Abstract
Aberrant regulation of the Wnt signaling pathway is a prevalent theme in cancer biology. From the earliest observation that Wnt overexpression could lead to malignant transformation of mouse mammary tissue to the most recent genetic discoveries gleaned from tumor genome sequencing, the Wnt pathway continues to evolve as a central mechanism in cancer biology. This article summarizes the evidence supporting a role for Wnt signaling in human cancer. This includes a review of the genetic mutations affecting Wnt pathway components, as well as some of epigenetic mechanisms that alter expression of genes relevant to Wnt. I also highlight some research on the cooperativity of Wnt with other signaling pathways in cancer. Finally, some emphasis is placed on laboratory research that provides a proof of concept for the therapeutic inhibition of Wnt signaling in cancer.
Genetic and epigenetic modifications lead to aberrant Wnt signaling and cancer progression. Wnt also cooperates with other signaling pathways (e.g., prostaglandin signaling) in cancer.
The starting point of the field was the discovery that mammary tumors arising in mice infected with the murine mammary tumor virus was often caused by the activation of the murine int-1 gene, later called Wnt1. It was later shown that the Wnt gene resembled the fly wingless gene, a secreted factor controlling a signaling cascade that included GSK3 and armadillo, the fly version of mammalian β-catenin. The importance of this pathway in human cancer became very clear when the human tumor suppressor adenomatous polyposis coli (APC) protein was found in association with β-catenin. The finding that APC could downregulate β-catenin and Wnt-1 could upregulate it, provided further support for the Wnt cancer connection. Ultimately, the TCF transcription factors that associated with β-catenin completed the understanding of a basic signaling pathway that could account for the potent tumorigenic effects of Wnt (reviewed by Klaus and Birchmeier 2008).
ONCOGENES AND TUMOR SUPPRESSORS
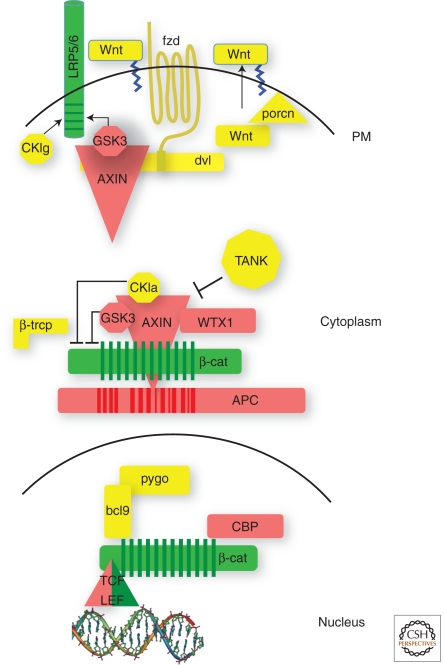
As in many other oncogenic signaling pathways, constituents of Wnt signaling can roughly be subdivided into positive and negatively acting components. By and large, the negatively acting, suppressing components are found mutated to a loss of function status in cancer, while the positive components are activated (Fig. 1). Among the suppressing components of Wnt signaling, APC stands as the most frequently mutated gene in human cancers. Genetic defects in APC are the cause of familial adenomatous polyosis, a heritable syndrome in which affected individuals develop hundreds of polyps in the large intestine at an early age and ultimately succumb to colorectal cancer (Clements et al. 2003). APC is also mutated in the vast majority of all sporadic colorectal cancers. Loss of function in both alleles is required for tumorigenesis and that loss is structurally linked to the protein's ability to regulate β-catenin protein stability (Polakis 2007).
Figure 1.
Tumor suppressors and oncogenes in the Wnt pathway. Diagram of a basic Wnt signaling pathway in which oncogenes are depicted in green and tumor suppressors in red.
Specifically, the truncating mutations in APC remove all binding sites for Axin, a scaffold that also binds β-catenin and recruits the protein kinases GSK3 and CKI, both essential for marking β-catenin for destruction facilitated by the E3 ubiquitin ligase β-TRCP (Fig 1). Axins I and II are also tumor suppressors found mutated in both sporadic cancers, particularly hepatocellar and some colorectal, as well as in some familial cancer syndromes (Lammi et al. 2004; Salahshor and Woodgett 2005; Marvin et al. 2011). Regulation of β-catenin also fails when β-catenin itself contains mutations that prevent it from being marked for destruction by the kinases (Polakis 2007). These mutations are found with significant frequency in hepatocellular cancers and medulloblastoma.
More recently, WTX has joined APC, Axin, and β-TrCP as a part of the so-called β-catenin destruction complex (Major et al. 2007). This is particularly intriguing as WTX is a tumor suppressor associated with the pediatric renal cancer Wilm’s tumor, which is also commonly associated with β-catenin mutations (Huff 2011). That both WTX and β-catenin mutations coexist in some Wilm’s tumors suggests the two genes are not strictly functionally redundant (Ruteshouser et al. 2008). In a recent developmental study in mice, germline inactivation of WTX resulted in the accumulation of multipotent mesenchymal precursor cells resulting from aberrant β-catenin activation (Moisan et al. 2011). Although these animals did not develop renal tumors, it was proposed that the expansion of these mesenchymal progenitors could increase the target population of cells susceptible to transformation by additional genetic insults.
One might expect the occurrence of inactivating mutations in the GSK3 genes as they are critical to β-catenin regulation. Although mutations in the alleles coding for GSK3α and β have not been associated with cancer, the Jamieson lab found an in-frame splice deletion affecting the kinase domain of GSK3β in chronic mylogenous leukemia (Abrahamsson et al. 2009). Aberrant splicing has also been described for the Wnt coreceptor LRP5 in parathyroid and breast cancers. Here missplicing deletes the region of LRP5 that interacts with the secreted Wnt signaling repressor DDK1 (Bjorklund et al. 2009).
Frameshift mutations in the 3′ region of the gene encoding the β-catenin-binding transcription factor TCF4 are extremely common in colorectal cancers with microsatellite instability (Cuilliere-Dartigues et al. 2006). These mutations were proposed to be activating as they ablate binding of the transcriptional repressor CtBP to TCF4. Paradoxically, mutations in TCF4, identified through genomic sequencing efforts of colorectal cancers, appear to be inactivating by compromising its ability to repress cell growth (Sjoblom et al. 2006; Tang et al. 2008). This is consistent with recent findings from the Cappechi laboratory in which TCF4 haploinsuffciency promotes cell proliferation in the mouse intestine and enhances tumorigenesis in an APC mutant mouse model (Angus-Hill et al. 2011). In a colorectal cancer genome sequencing effort, the Meyerson laboratory recently reported a rare, but recurring rearrangement in which the TCF4 gene TCFL72 was fused to VTI1A (Bass et al. 2011). The specific function of the VTI1A-TCF7L2 fusion was not elucidated, but knockdown of the transcript inhibited anchorage independent cell growth, indicative of a positive acting tumorigenic function.
Mutations in the gene coding for the CREB binding protein (CREBBP/CBP) have been identified in acute lypmhoblastic leukemia and B-cell lymphoma (Mullighan et al. 2011; Pasqualucci et al. 2011). This is of interest to Wnt signaling as CBP associates directly with the carboxy-terminal sequence in β-catenin and CBP/β-catenin mediated transcription has been proposed to be critical for stem cell/progenitor cell maintenance and proliferation (Teo and Kahn 2011). Accordingly, CBP is currently a clinical target for small molecule inhibition in Wnt-driven cancers. CBP is a transcriptional coactivator with histone acetyltransferase activity and associates with a wide variety of transcription factors in controlling facets of development and cellular homeostasis. Given this pleiotropy, the relevance of the CBP mutations to Wnt signaling, per se, is difficult to gauge. Moreover, the mutations identified in the hematopoietic cancers impair the histone acetyltransferase activity, which seem inconsistent with a role in facilitating active Wnt signaling. Table 1 summarizes some of the salient DNA and mRNA aberrations that potentially contribute to Wnt-driven tumorigenesis.
Table 1.
Genetic alterations in Wnt signaling in cancer
| Affected gene | DNA/mRNA alteration | Functional outcome | Cancer type | Reference |
|---|---|---|---|---|
| CTNNBl (b-catenin) | Missense/in-frame deletion | Enhanced protein stability | Hepatocellular/Medulloblastoma | Polakis 2007 |
| APC (APC) | Truncation | Reduced regulatory activity | Colorectal/gastric | Clements et al. 2003 |
| Axins (Axin I, Axin II) | Truncation/missense | Reduced regulatory activity | Hepatocellular/colorectal | Salahshor and Woodgett 2005 |
| CREBP(CBP) | Truncation/ missense | Inactive acetyltransferase | Lymphoma/leukemia | Teo and Kahn 2011 |
| GSK3b | Missplicing, in-frame deletion | Inactive kinase | Leukemia | Abrahamsson et al. 2009 |
| LRP5 | Missplicing, in-frame deletion | Loss of repression by DKK1 | Breast/parathyroid | Bjorklund et al. 2009 |
| TCF7L2 (TCF4) | Missense/deletion/truncation | Loss of repression | Colorectal | Cuilliere-Dartigues et al. 2006 |
| TCF7L2 (TCF4) | Fusion with VT11A gene | Unclear | Colorectal | Bass 2011 |
| FAB123B (WTX) | Truncation/deletion | Loss of function | Wilm’s tumor | Ruteshouser et al. 2008 |
EPIGENETIC MODULATION OF Wnt SIGNALING
Recurrent mutations in a gene that alter the products’ function in a relevant manner typically offer the clearest evidence for its involvement in oncogenic transformation. Nevertheless, it is apparent that epigenetic modifications in DNA and chromatin also play a role in tumor progression by modulating the suppressor or activator components of a signaling pathway (Rodriguez-Paredes and Esteller 2011). The impact of DNA methylation on Wnt-driven tumorigenesis was reported by Laird et al., who found that genetic depletion and pharmacological suppression of DNA methyl transferase effectively reduced the tumor burden in the APC min mouse (Laird et al. 1995). Subsequent studies with this model revealed that deficiency of the methyl–CpG binding repressor Mbd2 also severely repressed tumor formation by attenuating Wnt signaling (Sansom et al. 2003; Phesse et al. 2008). These studies point to the silencing of Wnt antagonists by DNA hypermethylation. Indeed, genome-wide analysis of promotor DNA methylation in human medulloblastoma, colon, and pancreatic cancers showed that Wnt pathway inhibitors are common targets of this mechanism of gene repression (Kongkham et al. 2010; Vincent et al. 2011; Suzuki et al. 2002). In particular, the promotor region of the gene coding for the secreted frizzled related protein 1 (sFRP1), that binds to and inhibits Wnt ligands, was commonly identified in these studies. Members of the sFRP family have also been reported to be silenced by DNA methylation in numerous other cancers including hepatocellular cancer, breast cancer, and melanoma (Suzuki et al. 2008; Takagi et al. 2008; Ekstrom et al. 2011).
In addition to the direct methylation of DNA, many Wnt signaling genes and targets undergo chromatin modifications that affect gene transcription. Considering that DNA hypermethylation of promotor regions is frequently accompanied by chromatin modifications, it is not surprising that the sSFRP1 gene is also a target of chromatin modifying factors (McGarvey et al. 2006). In the case of AML, it was proposed that chromatin modification resulted from the targeting of the sFRP1 gene by the RUNX-ETO fusion protein (Cheng et al. 2011b). A RUNX binding site was identified in the 5′ regulatory region of the sFRP1 gene and this region was also detected in chromatin immunoprecipitated with antibodies specific to ETO. The ETO portion of this fusion recruits multiple chromatin modifying factors that can repress gene activation and, in the case of sFRP1, drive Wnt signaling and cell proliferation.
An additional epigenetic mechanism underlying the suppression of numerous Wnt antagonists in hepatocellular cancer was reported by Cheng et al (2011a). In this study, chromatin immunoprecipitation/microarray analysis was used to show that many genes coding for Wnt inhibitors, including sFRP5, Axin2, and NKD2 were occupied by EZH2, a subunit of the polycomb repressor complex 2 (PRC2). EZH2 is a methyl transferase that deposits the repressive H3K27me3 marks on chromatin (Rodriguez-Paredes and Esteller 2011). Knockdown of EZH2 in HCC cells resulted in increased expression of NKD2 and a concomitant reduction in Wnt reporter signaling. This is particularly intriguing because somatic mutations in EZH2, as well as another H3k27 methyltransferase, UTX, have been detected in a variety of human cancers (van Haaften et al. 2009; Morin et al. 2011).
DACT3 is an additional inhibitor of Wnt signaling that is repressed in colon cancers by histone modification (Jiang et al. 2008). DACT3 is a member of the FDO/DPR gene family that inhibits Wnt signaling by interacting with DVL proteins. Derepression of DACT3 inhibited Wnt signaling and promoted apoptosis of colorectal cancer cells, an effect that could be rescued by knockdown of DACT3 mRNA. Unlike sFRP1 in colon cancer cells, the DACT3 DNA promotor region does not appear to be hypermethylated. Thus, DACT3 derepression might not require reversal of DNA methylation, but perhaps may respond singularly to treatment with an inhibitor of histone modification such as that represented by the Trichostatin A compound.
In contrast to gene silencing, modification of chromatin can also enable gene activation. The carboxyl terminus of β-catenin is known to recruit numerous chromatin-remodeling subunits, such as set1 and paf1, that mediate the H3K4me3 histone marks associated with highly active genes (Willert and Jones 2006). Although many of these sorts of modifiers are commonly associated with gene activation by transcription factors, the Clevers laboratory reported that the MLLT10/AF10-DOT1L complex was principally dedicated to Wnt signaling (Mahmoudi et al. 2011). They discovered that this leukemia-associated chromatin-modifying complex was recruited to Wnt target genes in a β-catenin/TCF4-dependent manner. DOT1L is a methyl transferase responsible for H3K79me3 modification associated with gene activation. The study showed a requirement for MLLT10/AF10-DOT1L in Wnt target gene expression in colon cancer cells and in normal intestinal homeostasis. MLLT10/AF10 expression was localized to the proliferative compartment of the intestine and its knockdown rescued defects in APC mutant zebrafish. On the basis of this specificity, the investigators concluded that the DOT1L enzyme might represent a colon cancer target.
COOPERATIVITY WITH OTHER SIGNALING PATHWAYS
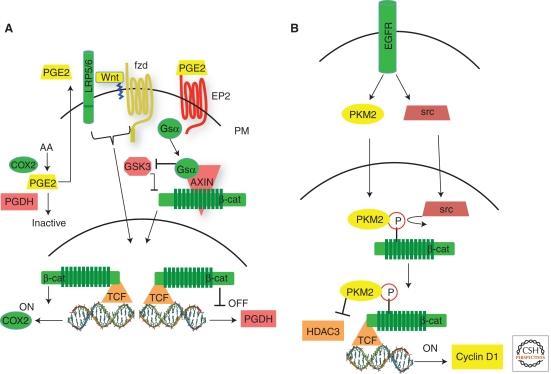
Studies involving the use genetically engineered mouse models of cancer, as well as expansive human studies on nonsteroidal anti-inflammatory drugs (NSAIDS) and cancer risk, have firmly established a role for prostaglandin signaling in colorectal tumorigenesis (Oshima and Taketo 2002; Chan et al. 2007). The cyclooxygenase 2 (COX2) enzyme, which catalyzes the production of PGE2 is targeted by NSAIDS and its deletion in the APCmin mouse negatively impacts tumor burden. Mechanistic proposals for the interaction between prostaglandin and Wnt signaling include COX2 as a direct target gene of Wnt signaling, PPARDelta as a convergent focal point for the two pathways, and stimulation of the Wnt pathway by crosstalk from the EP2 prostaglandin receptor (Araki et al. 2003; Wang et al. 2004; Castellone et al. 2005). The levels of PGE2 are downregulated by 15-Hydroxyprostaglandin dehydrogenase (15-PGDH), which catalyzes the rate-limiting step in prostaglandin degradation. Therefore, it acts opposite to COX2, and knockout of 15-PGDH in the APCmin mouse dramatically increases tumor burden, further fortifying the case for prostaglandins as intestinal tumor promotors (Myung et al. 2006). In human cancers, 15-PGDH is consistently downregulated, although it is highly expressed in normal human intestinal epithelium. In a recent study, the Paraskeva lab offered an additional mechanism for the interaction of the prostaglandin and Wnt signals by showing that the 15-PGDH gene is a target of β-catenin-dependent repression (Smartt et al. 2011). This study documented 15-PGDH gene promotor occupancy by β-catenin/TCF4, in vivo activation of 15-PDGH expression by conditional knockdown of β-catenin, and inhibition of 15-PGDH expression in cultured cells treated with Wnt ligand.
Cooperativity between the androgen receptor (AR) and Wnt signaling was proposed as a mechanism for the potentiation of EGF receptor tyrosine kinase family signaling in ER-/Her2+ breast cancers (Ni et al. 2011). In this study, the Wnt7b gene was occupied and activated by AR on stimulation of breast cancer cells with DHT. This led to activation of canonical Wnt signaling in which the stabilized β-catenin cooperated with AR to turn on the Her3 gene. Her3 is the ligand binding receptor that partners with the catalytically active Her2 tyrosine kinase to activate the Pi3K pathway. The paper showed that the upregulation of HER3, activation of HER2/HER3 signaling and enhancement of cell growth by AR activation was dependent on β-catenin.
The EGF receptor tyrosine kinase has had a long history of cooperating with Wnt signaling, resulting in a litany of mechanisms to account for their interaction. These include direct binding of β-catenin to EGFR, modulation of the cadherin-β-catenin interface by EGFR, activation of β-catenin-dependent signaling by EGFR through PI3K signaling as well as the EGFR gene itself as a Wnt target (review by Hu and Li 2011). However, in a very surprising finding by the Lu lab, the M2 splice form of pyruvate kinase (PKM2) was found to translocate to the nucleus in response to EGFR signaling, where it binds to tyrosine phosphorylated of β-catenin and activates β-catenin-dependent target genes (Fig. 2A) (Yang et al. 2011b). PKM2 has received considerable attention lately, although mainly because of a renewed interest in the role of metabolism and aerobic glycolysis in cancer (Christofk et al. 2008). However, in the Lu study, knockdown of PKM2 reduced tumor cell proliferation by EGFR, which correlated with a failure to activate the myc and cyclin D1 genes, both targets of Wnt signaling. Moreover, chromatin immunoprecipitation was used to show binding of PKM2 to the cyclin D1 promotor in a β-catenin-dependent manner. Nevertheless, PKM2 deficiency did interfere with the activation of cyclin D1 by Wnt ligands. Thus, EGF and Wnt receptors both employ β-catenin to activate some common target genes, yet engage independent effectors in route to the nucleus.
Figure 2.
Signaling pathways that cooperate with Wnt signaling. (A) Wnt and prostaglandin signaling. Activation of canonical Wnt signaling drives expression of COX2, which catalyzes production of PGE2, and repression of 15-PDGH, which catalyzes the inactivation of PGE2. PGE2 activates the prostaglandin GPCR receptor EP2 releasing the Gsα subunnit that displaces GSK3 form Axin, resulting in the stabilization of β-catenin. (B) Activation of b-catenin by EGFR. EGFR activity promotes the activation of src and the translocation of PKM2 into the nucleus, in which src phosphorylates residue Y333 on β-catenin. The phosphorylated β-catenin binds PKM2 and the resulting complex associates with TCF in which kinase-active PKM2 displaces HDAC3 to turn on the cyclin D1 target gene.
Another surprise from the Lu study was that the tyrosine phosphorylation of β-catenin resulting from EGFR activation was mediated by src and occurred on β-catenin residue Y-333. Previously, EGFR-mediated phoshorylation of β-catenin on Y-654 had been reported to affect its interaction with cadherins and also correlated with an increase in β-catenin-mediated transcriptional activity (Lilien and Balsamo 2005). The relevance of Y-654 phosphorylation was finally tested in vivo by the Smits laboratory, who corroborated its importance in binding of cadherin to β-catenin, but also found a positive impact on Wnt signaling (van Veelen et al. 2011). When the constitutive phosphorylation was mimicked by knocking in the Y654E mutation, the homozygotes were embryonic lethal, whereas 38% of the heterozygotes presented with intestinal tumors by 16–18 months of age. On the background of a germline APC mutation, the compound heterozygotes were embryonic lethal, suggesting that half of a dose of wildtype APC was not sufficient to regulate the excess signaling produced by a single Y654E β-catenin allele. An intestine-specific allele of the Y654E β-catenin mutant was then generated and when crossed to an APC mutant mouse, increased intestinal tumor multiplicity was observed in male animals.
Given the interplay between morphogenic signals in developing embryos, the interaction of these pathways might be expected in cancer. In a particularly intriguing example of Wnt and notch pathway cooperativity, Fre et al. (2009) showed that the expansion of intestinal progenitor cells by constitutive Notch signaling was dependent on Wnt. Notch and Wnt signaling also synergized to induce intestinal adenomas in compound mice, with a dramatic increase in colonic tumors, which are typically rare in mice deficient only in APC. The impact of Notch in the intestine was consistent with previous work showing that its activation or inhibition increased or decreased the population of proliferating/progenitor cells, respectively, at the expense of the differentiated population (Fre et al. 2005; van Es et al. 2005). The Srivastava laboratory recently offered an unexpected mechanism for cooperativity between Wnt and Notch signaling (Kwon et al. 2011). Here the ICD domain of Notch was shown to directly associate with and thereby titrate the unphosphorylated pool of active β-catenin, rendering it unavailable for transcriptional activity. Accordingly, the knockdown of Notch1 exacerbated the activation of Wnt target genes, whereas chemical inhibition of γ-secretase or expression of a noncleavable Notch1 mutant, was inhibitory to Wnt signaling. It was concluded that the active β-catenin associated with Notch was ultimately directed to the lysosome for destruction in a Numb-dependent manner.
EXPERIMENTAL EVIDENCE FOR A CRITICAL ROLE FOR Wnt IN HUMAN CANCER
A variety of preclinical experiments suggest that inhibition of Wnt signaling can thwart tumor cell growth and survival. One of the first such experiments involved the induced expression of wild type APC in a colorectal cancer cell line harboring only mutant alleles. On induction, the growth of cultured HT29 cells was inhibited by the wild-type APC, which was attributed to an increase in apoptosis (Morin et al. 1996). Similarly, ectopic expression of Axin1, an additional tumor suppressor in the Wnt pathway, promoted apoptosis in colorectal and hepatocellular cancer cell lines containing mutations in either β-catenin, APC or Axin1 (Satoh et al. 2000). Although encouraging, the reanimation of wild-type tumor suppressors is inferior to a proof of principle in which a positive acting element is interfered with. Early examples of this included the ectopic expression of a dominant negative TCF-4. This amino-terminally deleted TCF-4 binds Wnt target genes nonproductively as a result of its inability to associate with β-catenin. Consequently, inhibition of target gene activation in colorectal cancer cells expressing the dominant negative TCF4 resulted in their growth arrest (Tetsu and McCormick 1999; van de Wetering et al. 2002).
Based on the genetics of Wnt signaling in cancer, β-catenin would be considered a high value drug target. Two tumor suppressors, Axin and APC, negatively regulate β-catenin, and β-catenin itself is oncogenically activated by gain-of-function mutations. Accordingly, interfering with β-catenin, by various methods, produces antitumorigenic affects. Gottardi et al. (2001), exploited the mutually exclusive binding of β-catenin to E-cadherin and the TCF transcription factors by expressing cadherin chimeras that block the latter interaction. This was accompanied by growth inhibition of SW480 colorectal cancer cells. Suppression of β-catenin by antisense oligonucleotides was also found to inhibit tumorgenicity of colon cancer cells in vitro and in vivo (Green et al. 2001; Roh et al. 2001). Kim et al. (2002) employed a somatic cell gene targeting strategy to show that β-catenin expression was essential for clonal growth of the APC mutant and β-catenin mutant colorectal cancer lines, DLD-1 and SW48, respectively. Curiously, a second β-catenin mutant cell line, HCT116, was unaffected by the gene targeting.
The advent of interfering RNA technologies ushered in further validation of β-catenin as a potential cancer target. In one of the first examples of this, siRNA knockdown of β-catenin reduced β-catenin-dependent transcription and diminished the anchorage independent growth of colon cancer cells and their ability to generate tumors in mice (Verma et al. 2003). Unlike the gene targeting approach, the tumorigenecity of the HCT116 cell line was negatively impacted by the siRNA. In a separate study, knockdown of β-catenin by inducible shRNA strongly inhibited proliferation of HCT116 and Ls174t colorectal cancer cells, both of which harbor mutant β-catenin alleles (Mologni et al. 2010). However, induction of apoptosis was insignificant with knockdown of either β-catenin or K-ras individually, whereas the combination produced a strong effect. The effect of this combination on in vivo tumor growth was again far more effective than knockdown of either transcript alone.
In addition to its role in gene activation, β-catenin is essential to cadherin function in forming cell–cell contacts. To some extent this complicates the interpretation of β-catenin mRNA knockdown experiments. To overcome this, Cong et al. (2003), devised a fusion protein that artificially recruited the E3 ubiquitin ligase β-TrCP to β-catenin. This resulted in the selective destruction of the Wnt signaling pool, but not the cadherin-bound fraction. Colon cells expressing this construct lost their tumorigenic potential, including their ability to form tumors in mice.
Inhibitors of Wnt signaling might also be effective in hepatocellular cancers, where mutations in Wnt pathway components are relatively common. In the HepG2 human hepatoma cell line, RNAi mediated knockdown of β-catenin decreased cell viability, proliferation and growth in soft agar (Zeng et al. 2007). Although the HepG2 cell line carries a mutant β-catenin allele, a second cell line, Hep3b, is wild type, yet similar inhibitory effects were observed with it. Ashihara et al. (2009) provided an additional example in which interference with Wnt signaling impaired the tumorigencity of cancer cells lacking Wnt pathway mutations. Here, siRNA knockdown of β-catenin significantly reduced the tumor burden resulting from the inoculation of RPMI8226 myeloma cells in irradiated nude mice. Thus, interference with Wnt signaling in cancers lacking pathway mutations can be effective and suggests that aberrant activation of Wnt receptors should also be entertained as an oncogenic mechanism.
Mammary cancer is a prominent example in which Wnt pathway mutations are very rare, yet hyperactive signaling is apparent, particularly in cancers classified as basal-like or triple negative type (Lin et al. 2000; Chung et al. 2004; Howe and Brown 2004; Khramtsov et al. 2010; Geyer et al. 2011). Expression of the Wnt receptor FZD7 is characteristic of these types of breast cancer (Sorlie et al. 2003). Accordingly, Yang, et al. recently reported a proof of principle experiment in which knockdown of FZD7, in cell line models of triple negative breast cancer, reduced expression of Wnt target genes, inhibited tumorigenesis in vitro and greatly retarded the capacity of the MDA-MD-231 cell line to form tumors in mice (Yang et al. 2011a). This suggests that Wnt ligands might drive certain breast cancers and is consistent with previous work from the Hynes laboratory (Matsuda et al. 2009). Here the secreted frizzled related protein (sFRP1), which binds to and effectively competes with FZD receptors for Wnt ligands, was ectopically expressed in the MDA-MΒ-231 cell line. The sFRP1 expressing cells struggled to form tumors upon inoculation into the mammary fat pads of mice and their propensity to metastasize to lung was greatly impaired. The source of Wnt ligand in these studies could arise from either host cells and/or the cancer cells, themselves. The latter possibility was first made apparent by Bafico et al. (2004), when they reported autocrine Wnt signaling in a panel of breast and ovarian cancer cell lines, MDA-MΒ-231 among them. These cells were identified by the presence of a signaling pool of uncomplexed β-catenin, a hallmark of Wnt signaling. This pool of β-catenin, as well as the signal from a Wnt reporter gene, was reduced on expression or addition of the soluble Wnt inhibitors sFRP1 or DKK1. These investigators subsequently reported autocrine signaling in non-small cell lung cancers, as well (Akiri et al. 2009).
Work from the Massague laboratory encourages us to target Wnt signaling in lung cancer, which again typically lacks pathway mutations (Nguyen et al. 2009). In this study, a Wnt target gene signature was associated with metastatic lesions derived from human lung cancer models grown in mice. Expression of dominant negative TCF4 or TCF1 in the metastasis-prone derivatives of the PC9 and H2030 nonsmall lung cancer lines provided a proof-of-principle. The expression of these dominant negative transcription factors had little impact on the growth rate of the pulmonary tumors, but retarded their metastatic spread to bone and brain.
Although the knockdown of FZD7 or the ectopic expression of sFRP1 implicate Wnt ligands and in cancer, they still involve genetic manipulation, and thus remain one step removed from a pharmacological proof of concept. A more pharmacologically relevant approach was provided by De Almeida et al. (2007), whom injected tumor-bearing mice with a fusion protein consisting of the Fc region IgG fused to the extra cellular domain of the FZD8 (FZD8CRD) Wnt receptor. That the FZD8CRD molecule was active in vivo was initially shown by its dramatic inhibitory effect in the MMTV-Wnt-1 model of mammary cancer. Secondly, the FZD8CRD showed significant tumor inhibition of two nonengineered cancer cell lines, the N-TERA2 human testicular cancer and the PA1 human ovarian cancer cell line. The PA1 was among those cells identified by Bafico et al. (2004) as having autocrine Wnt signaling. In an additional pharmacological test, You et al. (2004), inhibited the growth of a human melanoma xenograft model by administering a monoclonal antibody to Wnt2.
The impact of β-catenin knockdown in cancers without identifiable genetic lesions in Wnt signaling could relate to the purported role of Wnt in the establishment and maintenance of so-called cancer stem cells (CSCs) (Reya and Clevers 2005; Malanchi and Huelsken 2009). There is considerable evidence for this in hematopoetic cancers, in which evidence for a cellular hierarchy is well established. Granulocytic-macrophage progenitors isolated from CML patients show attributes of excessive Wnt signaling relative to normal progenitors and the introduction of the Wnt pathway inhibitor Axin decreased their capacity for proliferation and self-renewal (Jamieson et al. 2004). In a mouse model of CML, introduction of BCR-ABL into marrow cells from conditional β-catenin−/− knockout animals significantly reduced the onset of CML-like disease relative to that observed with wild-type marrow cells (Zhao et al. 2007). Nevertheless, recipients of the transformed β-catenin−/− cells went on to develop disease resembling acute lymphoblastic leukemia. In another mouse model of leukemia, initiated by expression of a mixed lineage leukemia (MLL) fusion protein, the conditional ablation of the β-catenin gene prevented preleukemic stem cells from inducing leukemia in grafted animals (Wang et al. 2010). In this same study, shRNA knock down of β-catenin in leukemia stem cells severely delayed the onset of leukemia in recipient hosts.
Recent evidence also points to a role for Wnt signaling in cutaneous cancer stem cells. The Huelsken laboratory found many similarities between normal skin stem cells and cancer stem cells isolated from murine epidermal tumors induced by carcinogens (Malanchi et al. 2008). The cancer stem cells appeared enriched for Wnt signaling and conditional knockdown of β-catenin effectively eliminated this population of cells resulting in complete regression of the epidermal tumors. Although this shows that a mouse epidermal tumor is indeed impacted by inhibition of Wnt signaling, the investigators also provided some evidence that Wnt signaling is exacerbated in human squamous cell carcinomas.
SUMMARY
Wnt signaling clearly contributes to human tumor progression. This is best defined by germline and somatic mutations in Wnt pathway components that deregulate signaling. The rapidly evolving field of epigenetics also continues to implicate Wnt signaling in cancer by identifying suppressors and activators whose expression is modulated by DNA and chromatin modifications. New mechanisms of cooperativity between Wnt signaling and other pathways underscore the flexibility available to cancer cells for maintaining growth and survival. Finally, a large body of experimental evidence strongly encourages us to develop therapies that specifically interfere with Wnt signaling in the cancer clinic.
Footnotes
Editors: Roel Nusse, Xi He, and Renee van Amerongen
Additional Perspectives on Wnt Signaling available at www.cshperspectives.org
REFERENCES
- Abrahamsson AE, Geron I, Gotlib J, Dao KH, Barroga CF, Newton IG, Giles FJ, Durocher J, Creusot RS, Karimi M, et al. 2009. Glycogen synthase kinase 3β missplicing contributes to leukemia stem cell generation. Proc Natl Acad Sci 106: 3925–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiri G, Cherian MM, Vijayakumar S, Liu G, Bafico A, Aaronson SA 2009. Wnt pathway aberrations including autocrine Wnt activation occur at high frequency in human non-small-cell lung carcinoma. Oncogene 28: 2163–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angus-Hill ML, Elbert KM, Hidalgo J, Capecchi MR 2011. T-cell factor 4 functions as a tumor suppressor whose disruption modulates colon cell proliferation and tumorigenesis. Proc Natl Acad Sci 108: 4914–4919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y, Okamura S, Hussain SP, Nagashima M, He P, Shiseki M, Miura K, Harris CC 2003. Regulation of cyclooxygenase-2 expression by the Wnt and ras pathways. Cancer Res 63: 728–734 [PubMed] [Google Scholar]
- Ashihara E, Kawata E, Nakagawa Y, Shimazaski C, Kuroda J, Taniguchi K, Uchiyama H, Tanaka R, Yokota A, Takeuchi M, et al. 2009. β-catenin small interfering RNA successfully suppressed progression of multiple myeloma in a mouse model. Clin Cancer Res 15: 2731–2738 [DOI] [PubMed] [Google Scholar]
- Bafico A, Liu G, Goldin L, Harris V, Aaronson SA 2004. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell 6: 497–506 [DOI] [PubMed] [Google Scholar]
- Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, Sougnez C, Voet D, Saksena G, Sivachenko A, et al. 2011. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet 43: 964–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund P, Svedlund J, Olsson AK, Akerstrom G, Westin G 2009. The internally truncated LRP5 receptor presents a therapeutic target in breast cancer. PLoS ONE 4: e4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellone MD, Teramoto H, Williams BO, Druey KM, Gutkind JS 2005. Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-β-catenin signaling axis. Science 310: 1504–1510 [DOI] [PubMed] [Google Scholar]
- Chan AT, Ogino S, Fuchs CS 2007. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N Engl J Med 356: 2131–2142 [DOI] [PubMed] [Google Scholar]
- Cheng AS, Lau SS, Chen Y, Kondo Y, Li MS, Feng H, Ching AK, Cheung KF, Wong HK, Tong JH, et al. 2011a. EZH2-mediated concordant repression of Wnt antagonists promotes β-catenin-dependent hepatocarcinogenesis. Cancer Res 71: 4028–4039 [DOI] [PubMed] [Google Scholar]
- Cheng CK, Li L, Cheng SH, Ng K, Chan NP, Ip RK, Wong RS, Shing MM, Li CK, Ng MH 2011b. Secreted-frizzled related protein 1 is a transcriptional repression target of the t(8;21) fusion protein in acute myeloid leukemia. Blood 118: 6638–6648 [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC 2008. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452: 230–233 [DOI] [PubMed] [Google Scholar]
- Chung GG, Zerkowski MP, Ocal IT, Dolled-Filhart M, Kang JY, Psyrri A, Camp RL, Rimm DL 2004. β-Catenin and p53 analyses of a breast carcinoma tissue microarray. Cancer 100: 2084–2092 [DOI] [PubMed] [Google Scholar]
- Clements WM, Lowy AM, Groden J 2003. Adenomatous polyposis coli/β-catenin interaction and downstream targets: Altered gene expression in gastrointestinal tumors. Clin Colorectal Cancer 3: 113–120 [DOI] [PubMed] [Google Scholar]
- Cong F, Zhang J, Pao W, Zhou P, Varmus H 2003. A protein knockdown strategy to study the function of β-catenin in tumorigenesis. BMC Mol Biol 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuilliere-Dartigues P, El-Bchiri J, Krimi A, Buhard O, Fontanges P, Flejou JF, Hamelin R, Duval A 2006. TCF-4 isoforms absent in TCF-4 mutated MSI-H colorectal cancer cells colocalize with nuclear CtBP and repress TCF-4-mediated transcription. Oncogene 25: 4441–4448 [DOI] [PubMed] [Google Scholar]
- DeAlmeida VI, Miao L, Ernst JA, Koeppen H, Polakis P, Rubinfeld B 2007. The soluble Wnt receptor Frizzled8CRD-hFc inhibits the growth of teratocarcinomas in vivo. Cancer Res 67: 5371–5379 [DOI] [PubMed] [Google Scholar]
- Ekstrom EJ, Sherwood V, Andersson T 2011. Methylation and loss of Secreted Frizzled-Related Protein 3 enhances melanoma cell migration and invasion. PLoS ONE 6: e18674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S 2005. Notch signals control the fate of immature progenitor cells in the intestine. Nature 435: 964–968 [DOI] [PubMed] [Google Scholar]
- Fre S, Pallavi SK, Huyghe M, Lae M, Janssen KP, Robine S, Artavanis-Tsakonas S, Louvard D 2009. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci 106: 6309–6314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer FC, Lacroix-Triki M, Savage K, Arnedos M, Lambros MB, MacKay A, Natrajan R, Reis-Filho JS 2011. β-Catenin pathway activation in breast cancer is associated with triple-negative phenotype but not with CTNNB1 mutation. Mod Pathol 24: 209–231 [DOI] [PubMed] [Google Scholar]
- Gottardi CJ, Wong E, Gumbiner BM 2001. E-cadherin suppresses cellular transformation by inhibiting β-catenin signaling in an adhesion-independent manner. J Cell Biol 153: 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green DW, Roh H, Pippin JA, Drebin JA 2001. β-catenin antisense treatment decreases β-catenin expression and tumor growth rate in colon carcinoma xenografts. J Surg Res 101: 16–20 [DOI] [PubMed] [Google Scholar]
- Howe LR, Brown AM 2004. Wnt signaling and breast cancer. Cancer Biol Ther 3: 36–41 [DOI] [PubMed] [Google Scholar]
- Hu T, Li C 2011. Convergence between Wnt-β-catenin and EGFR signaling in cancer. Mol Cancer 9: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff V 2011. Wilms’ tumours: About tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer 11: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, et al. 2004. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med 351: 657–667 [DOI] [PubMed] [Google Scholar]
- Jiang X, Tan J, Li J, Kivimae S, Yang X, Zhuang L, Lee PL, Chan MT, Stanton LW, Liu ET, et al. 2008. DACT3 is an epigenetic regulator of Wnt/β-catenin signaling in colorectal cancer and is a therapeutic target of histone modifications. Cancer Cell 13: 529–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khramtsov AI, Khramtsova GF, Tretiakova M, Huo D, Olopade OI, Goss KH 2010. Wnt/β-catenin pathway activation is enriched in basal-like breast cancers and predicts poor outcome. Am J Pathol 176: 2911–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Crooks H, Foxworth A, Waldman T 2002. Proof-of-principle: oncogenic β-catenin is a valid molecular target for the development of pharmacological inhibitors. Mol Cancer Ther 1: 1355–1359 [PubMed] [Google Scholar]
- Klaus A, Birchmeier W 2008. Wnt signalling and its impact on development and cancer. Nat Rev Cancer 8: 387–398 [DOI] [PubMed] [Google Scholar]
- Kongkham PN, Northcott PA, Croul SE, Smith CA, Taylor MD, Rutka JT 2010. The SFRP family of WNT inhibitors function as novel tumor suppressor genes epigenetically silenced in medulloblastoma. Oncogene 29: 3017–3024 [DOI] [PubMed] [Google Scholar]
- Kwon C, Cheng P, King IN, Andersen P, Shenje L, Nigam V, Srivastava D 2011. Notch post-translationally regulates β-catenin protein in stem and progenitor cells. Nat Cell Biol 13: 1244–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E, Weinberg RA, Jaenisch R 1995. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 81: 197–205 [DOI] [PubMed] [Google Scholar]
- Lammi L, Arte S, Somer M, Jarvinen H, Lahermo P, Thesleff I, Pirinen S, Nieminen P 2004. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am J Hum Genet 74: 1043–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilien J, Balsamo J 2005. The regulation of cadherin-mediated adhesion by tyrosine phosphorylation/dephosphorylation of β-catenin. Curr Opin Cell Biol 17: 459–465 [DOI] [PubMed] [Google Scholar]
- Lin SY, Xia W, Wang JC, Kwong KY, Spohn B, Wen Y, Pestell RG, Hung MC 2000. β-catenin, a novel prognostic marker for breast cancer: Its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci 97: 4262–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudi T, Boj SF, Hatzis P, Li VS, Taouatas N, Vries RG, Teunissen H, Begthel H, Korving J, Mohammed S, et al. 2011. The leukemia-associated Mllt10/Af10-Dot1l are Tcf4/β-catenin coactivators essential for intestinal homeostasis. PLoS Biol 8: e1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major MB, Camp ND, Berndt JD, Yi X, Goldenberg SJ, Hubbert C, Biechele TL, Gingras AC, Zheng N, Maccoss MJ, et al. 2007. Wilms tumor suppressor WTX negatively regulates WNT/β-catenin signaling. Science 316: 1043–1046 [DOI] [PubMed] [Google Scholar]
- Malanchi I, Huelsken J 2009. Cancer stem cells: Never Wnt away from the niche. Curr Opin Oncol 21: 41–46 [DOI] [PubMed] [Google Scholar]
- Malanchi I, Peinado H, Kassen D, Hussenet T, Metzger D, Chambon P, Huber M, Hohl D, Cano A, Birchmeier W, et al. 2008. Cutaneous cancer stem cell maintenance is dependent on β-catenin signalling. Nature 452: 650–653 [DOI] [PubMed] [Google Scholar]
- Marvin ML, Mazzoni SM, Herron CM, Edwards S, Gruber SB, Petty EM 2011. AXIN2-associated autosomal dominant ectodermal dysplasia and neoplastic syndrome. Am J Med Genet A 155A: 898–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda Y, Schlange T, Oakeley EJ, Boulay A, Hynes NE 2009. WNT signaling enhances breast cancer cell motility and blockade of the WNT pathway by sFRP1 suppresses MDA-MB-231 xenograft growth. Breast Cancer Res 11: R32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB 2006. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res 66: 3541–3549 [DOI] [PubMed] [Google Scholar]
- Moisan A, Rivera MN, Lotinun S, Akhavanfard S, Coffman EJ, Cook EB, Stoykova S, Mukherjee S, Schoonmaker JA, Burger A, et al. 2011. The WTX tumor suppressor regulates mesenchymal progenitor cell fate specification. Dev Cell 20: 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mologni L, Dekhil H, Ceccon M, Purgante S, Lan C, Cleris L, Magistroni V, Formelli F, Gambacorti-Passerini CB 2010. Colorectal tumors are effectively eradicated by combined inhibition of {β}-catenin, KRAS, and the oncogenic transcription factor ITF2. Cancer Res 70: 7253–7263 [DOI] [PubMed] [Google Scholar]
- Morin PJ, Vogelstein B, Kinzler KW 1996. Apoptosis and APC in colorectal tumorigenesis. Proc Natl Acad Sci 93: 7950–7954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. 2011. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet 42: 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, Heatley SL, Holmfeldt L, Collins-Underwood JR, Ma J, et al. 2011. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature 471: 235–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung SJ, Rerko RM, Yan M, Platzer P, Guda K, Dotson A, Lawrence E, Dannenberg AJ, Lovgren AK, Luo G, et al. 2006. 15-Hydroxyprostaglandin dehydrogenase is an in vivo suppressor of colon tumorigenesis. Proc Natl Acad Sci 103: 12098–12102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massague J 2009. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell 138: 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Chen Y, Lim E, Wimberly H, Bailey ST, Imai Y, Rimm DL, Liu XS, Brown M 2011. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell 20: 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima M, Taketo MM 2002. COX selectivity and animal models for colon cancer. Curr Pharm Des 8: 1021–1034 [DOI] [PubMed] [Google Scholar]
- Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, et al. 2011. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature 471: 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phesse TJ, Parry L, Reed KR, Ewan KB, Dale TC, Sansom OJ, Clarke AR 2008. Deficiency of Mbd2 attenuates Wnt signaling. Mol Cell Biol 28: 6094–6103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakis P 2007. The many ways of Wnt in cancer. Curr Opin Genet Dev 17: 45–51 [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H 2005. Wnt signalling in stem cells and cancer. Nature 434: 843–850 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Paredes M, Esteller M 2011. Cancer epigenetics reaches mainstream oncology. Nat Med 17: 330–339 [DOI] [PubMed] [Google Scholar]
- Roh H, Green DW, Boswell CB, Pippin JA, Drebin JA 2001. Suppression of β-catenin inhibits the neoplastic growth of APC-mutant colon cancer cells. Cancer Res 61: 6563–6568 [PubMed] [Google Scholar]
- Ruteshouser EC, Robinson SM, Huff V 2008. Wilms tumor genetics: Mutations in WT1, WTX, and CTNNB1 account for only about one-third of tumors. Genes Chromosomes Cancer 47: 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahshor S, Woodgett JR 2005. The links between axin and carcinogenesis. J Clin Pathol 58: 225–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom OJ, Berger J, Bishop SM, Hendrich B, Bird A, Clarke AR 2003. Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nat Genet 34: 145–147 [DOI] [PubMed] [Google Scholar]
- Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, Kawasoe T, Ishiguro H, Fujita M, Tokino T, et al. 2000. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet 24: 245–250 [DOI] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. 2006. The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274 [DOI] [PubMed] [Google Scholar]
- Smartt HJ, Greenhough A, Ordonez-Moran P, Talero E, Cherry CA, Wallam CA, Parry L, Al Kharusi M, Roberts HR, Mariadason JM, et al. 2011. β-catenin represses expression of the tumour suppressor 15-prostaglandin dehydrogenase in the normal intestinal epithelium and colorectal tumour cells. Gut 10.1136/gutjnl-2011-300817 [DOI] [PubMed] [Google Scholar]
- Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, et al. 2003. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci 100: 8418–8423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Gabrielson E, Chen W, Anbazhagan R, van Engeland M, Weijenberg MP, Herman JG, Baylin SB 2002. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet 31: 141–149 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Toyota M, Carraway H, Gabrielson E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T, et al. 2008. Frequent epigenetic inactivation of Wnt antagonist genes in breast cancer. Br J Cancer 98: 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi H, Sasaki S, Suzuki H, Toyota M, Maruyama R, Nojima M, Yamamoto H, Omata M, Tokino T, Imai K, et al. 2008. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol 43: 378–389 [DOI] [PubMed] [Google Scholar]
- Tang W, Dodge M, Gundapaneni D, Michnoff C, Roth M, Lum L 2008. A genome-wide RNAi screen for Wnt/β-catenin pathway components identifies unexpected roles for TCF transcription factors in cancer. Proc Natl Acad Sci 105: 9697–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo JL, Kahn M 2011. The Wnt signaling pathway in cellular proliferation and differentiation: A tale of two coactivators. Adv Drug Deliv Rev 62: 1149–1155 [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F 1999. β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398: 422–426 [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP, et al. 2002. The β-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111: 241–250 [DOI] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, et al. 2005. Notch/γ-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature 435: 959–963 [DOI] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O’Meara S, Teague J, et al. 2009. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41: 521–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Veelen W, Le NH, Helvensteijn W, Blonden L, Theeuwes M, Bakker ER, Franken PF, van Gurp L, Meijlink F, van der Valk MA, et al. 2011. β-catenin tyrosine 654 phosphorylation increases Wnt signalling and intestinal tumorigenesis. Gut 60: 1204–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB 2003. Small interfering RNAs directed against β-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin Cancer Res 9: 1291–1300 [PubMed] [Google Scholar]
- Vincent A, Omura N, Hong SM, Jaffe A, Eshleman J, Goggins M 2011. Genome-wide analysis of promoter methylation associated with gene expression profile in pancreatic adenocarcinoma. Clin Cancer Res 17: 4341–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Mann JR, DuBois RN 2004. WNT and cyclooxygenase-2 cross-talk accelerates adenoma growth. Cell Cycle 3: 1512–1515 [DOI] [PubMed] [Google Scholar]
- Wang Y, Krivtsov AV, Sinha AU, North TE, Goessling W, Feng Z, Zon LI, Armstrong SA 2010. The Wnt/β-catenin pathway is required for the development of leukemia stem cells in AML. Science 327: 1650–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willert K, Jones KA 2006. Wnt signaling: Is the party in the nucleus? Genes Dev 20: 1394–1404 [DOI] [PubMed] [Google Scholar]
- Yang L, Wu X, Wang Y, Zhang K, Wu J, Yuan YC, Deng X, Chen L, Kim CC, Lau S, et al. 2011a. FZD7 has a critical role in cell proliferation in triple negative breast cancer. Oncogene 43: 4437–4446 [DOI] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z 2011b. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature 480: 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You L, He B, Xu Z, Uematsu K, Mazieres J, Mikami I, Reguart N, Moody TW, Kitajewski J, McCormick F, et al. 2004. Inhibition of Wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene 23: 6170–6174 [DOI] [PubMed] [Google Scholar]
- Zeng G, Apte U, Cieply B, Singh S, Monga SP 2007. siRNA-mediated β-catenin knockdown in human hepatoma cells results in decreased growth and survival. Neoplasia 9: 951–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Blum J, Chen A, Kwon HY, Jung SH, Cook JM, Lagoo A, Reya T 2007. Loss of β-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell 12: 528–541 [DOI] [PMC free article] [PubMed] [Google Scholar]