMed] [Google Scholar]
- Kouba T, Rutkai E, Karaskova M, Valasek LS 2011. The eIF3c/NIP1 PCI domain interacts with RNA and RACK1/ASC1 and promotes assembly of translation preinitiation complexes. Nucleic Acids Res 10.1093/nar/gkr1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe T, Garcia-Gomez JJ, de la Cruz J, Roser D, Hurt E, Linder P, Kressler D 2009. Linear ubiquitin fusion to Rps31 and its subsequent cleavage are required for the efficient production and functional integrity of 40S ribosomal subunits. Mol Microbiol 72: 69–84 [DOI] [PubMed] [Google Scholar]
- Lecompte O, Ripp R, Thierry JC, Moras D, Poch O 2002. Comparative analysis of ribosomal proteins in complete genomes: An example of reductive evolution at the domain scale. Nucleic Acids Res 30: 5382–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung EK, Suslov N, Tuttle N, Sengupta R, Piccirilli JA 2011. The mechanism of peptidyl transfer catalysis by the ribosome. Annu Rev Biochem 80: 527–555 [DOI] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV 2003. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev 17: 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV 2006. The fidelity of translation initiation: Reciprocal activities of eIF1, IF3 and YciH. EMBO J 25: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lorsch JR 2012. Translational control: Pathway, mechanism of protein synthesis. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a011544 [DOI] [Google Scholar]
- Lu J, Deutsch C 2008. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol 384: 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Kobertz WR, Deutsch C 2007. Mapping the electrostatic potential within the ribosomal exit tunnel. J Mol Biol 371: 1378–1391 [DOI] [PubMed] [Google Scholar]
- Menne TF, Goyenechea B, Sanchez-Puig N, Wong CC, Tonkin LM, Ancliff PJ, Brost RL, Costanzo M, Boone C, Warren AJ 2007. The Shwachman–Bodian–Diamond syndrome protein mediates translational activation of ribosomes in yeast. Nat Genet 39: 486–495 [DOI] [PubMed] [Google Scholar]
- Meyuhas O 2008. Physiological roles of ribosomal protein S6: One of its kind. Int Rev Cell Mol Biol 268: 1–37 [DOI] [PubMed] [Google Scholar]
- Morgan DG, Menetret JF, Radermacher M, Neuhof A, Akey IV, Rapoport TA, Akey CW 2000. A comparison of the yeast and rabbit 80 S ribosome reveals the topology of the nascent chain exit tunnel, inter-subunit bridges and mammalian rRNA expansion segments. J Mol Biol 301: 301–321 [DOI] [PubMed] [Google Scholar]
- Muhs M, Yamamoto H, Ismer J, Takaku H, Nashimoto M, Uchiumi T, Nakashima N, Mielke T, Hildebrand PW, Nierhaus KH, et al. 2011. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res 39: 5264–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda JS, Cheung YN, Takacs JE, Martin-Marcos P, Saini AK, Hinnebusch AG, Lorsch JR 2009. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J Mol Biol 394: 268–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J, Sengupta J, Frank J, Nissen P 2004. Regulation of eukaryotic translation by the RACK1 protein: A platform for signalling molecules on the ribosome. EMBO Rep 5: 1137–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930 [DOI] [PubMed] [Google Scholar]
- Noble CG, Song H 2008. Structural studies of elongation and release factors. Cell Mol Life Sci 65: 1335–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V 2005. Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177 [DOI] [PubMed] [Google Scholar]
- Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG 2003. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J 22: 193–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50 [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV 2010. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell 37: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool MR 2009. A trans-membrane segment inside the ribosome exit tunnel triggers RAMP4 recruitment to the Sec61p translocase. J Cell Biol 185: 889–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science 331: 730–736 [DOI] [PubMed] [Google Scholar]
- Reibarkh M, Yamamoto Y, Singh CR, del Rio F, Fahmy A, Lee B, Luna RE, Ii M, Wagner G, Asano K 2008. Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J Biol Chem 283: 1094–1103 [DOI] [PubMed] [Google Scholar]
- Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG 2010. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNAiMet binding to the ribosome. Genes Dev 24: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savelsbergh A, Rodnina MV, Wintermeyer W 2009. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA 15: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, et al. 2000. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102: 615–623 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Moore PB, Steitz TA 2003. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA 9: 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Ramakrishnan V 2011. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat Struct Mol Biol 18: 432–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM 2006. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat Struct Mol Biol 13: 1092–1096 [DOI] [PubMed] [Google Scholar]
- Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310: 827–834 [DOI] [PubMed] [Google Scholar]
- Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache JP, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, et al. 2009. Structural insight into nascent polypeptide chain–mediated translational stalling. Science 326: 1412–1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FV IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, Fasiolo F 2001. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol Cell 8: 1363–1373 [DOI] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Spahn CM, Nissen P, Frank J 2004. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol 11: 957–962 [DOI] [PubMed] [Google Scholar]
- Sengupta J, Nilsson J, Gursky R, Kjeldgaard M, Nissen P, Frank J 2008. Visualization of the eEF2–80S ribosome transition-state complex by cryo–electron microscopy. J Mol Biol 382: 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DH, Brandsen J, Jancarik J, Yokota H, Kim R, Kim SH 2004. Structural analyses of peptide release factor 1 from Thermotoga maritima reveal domain flexibility required for its interaction with the ribosome. J Mol Biol 341: 227–239 [DOI] [PubMed] [Google Scholar]
- Shin BS, Kim JR, Acker MG, Maher KN, Lorsch JR, Dever TE 2009. rRNA suppressor of a eukaryotic translation initiation factor 5B/initiation factor 2 mutant reveals a binding site for translational GTPases on the small ribosomal subunit. Mol Cell Biol 29: 808–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A, Marzi S, Jenner L, Myasnikov A, Romby P, Yusupova G, Klaholz BP, Yusupov M 2009. A structural view of translation initiation in bacteria. Cell Mol Life Sci 66: 423–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovic M, Steitz TA 2009. A structural view on the mechanism of the ribosome-catalyzed peptide bond formation. Biochim Biophys Acta 1789: 612–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310: 1513–1515 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D 2000. The crystal structure of human eukaryotic release factor eRF1—Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100: 311–321 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J 2001a. Structure of the 80S ribosome from Saccharomyces cerevisiae–tRNA-ribosome and subunit–subunit interactions. Cell 107: 373–386 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J 2001b. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science 291: 1959–1962 [DOI] [PubMed] [Google Scholar]
- Spahn CM, Gomez-Lorenzo MG, Grassucci RA, Jorgensen R, Andersen GR, Beckmann R, Penczek PA, Ballesta JP, Frank J 2004a. Domain movements of elongation factor eEF2 and the eukaryotic 80S ribosome facilitate tRNA translocation. EMBO J 23: 1008–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J 2004b. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell 118: 465–475 [DOI] [PubMed] [Google Scholar]
- Srivastava S, Verschoor A, Frank J 1992. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit–subunit interface. J Mol Biol 226: 301–304 [DOI] [PubMed] [Google Scholar]
- Sweeney R, Chen L, Yao MC 1994. An rRNA variable region has an evolutionarily conserved essential role despite sequence divergence. Mol Cell Biol 14: 4203–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B, Rutkai E, Cuchalova L, Munzarova V, Herrmannova A, Nielsen KH, Burela L, Hinnebusch AG, Valasek L 2008. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev 22: 2414–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Nilsson J, Merrill AR, Andersen GR, Nissen P, Frank J 2007. Structures of modified eEF2 80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J 26: 2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DJ, Devkota B, Huang AD, Topf M, Narayanan E, Sali A, Harvey SC, Frank J 2009. Comprehensive molecular structure of the eukaryotic ribosome. Structure 17: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T, Ehrenberg M 2002. Regulatory nascent peptides in the ribosomal tunnel. Cell 108: 591–594 [DOI] [PubMed] [Google Scholar]
- Triana-Alonso FJ, Chakraburtty K, Nierhaus KH 1995. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J Biol Chem 270: 20473–20478 [DOI] [PubMed] [Google Scholar]
- Uchiumi T, Hori K, Nomura T, Hachimori A 1999. Replacement of L7/L12.L10 protein complex in Escherichia coli ribosomes with the eukaryotic counterpart changes the specificity of elongation factor binding. J Biol Chem 274: 27578–27582 [DOI] [PubMed] [Google Scholar]
- Uchiumi T, Honma S, Nomura T, Dabbs ER, Hachimori A 2002. Translation elongation by a hybrid ribosome in which proteins at the GTPase center of the Escherichia coli ribosome are replaced with rat counterparts. J Biol Chem 277: 3857–3862 [DOI] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV 2004. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev 18: 3078–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Mathew AA, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG 2003. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev 17: 786–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J 2003. Locking and unlocking of ribosomal motions. Cell 114: 123–134 [DOI] [PubMed] [Google Scholar]
- Vestergaard B, Van LB, Andersen GR, Nyborg J, Buckingham RH, Kjeldgaard M 2001. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol Cell 8: 1375–1382 [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V 2009. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 16: 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V 2010. The mechanism for activation of GTP hydrolysis on the ribosome. Science 330: 835–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss NR, Gerstein M, Steitz TA, Moore PB 2006. The geometry of the ribosomal polypeptide exit tunnel. J Mol Biol 360: 893–906 [DOI] [PubMed] [Google Scholar]
- Wang DO, Martin KC, Zukin RS 2010. Spatially restricting gene expression by local translation at synapses. Trends Neurosci 33: 173–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN 2011. On the specificity of antibiotics targeting the large ribosomal subunit. Ann NY Acad Sci 1241: 1–16 [DOI] [PubMed] [Google Scholar]
- Wilson DN, Beckmann R 2011. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr Opin Struct Biol 21: 274–282 [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V 2000. Structure of the 30S ribosomal subunit. Nature 407: 327–339 [DOI] [PubMed] [Google Scholar]
- Woolhead CA, McCormick PJ, Johnson AE 2004. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 116: 725–736 [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Suzuki T 2008. Ribosomal RNAs are tolerant toward genetic insertions: Evolutionary origin of the expansion segments. Nucleic Acids Res 36: 3539–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Marintchev A, Kolupaeva VG, Unbehaun A, Veryasova T, Lai SC, Hong P, Wagner G, Hellen CU, Pestova TV 2009. Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res 37: 5167–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896 [DOI] [PubMed] [Google Scholar]
- Zavialov AV, Buckingham RH, Ehrenberg M 2001. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 107: 115–124 [DOI] [PubMed] [Google Scholar]