Abstract
Human glutaredoxin 3 (Glrx3) is an essential [2Fe-2S]-binding protein with ill-defined roles in immune cell response, embryogenesis, cancer cell growth, and regulation of cardiac hypertrophy. Similar to other members of the CGFS monothiol glutaredoxin (Grx) family, human Glrx3 forms homodimers bridged by two [2Fe-2S] clusters that are ligated by the conserved CGFS motifs and glutathione (GSH). We recently demonstrated that the yeast homologues of human Glrx3 and the yeast BolA-like protein Fra2 form [2Fe-2S]-bridged heterodimers that play a key role in signaling intracellular iron availability. Herein we provide biophysical and biochemical evidence that the two tandem Grx-like domains in human Glrx3 form similar [2Fe-2S]-bridged complexes with human BolA2. UV-visible absorption and CD, resonance Raman, and EPR spectroscopic analyses of recombinant [2Fe-2S] Glrx3 homodimers and [2Fe-2S] Glrx3-BolA2 complexes indicate that the Fe-S coordination environments in these complexes are virtually identical to the analogous complexes in yeast. Furthermore, we demonstrate that apo BolA2 binds to each Grx domain in the [2Fe-2S] Glrx3 homodimer forming a [2Fe-2S] BolA2-Glrx3 heterotrimer. Taken together, these results suggest that the unusual [2Fe-2S]-bridging Grx-BolA interaction is conserved in higher eukaryotes and may play a role in signaling cellular iron status in humans.
CGFS monothiol glutaredoxins (Grx) represent a subclass of Grxs present in all branches on the evolutionary tree. Both prokaryotes and eukaryotes typically possess single domain monothiol Grxs with the signature CGFS active site. Eukaryotes also have multi-domain CGFS-type Grxs with one monothiol thioredoxin (Trx)-like domain at the N-terminus and one or more monothiol Grx-like domains at the C-terminus (Fig. 1A). The specific molecular functions of CGFS Grxs are probably best characterized in yeast, for which emerging evidence suggests that they play critical roles in iron metabolism via formation of [2Fe-2S]-bridged homodimers (1, 2). The Fe-S ligands in CGFS Grxs are provided by the two Cys ligands from the Grx active sites and two glutathione molecules (GSH, or γ-glutamylcysteinylglycine) (3, 4). Eukaryotic single domain CGFS Grxs, such as S. cerevisiae and human Grx5, are localized to mitochondria where they are proposed to facilitate transfer of nascent Fe-S clusters to target proteins during maturation of Fe-S cluster-containing proteins (5-7). Multi-domain CGFS Grxs (e.g. yeast Grx3 and Grx4) are localized to the cytosol and/or nucleus. In yeast, [2Fe-2S]-bridged Grx3/4 homodimers are proposed to traffic iron from cytosolic pools to iron-dependent enzymes and the mitochondrion (2). In addition, yeast Grx3 and Grx4 help regulate cellular iron homeostasis by inhibiting the activity of the two Fe-responsive transcription factors (Aft1 and Aft2) that activate transcription of iron uptake and storage genes (2, 8-10). Both trafficking and regulation functions for Grx3/4 require binding of the GSH-ligated [2Fe-2S] cluster via the CGFS active site (2, 8).
Figure 1.
Domain structure and amino acid sequence alignment of yeast (y) and human (h) cytosolic CGFS-type Grxs. (A) Domain structure of yeast Grx3/Grx4 and human Glrx3. (B) Multiple sequence alignment of the two Grx-like domains (GrxA and GrxB) in human Glrx3 with the single Grx-like domains in yeast Grx3 and Grx4 using ClustalW2 (39). Identical residues are highlighted in black and similar residues are shown in gray. Red asterisks indicate the location of the signature CGFS active site.
Humans have one multi-domain CGFS Grx (Glrx3) that participates in a variety of signaling pathways and processes. Glrx3 was initially identified in T lymphocytes as a negative regulator of Protein Kinase C-θ and hence is frequently referred to in the literature as the PKC-interacting cousin of thioredoxin (PICOT) protein (13). Studies in rodents demonstrate that Glrx3 is also a negative regulator of cardiomyocyte hypertrophy and is essential for viability (14, 15). In addition, Glrx3 is overexpressed in certain human cancers and plays a role in regulating tumor growth and metastasis (16, 17). Interestingly, a recent study indicates that human Glrx3 binds iron in vivo and forms [2Fe-2S]-bridged homodimers in a similar manner to its yeast homologues (18). However, while yeast Grx3 and Grx4 each have one Grx-like domain, human Glrx3 has two tandem Grx-like domains (Fig. 1A) that both form a [2Fe-2S] bridge with another Glrx3 monomer. The two Grx-like domains in hGlrx3 share a high degree of sequence identity with each other (60%) and with the Grx-like domains in yeast Grx3 and Grx4 (~48–56%) (Fig. 1B). In contrast, the Trx-like domains in yeast and human CGFS-type Grxs are less well conserved (< 27% sequence identity) (19). The Trx-like domain is not involved in [2Fe-2S] binding since removal of this domain does not disrupt [2Fe-2S]-bridged homodimer formation in either yeast or human CGFS-type Grxs (1, 18). However, the Trx-like domain is required for the essential function of yeast Grx3/4 in both intracellular iron trafficking and regulation of iron metabolism, possibly via mediation of protein-protein interactions (20). Despite the identification of human Glrx3 as an [2Fe-2S]-binding protein and the well-documented role of other monothiol Grxs in iron metabolism, no studies have yet reported a specific connection between mammalian Glrx3 and iron metabolism.
Several lines of evidence, including affinity purification, yeast two-hybrid studies, and gene co-occurrence analysis, indicate that the monothiol Grxs functionally and physically interact with another widely conserved protein family, the BolA-like proteins (11). Genetic studies in yeast demonstrate that the Grx-BolA interaction is required for efficient iron-dependent inhibition of the Aft1/2 transcriptional activators (10). We recently unveiled the molecular details of the Grx-BolA interaction, demonstrating that the yeast BolA-like protein Fra2 forms a [2Fe-2S]-bridged heterodimeric complex with yeast Grx3 or Grx4, with iron ligands provided by a cysteine from Grx3/4, a histidine from Fra2, and GSH (1, 12). Under iron replete conditions, this [2Fe-2S]-bridged complex is proposed to inhibit Aft1/2 activity leading to deactivation of the iron regulon. These studies thus established the ubiquitous CGFS monothiol Grxs and BolA-like proteins as a novel type of Fe-S cluster binding regulatory complex.
Since the monothiol Grx-BolA interaction plays an important role in yeast iron metabolism, the purpose of this study was to determine whether human Glrx3 interacts with a human BolA-like protein in a similar manner. Using biochemical, spectroscopic, and analytical techniques, we demonstrate that each Grx-like domain in Glrx3 forms a [2Fe-2S]-bridged complex with human BolA2. Our results indicate that the coordination environment of the [2Fe-2S] cluster in these complexes is identical to their yeast homologues with both cysteinyl and histidyl ligation. Furthermore, we demonstrate that apo-BolA2 binds to the two tandem Grx domains in the [2Fe-2S] Glrx3 homodimer to form a [2Fe-2S] Glrx3-BolA2 heterotrimeric complex. Overall, these results suggest that the [2Fe-2S]-bridged Grx-BolA interaction is conserved in higher eukaryotes and may play a role in human iron metabolism.
EXPERIMENTAL PROCEDURES
Plasmid Construction
The full length cDNA for human Glrx3 (Glrx3) (Open Biosystems) with the Trx-like domain and both Grx-like domains was PCR-amplified and subcloned into pRSFDuet-1 (Novagen) using NdeI and KpnI restriction sites to generate pRSFDuet-1-Glrx3. For expression of one or both Grx-like domains (domains A and B, see Figure 1A), deletion mutants of human Glrx3, Glrx3130–335 (expressing Glrx3(A,B)) and Glrx3234–335 (expressing Glrx3(B)) were constructed by site-directed mutagenesis of pRSFDuet-1-Glrx3 (QuikChange Site-Directed Mutagenesis Kit; Stratagene) using primers listed in Supplementary Table 1 to generate pRSFDuet-1-Glrx3(K130M) and pRSFDuet-1-Glrx3(K234M), followed by digestion with NdeI and religation to create pRSFDuet-1-Glrx3130–335 and pRSFDuet-1-Glrx3234–335, respectively. pRSFDuet-1-Glrx3130–230 (expressing Glrx3(A)) was constructed through PCR amplification using pRSFDuet-1-Glrx3 as the template and primers listed in Supplementary Table 1, followed by insertion into NdeI and KpnI restriction sites in pRSFDuet-1. The cDNA for human BolA2 lacking the N-terminal 66 amino acids (Open Biosystems) was amplified by PCR and subcloned into NcoI and BamHI restriction sites in the first multiple cloning site of pETDuet-1 (Novagen) to generate pETDuet-1-BolA2Δ1–66. The sequence integrity of all plasmids was confirmed by double-stranded DNA sequencing (Environmental Genomics Facility, University of South Carolina School of Public Health).
Protein Expression and Purification
Human Glrx3 lacking the Trx-like domain but including both Grx domains in tandem (denoted Glrx3(A,B)) was overexpressed in E. coli strain BL21(DE3) and grown in LB at 30 °C until OD600 = 0.6–0.8 followed by induction with 1 mM isopropyl β-D-thiogalactoside (IPTG). The cells were collected 18 h after induction, resuspended in 50 mM Tris/MES, pH 8.0, 5 mM GSH, sonicated, and centrifuged to remove cell debris. The cell-free extract was loaded onto a Q Sepharose anion-exchange column (GE Healthcare) pre-equilibrated with 50 mM Tris/MES, pH 8.0, 5 mM GSH, and the protein was eluted with a 0–1 M NaCl gradient. The reddish-brown fractions containing [2Fe-2S] Glrx3(A,B) were pooled and (NH4)2SO4 was added to these fractions to reach a final concentration of 1 M. The protein was loaded onto a Phenyl-Sepharose column (GE Healthcare) pre-equilibrated with 50 mM Tris/MES, pH 8.0, 100 mM NaCl, 1 M (NH4)2SO4, 5 mM GSH and eluted with a decreasing (NH4)2SO4 gradient. The fractions containing apo- and holo-Glrx3(A,B) as judged by SDS-PAGE and UV-visible spectroscopy were concentrated with the addition of 5% glycerol (v/v) and stored at −80 °C.
Single domains Glrx3(A) and Glrx3(B) were overexpressed in BL21(DE3) and grown at 30 °C in LB until OD600 = 0.6–0.8. The cultures were induced with 1 mM IPTG and grown for 18 hours at 16 °C. Purification of Glrx3(A) and Glrx3(B) was performed using the same procedures described above for Glrx3(A,B), except that both of these constructs were collected in the flow-through of the Phenyl-Sepharose column as both apo- and holo-forms. We note here that Glrx3(A) purification resulted in low but measurable levels of the Fe-S-bound form. To facilitate further biochemical analysis of the holo-form, Glrx3(A) was subjected to anaerobic cysteine desulfurase-mediated Fe-S cluster reconstitution and repurification in the presence of GSH, ferrous ammonium sulfate, and L-cysteine as described previously (1). The spectroscopic features of this reconstituted form were identical to the as-purified Fe-S-bound form.
Co-expression of Glrx3 constructs Glrx3(A,B), Glrx3(A), and Glrx3(B) with BolA2 was accomplished by co-transformation of BL21(DE3) cells with one of the pRSFDuet-1-Glrx3 expression vectors and pETDuet-1-BolA2Δ1–66. Glrx3-BolA2 complexes were purified following the same protocol as above for Glrx3(A) and Glrx3(B). The fractions containing Glrx3-BolA2 were present in the Phenyl-Sepharose flow-through and further concentrated and loaded onto a HiLoad Superdex 75 gel filtration column (GE Healthcare) pre-equilibrated with 50 mM Tris/MES, pH 8.0, 150 mM NaCl, 5 mM GSH. The purest fractions of Glrx3-BolA2 complexes as judged by SDS-PAGE and UV-visible spectroscopy were collected, concentrated with the addition of 5% glycerol (v/v), and stored at −80 °C. For all the protocols described above, the purification steps using Q Sepharose and Phenyl-Sepharose columns were performed under anaerobic conditions (O2 < 5 ppm) in a glovebox (Coy Laboratory Products, Inc.) to preserve the Fe-S cluster, while the steps using the HiLoad Superdex 75 gel filtration column was done aerobically with N2-purged buffers.
For purification of BolA2 alone, BL21(DE3) cells were transformed with pETDuet-1-BolA2Δ1–66 and grown in 1 L of LB media at 30 °C until OD600 = 0.6–0.8. After an 18-h induction with 1 mM IPTG, the cells were collected, resuspended in 25 mM MES-NaOH, pH 6.0, sonicated, and centrifuged to remove cell debris. The cell-free extract was then loaded onto an SP FF column (GE Healthcare) pre-equilibrated with 25 mM MES-NaOH, pH 6.0, and the protein eluted with a 0–1 M NaCl gradient. BolA2-containing fractions were concentrated and loaded onto a HiLoad Superdex 75 gel filtration column (GE Healthcare) pre-equilibrated with 50 mM Tris/MES, pH 8.0, 150 mM NaCl. The purest fractions of BolA2 as judged by SDS-PAGE were collected and concentrated with the addition of 5% glycerol (v/v) and stored at −80 °C.
Biochemical and Spectroscopic Methods
The calculated extinction coefficient for each apo protein studied was used to standardize the Bradford assay (Bio-Rad). Subsequently, protein concentrations were routinely determined by the Bradford assay using BSA as the calibration standard. Iron concentrations were determined using the colorimetric ferrozine assay (21). Acid-labile sulfur concentrations were determined using published methods (22, 23). GSH measurements of the purified Fe-S protein complexes were described previously (1). Analytical gel filtration analyses were performed on a Superdex 75 10/300 GL column (GE Healthcare) pre-equilibrated with 50 mM Tris/MES, pH 8.0, 150 mM NaCl, 5 mM GSH and calibrated with the Low Molecular Weight Gel Filtration Calibration kit (GE Healthcare) as previously described (1). UV−visible absorption, circular dichroism (CD), resonance Raman, and X-band (9.6 GHz) EPR spectra were recorded as previously described (12). The samples for spectroscopic studies were handled under anaerobic conditions in a glove box (< 2 ppm O2).
Titration of [2Fe-2S]2+ Cluster-Bound Glrx3 with BolA2
The titration of [2Fe-2S]2+-bound Glrx3(A), Glrx3(B), or Glrx3(A,B) with apo-BolA2 was monitored under anaerobic conditions at room temperature using UV-visible CD spectroscopy. Reactions were carried out in 50 mM Tris/MES, pH 8.0, 5 mM GSH, with the [2Fe-2S]2+ cluster concentration kept constant at ~35 µM, and BolA2:[2Fe-2S] ratios varying from 0 to 2.5. Samples were equilibrated for 5 min at room temperature after addition of BolA2 prior to recording CD spectra. Similar saturation binding curves were obtained for more concentrated samples (200 µM [2Fe-2S]2+, 25–400 µM BolA2) indicating stoichiometric conditions at both [2Fe-2S]2+ concentrations tested.
RESULTS AND DISCUSSION
Glrx3 and BolA2 Form an Fe-S Cluster-Containing Complex
Given the importance of the [2Fe-2S] Fra2-Grx3/4 complex in yeast iron sensing, we sought to test whether human Glrx3 forms a similar complex with the human homologue of Fra2. Mammalian cells have three BolA homologues denoted BolA1, BolA2, and BolA3. The solution structure of mouse BolA2 has been determined (24); however, no specific molecular function has been assigned to any of the three human BolA proteins. A recent study demonstrates that a mutation in mitochondria-localized human BolA3 results in defects in mitochondrial Fe-S enzymes, suggesting a role in maturation of mitochondrial Fe-S proteins (25). Human BolA2 is the closest homologue to yeast Fra2 in the BolA phylogenetic tree (35% identical, 56% similar) (24, 26) and lacks a predicted mitochondrial targeting signal, thus it is the most likely partner for human Glrx3. To determine whether human Glrx3 interacts with BolA2, we cloned both sequences and expressed the recombinant proteins individually or together in E. coli. Untagged, full-length Glrx3 and BolA2 were not visibly detectable by SDS-PAGE analysis when expressed alone or together, indicating low expression levels or protein instability (data not shown). To overcome this problem, truncated constructs of both proteins were made to optimize expression of soluble protein. Glrx3 was successfully expressed with the two tandem Grx-like domains intact (residues 130–335), but lacking the N-terminal Trx-like domain (designated Glrx3(A,B)). In addition, the individual Grx-like domains were expressed separately (designated Glrx3(A) and Glrx3(B)). Improved BolA2 expression was achieved by removing the non-conserved N-terminus (residues 1–66).
Each Glrx3 construct (Glrx3(A), Glrx3(B), Glrx3(A,B)) was purified as apo and reddish-brown [2Fe-2S] bound complexes, in agreement with a previous report (18). The apparent molecular masses and the iron and acid-labile sulfide content of the holo complexes were consistent with the formation of approximately one bridging [2Fe-2S]2+ cluster for each single Grx-like domain complex (~0.90 [2Fe-2S]2+ clusters per Glrx3(A) homodimer and ~0.85 [2Fe-2S]2+ clusters per Glrx3(B) homodimer) and two bridging [2Fe-2S] clusters for the two-domain construct (~1.90 [2Fe-2S]2+ clusters per Glrx3(A,B) homodimer) (Fig. 2, Table 1). Thus, as in yeast (1) and as previously demonstrated for human Glrx3 (18), removal of the Trx-like domain does not impact the [2Fe-2S]-bridging homodimeric interaction. Upon co-expression with BolA2, each Glrx3 construct co-eluted with BolA2 as a reddish-brown complex as well. The calibrated molecular masses of Glrx3(A)-BolA2 and Glrx3(B)-BolA2 indicate that the single domain Glrx3(A) and Glrx3(B) constructs each form heterodimeric complexes with BolA2. Iron and acid-labile sulfide analyses of these samples indicate ~0.65 [2Fe-2S]2+ clusters per Glrx3(A)-BolA2 heterodimer and ~0.75 [2Fe-2S]2+ clusters per Glrx3(B)-BolA2 heterodimer. The apparent molecular mass of Glrx3(A,B)-BolA2 is essentially similar to that of holo Glrx3(A,B), and also the sum of the calibrated masses of Glrx3(A)-BolA2 and Glrx3(B)-BolA2, which suggests that BolA2 and Glrx3(A,B) form a 2:1 heterotrimeric complex. The Fe and S content of this form is consistent with ~1.5 [2Fe-2S]2+ clusters per Glrx3(A,B)-BolA2 heterotrimer (Table 1). Overall, the analytical and biochemical evidence clearly demonstrate that each Grx-like domain in the Glrx3 monomer is able to coordinate a [2Fe-2S]2+ cluster with either a Grx-like domain from another Glrx3 monomer or from BolA2. This result parallels our finding in yeast that Grx3 and Grx4 can form either [2Fe-2S] bridged homodimers or [2Fe-2S]-bridged heterodimers with the yeast BolA-like protein Fra2 (1). The Fe-S clusters in human Glrx3 homodimers are oxidatively labile and are gradually degraded in air over a period of ~1 hr. In contrast, the Glrx3-BolA heterocomplexes are stable in air for at least 3 hours (data not shown). Thus BolA2 binding stabilizes the [2Fe-2S]2+ clusters against oxidative degradation as previously demonstrated for yeast [2Fe-2S] Fra2-Grx3/4 complexes (1).
Figure 2.
The Grx-like domains of Glrx3 co-purify with BolA2 as [2Fe-2S]-bridged complexes. (A) SDS-PAGE analysis of purified Glrx3-BolA2 [2Fe-2S]-bound complexes. – indicates protein samples obtained by expression and purification of Glrx3 constructs alone, + indicates samples derived from co-expression and co-purification of Glrx3 constructs with BolA2. The gel indicates that all purified complexes are >95% pure. (B) Gel filtration chromatograms of apo (upper chromatogram) and [2Fe-2S] forms (bottom chromatogram) of Glrx3 and BolA2 (0.5–1 mg loaded). The elution positions of the molecular weight standards used for column calibration are shown at the top of the chromatograms.
Table 1.
Molecular mass, iron, and acid-labile sulfide analyses of Glrx3 and BolA2 complexes. Experimental mass (shown in Da) is from gel filtration analyses in Fig. 2B. Fe and S measurements are the averages of three independent samples. The column labeled “Model“ depicts the predicted domain arrangement and oligomeric state of the apo and [2Fe-2S]-bridged Glrx3 and BolA2 complexes.
| Sample | Experimental mass |
Theoretical mass |
Fea | Sa | Model |
|---|---|---|---|---|---|
| Apo Glrx3(A) | 21,200 | 11,534 (monomer) |
NDb | ND | |
| Apo Glrx3(B) | 23,100 | 11,623 (monomer) |
ND | ND | |
| Apo Glrx3(A,B) | 37,700 | 23,433 (monomer) |
ND | ND | |
| Apo BolA2 | 12,900 | 10,116 (monomer) |
ND | ND | |
| [2Fe-2S] Glrx3(A) | 34,500 | 23,068 (homodimer) |
1.8 ± 0.1 | 1.9 ± 0.1 |  |
| [2Fe-2S] Glrx3(B) | 35,200 | 23,246 (homodimer) |
1.7 ± 0.1 | 1.7 ± 0.1 |  |
| [2Fe-2S] Glrx3(A,B) | 51,400 | 46,866 (homodimer) |
3.8 ± 0.1 | 4.0 ± 0.1 |  |
| [2Fe-2S] Glrx3(A)-BolA2 | 28,400 | 21,650 (heterodimer) |
1.3 ± 0.1 | 1.4 ± 0.1 |  |
| [2Fe-2S] Glrx3(B)-BolA2 | 28,400 | 21,739 (heterodimer) |
1.5 ± 0.1 | 1.6 ± 0.1 |  |
| [2Fe-2S] Glrx3(A,B)-BolA2 | 51,900 | 43,665 (heterotrimer) |
3.0 ± 0.1 | 3.2 ± 0.1 |  |
Fe and acid-labile S measurements are reported as mol Fe or S per mol of complex (dimer or trimer).
Not determined
UV-Visible Absorption and CD Spectroscopy Reveal Differences in Cluster Coordination Environments in Human [2Fe-2S] Glrx3 and [2Fe-2S] Glrx3-BolA2 Complexes
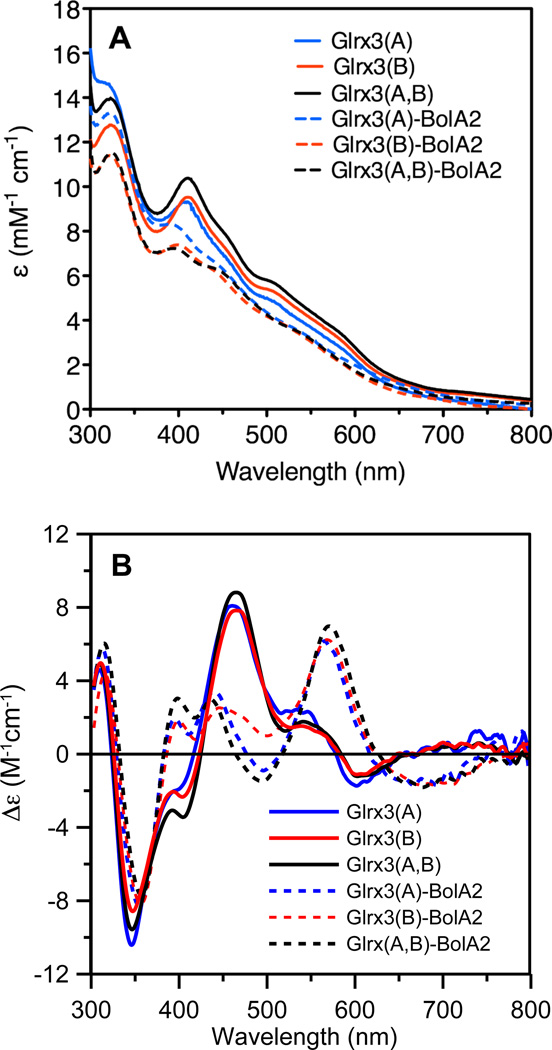
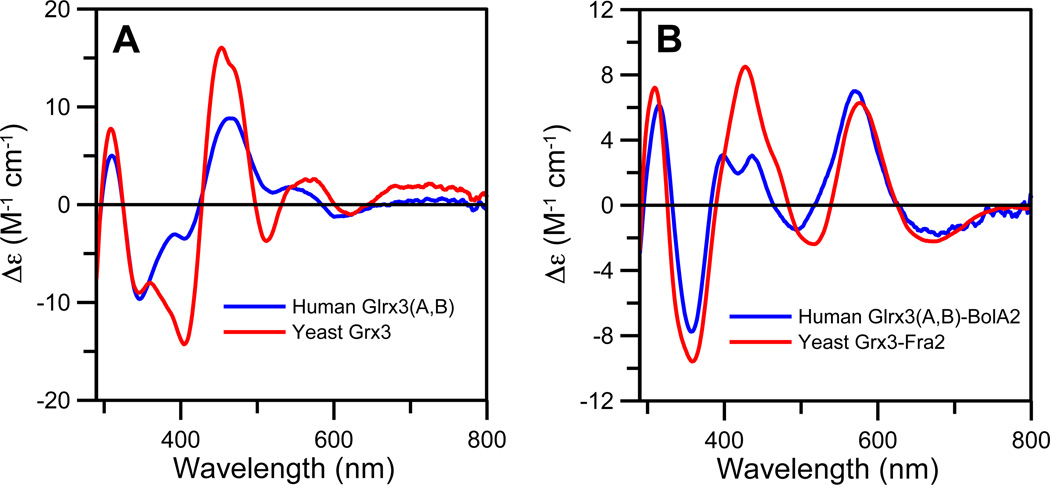
The UV-visible absorption spectra of the holodimeric forms of all three Glrx3 constructs are nearly identical with dominant absorption peaks at 320 and 410 nm similar to [2Fe-2S]2+-bridged monothiol Grx homodimers from other organisms (1, 27, 28) (Fig. 3). In contrast, the UV-visible CD spectra of [2Fe-2S]2+-bridged monothiol Grx homodimers display considerable variability (1, 27,29). This variability is illustrated by comparing the CD spectra of human Glrx3(A,B) and S.cerevisiae Grx3, which represent the extremes in the range of observed spectra (Fig. 4A). Notably, the spectra differ in terms of the magnitude of CD intensity rather than the energies of discrete electronic transitions. Taken together with near-identical UV-visible absorption and resonance Raman spectra, vide infra, this indicates changes in the asymmetry of the cluster protein environment, rather than significant changes in the cluster coordination environment. Such changes in the cluster protein environment presumably reflect conformational or oligomeric state differences resulting from concentration or medium effects. Indeed in the case of plant GrxS14, addition of DTT resulted in conversion from one form to the other (27). More structural work is clearly required to address the origin of the differences in the CD spectra of [2Fe-2S]2+-bridged monothiol Grxs.
Figure 3.
(A) UV-visible absorption and (B) CD spectra of [2Fe-2S]-bridged Glrx3 homodimers (solid lines) and Glrx3-BolA2 complexes (dashed lines). ε and Δε values are based on the [2Fe-2S] cluster concentration.
Figure 4.
Comparison of the UV-visible CD spectra of [2Fe-2S]-bridged human Glrx3(A,B) and S. cerevisiae Grx3 homodimers (A) and [2Fe-2S]-bridged human Glrx3(A,B)-BolA2 and S. cerevisiae Grx3-Fra2 complexes (B). The S. cerevisiae Grx3 and Grx3-Fra2 spectra are taken from ref. (1) and Δε values are based on the [2Fe-2S] cluster concentration.
Both the UV-visible absorption and CD spectra of the [2Fe-2S]2+ centers in all Glrx3-BolA2 complexes are significantly different to those of the [2Fe-2S]2+ centers in the holodimer forms of the three Glrx3 constructs (Fig. 3). In particular, the dominant visible absorption bands in the 375–500 nm region in [2Fe-2S]2+ Glrx3 homodimers are blue-shifted by 15–20 nm in all [2Fe-2S]2+ Glrx3-BolA2 complexes, with concomitant major changes in the intensity and wavelengths of the visible CD bands. Overall, these results demonstrate distinct differences in cluster ligation and asymmetry of the cluster environments for the [2Fe-2S]2+ centers in Glrx3-BolA2 complexes compared to Glrx3 homodimers. Moreover, both the UV-visible absorption and CD spectra of the human Glrx3-BolA2 complexes are similar to that of the S. cerevisiae Grx3-Fra2 heterodimer complex (see Figure 4B for a comparison of the CD spectra), which has been shown to involve a subunit-bridging [2Fe-2S]2+ cluster that is ligated by cysteines from Grx3 and GSH, and a conserved histidine in Fra2.
Resonance Raman and EPR Studies of Human Glrx3 and Glrx3-BolA2 complexes
The resonance Raman spectra of [2Fe-2S]2+ centers in the Fe-S stretching region are exquisitely sensitive to changes in cluster ligation and the Fe-S-C-C dihedral angles of coordinated cysteine residues (30-32). Hence the observation of almost identical resonance Raman spectra for the [2Fe-2S]2+ centers in each of the three human Glrx3 and S. cerevisiae Grx3 homodimers (Fig. 5A) and in plant chloroplast GrxS14 (27) demonstrates identical [2Fe-2S]2+ clusters with analogous coordination environments. In each case, the number and frequencies of the observed Fe-S stretching modes are characteristic of all-cysteinyl ligation and therefore in accord with subunit-bridging [2Fe-2S]2+ clusters ligated by the active-site cysteines of the two Grxs and two GSHs as characterized by crystallography (3, 4).
Figure 5.
Comparison of the resonance Raman spectra of [2Fe-2S]-bridged human Glrx3 and S. cerevisiae Grx3 homodimers (A) and human Glrx3-BolA2 and S. cerevisiae Grx3-Fra2 complexes (B) using 457.9-nm laser excitation. Samples were ~ 2 mM in [2Fe–2S] cluster and were in the form of a frozen droplet at 17 K. Each spectrum is the sum of 100 scans, with each scan involving photon counting for 1 s at 0.5 cm−1increments with 6 cm−1spectral resolution. Bands due to lattice modes of ice have been subtracted from all spectra. The S. cerevisiae Grx3 and Grx3-Fra2 spectra are taken from ref. (1).
Our previous spectroscopic studies of the S. cerevisiae Grx3-Fra2 complex revealed that heterodimeric complex formation is accompanied by dramatic changes in the [2Fe-2S] cluster ligation, environment, and redox properties (1). For example, the resonance Raman spectrum reveals frequency shifts, particularly in the low energy region, that are indicative of partial histidyl cluster ligation based on comparison with published data for Rieske-type proteins (two His ligands at one Fe site) (31, 33, 34), His-to-Cys Rieske-type protein variants (one His ligand) (35) and mitoNEET (one His ligand) (36). Mixing of the Fe-N(His) and Fe-S(Cys) stretching modes results in splitting of the out-of-phase predominantly Fe-S(Cys) stretching mode at 288 cm−1 into bands at 275 and 300 cm−1 (Fig. 5). Moreover, both EXAFS and 14N ENDOR studies confirmed the presence of a single His ligand for the [2Fe-2S]2+, + cluster in the S. cerevisiae Grx3-Fra2 complex (1) and subsequent mutagenesis studies identified the His ligand as His103 in Fra2 (12), which is highly conserved in all BolA-type proteins, including human BolA2. Comparison of the resonance Raman spectra of [2Fe-2S]2+-bridging human Glrx3 homodimers and Glrx3-BolA2 complexes (Fig. 5) clearly demonstrates analogous frequency shifts to those observed for the [2Fe-2S]2+-bridging S. cerevisiae Grx3 homodimers and Grx3-Fra2 heterodimers, indicating analogous cluster coordination in each of the human Glrx3-BolA2 complexes. In particular, the out-of-phase predominantly Fe-S(Cys) stretching mode at 288 cm−1 is split into two bands at 271 and 296 cm−1, indicating a single His ligand for the [2Fe-2S]2+ cluster. Overall, the frequencies and relative intensities of the bands in the resonance Raman spectra of the human Glrx3-BolA2 complexes are very similar to those observed for the S. cerevisiae Grx3-Fra2 complex except for frequencies downshifts of up to 7 cm−1, indicating slightly weaker bonding for the [2Fe-2S]2+ centers in the human Glrx3-BolA2 complexes.
EPR and absorption studies of cluster-bound homodimeric S. cerevisiae Grx3 have shown that the S = 0 [2Fe-2S]2+ center is irreversibly degraded, via a transient S = 1/2 [2Fe-2S]+ cluster, upon reduction with sodium dithionite under anaerobic conditions (1, 27). Rapid freezing within 5 s of anaerobic addition of stoichiometric dithionite revealed a slow relaxing axial S = 1/2 resonance (g∥ = 2.03, g⊥ = 1.94, gav ~ 1.97) accounting for 0.2 spins/[2Fe-2S] cluster, which was not observed in samples treated with 10-fold excess of dithionite and incubated for 10 min before freezing. Analogous behavior was observed for the [2Fe-2S]2+ cluster-bound human Glrx3(A) homodimer (Figure 6A), which exhibits a very similar axial resonance from a transient S = 1/2 [2Fe-2S]+ (g∥ = 2.02, g⊥ = 1.94, gav ~ 1.97), that is still observed without significant broadening at 70 K and accounts for 0.1 spins/[2Fe-2S] cluster, upon reduction with stoichiometric dithionite and rapid freezing. The additional feature at g = 2.0 is attributed to an isotropic radical species based on variable temperature EPR studies. Hence the [2Fe-2S]2+ center in the Glrx3(A) homodimer is both oxidatively and reductively labile. In contrast, the [2Fe-2S]+ centers in dithionite-reduced samples of human Glrx3(B) and Glrx3(A,B) are stable and give rise to slow relaxing rhombic resonances (g = 2.01, 1.97, 1.92, gav = 1.97 and g = 2.01, 1.95, 1.91, gav = 1.96, respectively), see Figure 6A, that each account for 1.0 spins/[2Fe-2S] cluster. The latter is likely to be a composite resonance from two non-interacting [2Fe-2S]+ clusters involving a slightly modified and stabilized form of the Glrx(A) [2Fe-2S]+ cluster, since no half-field signal indicative of weak magnetic interaction between the two [2Fe-2S]+ clusters was observed. This is in accord with the proposed structure for the [2Fe-2S]-bridged human Glrx3 homodimer, which has the two clusters separated by at least 25 Å (18). While the origin of the observed differences in the stabilities of the [2Fe-2S]+ centers and the anisotropies of the EPR resonances from the individual A and B domains and in the combined (A,B) domains of human Glrx3 is unknown at present, the gav values (1.96–1.97) are consistent with complete cysteinyl ligation being preserved on reduction in all cases (1).
Figure 6.
X-band EPR spectra of the S = 1/2 [2Fe-2S]+ centers in dithionite-reduced human Glrx3 and S. cerevisiae Grx3 (A) and human Glrx3-BolA2 and S. cerevisiae Grx3-Fra2 complexes (B). Samples were reduced under anaerobic conditions by addition of stoichiometric sodium dithionite (i.e. a 2-fold excess of reducing equivalents) and frozen within 5 s in liquid nitrogen. EPR conditions: microwave frequency, 9.60 GHz; modulation frequency, 100 KHz; modulation amplitude, 0.65 mT; microwave power; 1 mW; temperature, 26 K. The S. cerevisiae Grx3 and Grx3-Fra2 spectra are taken from ref. (1).
Evidence that the single histidyl ligand for the [2Fe-2S]2+ centers in the human Glrx3-BolA complexes is retained in the reduced cluster was provided by EPR spectroscopy (Fig. 6B). The [2Fe-2S] center in the S. cerevisiae Grx3-Fra2 complex was found to undergo reversible redox cycling and to exhibit a stable S = 1/2 [2Fe-2S]+ center with a near- axial EPR signal (g = 2.01, 1.92, ~1.87, gav ~ 1.93) accounting for 1.0 spins/[2Fe-2S] cluster on dithionite reduction (1). The low gav value, compared to [2Fe-2S]+ centers with complete cysteinyl ligation (typically gav ~ 1.97), is characteristic of [2Fe-2S]+ centers with one histidyl ligand and this was confirmed by 14N ENDOR spectroscopy (1). Hence the observation of analogous, stable S = 1/2 [2Fe-2S]+ centers (g = 2.01, 1.91, 1.88, gav ~ 1.93) accounting for 1.0 spins/[2Fe-2S] cluster in dithionite-reduced samples of each of the human Glrx3-BolA2 complexes provides compelling evidence for analogous histidyl-ligated [2Fe-2S]+ clusters (Fig. 6B). In addition, the observation that the EPR spectrum of the two [2Fe-2S]+ centers in human Glrx3(A,B)-BolA2 has the same line shape and is not significantly broadened compared to that of the single [2Fe-2S]+ centers in human Glrx3(A)-BolA2 and Glrx3(B)-BolA2, indicates no significant magnetic interaction between the clusters in human Glrx3(A,B)-BolA2, suggesting that the clusters are separated by at least 20 Å.
BolA2 Binds to [2Fe-2S] Glrx3 Forming a [2Fe-2S] BolA2-Glrx3 Heterotrimer
To determine the stoichiometry and nature of the binding interaction between BolA2 and [2Fe-2S] Glrx3, we monitored changes in the CD spectrum of the [2Fe-2S] Glrx3(A,B) homodimer upon titration with apo-BolA2. As shown in Figure 7A, increasing concentrations of BolA2 lead to changes in the CD spectrum suggesting increased formation of the [2Fe-2S] Glrx3(A,B)-BolA2 heterotrimer. The CD spectral changes were plotted as a function of the BolA2:[2Fe-2S] ratio to evaluate the binding interaction (Fig. 7A, bottom). The saturation binding experiment suggests that the stoichiometry of the complex is 0.6 ~ 0.7 BolA2 per [2Fe-2S] cluster (or 1.2 ~ 1.4 BolA2 per Glrx3(A,B) homodimer). This result differs from our parallel studies in yeast demonstrating that yeast Fra2 binds [2Fe-2S] Grx3 with 1:1 stoichiometry (12). Gel filtration chromatography and SDS-PAGE analysis of the titration samples confirmed formation of the Glrx3(A,B)-BolA2 heterotrimer (data not shown); however, residual [2Fe-2S] Glrx3(A,B) homodimer was also isolated, indicating incomplete conversion from homodimer to heterotrimer. This interpretation is consistent with the final titration CD spectrum that suggests a mixture of [2Fe-2S] Glrx3(A,B) and [2Fe-2S] Glrx3(A,B)-BolA2 (compare red line in Figure 7A to as-purified spectra of [2Fe-2S] Glrx3(A,B) and [2Fe-2S] Glrx3(A,B)-BolA2 in Fig. 3). Given the presence of two tandem Grx-like domains in Glrx3, it is possible that binding of BolA2 to one domain may preclude BolA2 binding to the second domain, or that BolA2 may have different affinities for the two Grx-like domains. However, we performed similar BolA2 titration studies on single domain [2Fe-2S] Glrx3(A) and [2Fe-2S] Glrx3(B) homodimers, and found similar stoichiometries (0.5–0.7 BolA2 per [2Fe-2S]) and CD spectral changes to the [2Fe-2S] Glrx3(A,B) homodimer (Fig. 7B,C). Thus, the two Grx-like domains do not have significant differences in BolA2 binding ability and the incomplete conversion is not due to the presence of the second domain. Overall, these results indicate that BolA2 can partially displace Glrx3 monomers to form [2Fe-2S]-bridged complexes with the Grx-like domains of Glrx3. However, the data also suggest that a fraction of both the single and double domain Glrx3 homodimers is in a conformation that is nonfunctional for interaction with BolA2.
Figure 7.
Titration of [2Fe-2S]2+-Glrx3(A,B), -Glrx3(A), and -Glrx3(B) homodimers with apo BolA2 monitored by UV-visible CD spectroscopy. The top panels show the CD spectra for [2Fe-2S]2+ cluster-bound Glrx3 homodimers (blue lines) titrated with 0.08 – 2.5 fold excess BolA2 (black lines). (A), (B), and (C) correspond to Glrx3(A,B), Glrx3(A), and Glrx3(B), respectively. The red lines correspond to 2–2.5-fold excess BolA2 and the arrows at selected wavelengths indicate the direction of intensity change with increasing BolA2 concentration. Δε values are based on the [2Fe-2S]2+ cluster concentration (36 µM for Glrx3(A,B), 32 µM for Glrx3(A) and 38 µM for Glrx3(B)). (A)–(C), Bottom, Plots of the maximum difference in CD intensity (at 465 and 400 nm) as a function of the BolA2:[2Fe-2S]2+ cluster ratio. The vertical dotted lines at 0.63 BolA2:[2Fe-2S]2+ in (A), 0.68 in (B), and 0.51 in (C) are drawn at the intercept of the two solid lines.
In summary, our studies demonstrate that the [2Fe-2S]-bridging Grx-BolA interaction first detected in yeast is conserved in humans. Whether human Glrx3-BolA complexes are also involved in human iron metabolism remains unknown, but the available information does provide several clues linking human Glrx3 and Grx-BolA complexes with iron metabolism. Firstly, human Glrx3 binds iron in vivo (18), indicating its involvement in iron metabolism. Secondly, the human mitochondrial BolA homologue (BolA3) plays an essential role in maturation of mitochondrial Fe-S proteins, presumably via complex formation with the mitochondrial CGFS-type glutaredoxin Grx5 (25). Thirdly, human Glrx3 also interacts with the Fe-S binding protein CIAPIN1 (37), which is required for cytosolic Fe-S cluster biogenesis (38). Finally, human Glrx3 partially rescues the growth defects and iron accumulation in some S. cerevisiae grx3Δgrx4Δ mutants, suggesting that human Glrx3 can partially complement the functions of yeast Grx3/4 in iron homeostasis (15). Taken together with the findings presented herein, these results suggest that human Glrx3 and BolA2 are likely to play important roles in iron metabolism via formation of [2Fe-2S]-bridged complexes. However, more work will be needed to clarify the roles of [2Fe-2S] Glrx3 vs. [2Fe-2S] Glrx3-BolA2 complexes in vivo, as well as the specific function of the Trx-like domain.
Supplementary Material
ABBREVIATIONS
- Grx
glutaredoxin
- Trx
thioredoxin
- GSH
glutathione
- CD
circular dichroism
- EPR
electron paramagnetic resonance
- EXAFS
extended X-ray absorption fine structure
- ENDOR
electron-nuclear double resonance spectroscopy
Footnotes
This work was supported by NIH grants ES13780 (to C.E.O.) and GM62524 (to M.K.J.).
SUPPORTING INFORMATION AVAILABLE
Supplementary Table S1 shows the primers used for cloning and mutagenesis. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Li H, Mapolelo DT, Dingra NN, Naik SG, Lees NS, Hoffman BM, Riggs-Gelasco PJ, Huynh BH, Johnson MK, Outten CE. The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry. 2009;48:9569–9581. doi: 10.1021/bi901182w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muhlenhoff U, Molik S, Godoy JR, Uzarska MA, Richter N, Seubert A, Zhang Y, Stubbe J, Pierrel F, Herrero E, Lillig CH, Lill R. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 2010;12:373–385. doi: 10.1016/j.cmet.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwema T, Picciocchi A, Traore DA, Ferrer JL, Chauvat F, Jacquamet L. Structural basis for delivery of the intact [Fe2S2] cluster by monothiol glutaredoxin. Biochemistry. 2009;48:6041–6043. doi: 10.1021/bi900440m. [DOI] [PubMed] [Google Scholar]
- 4.Johansson C, Roos AK, Montano SJ, Sengupta R, Filippakopoulos P, Guo K, von Delft F, Holmgren A, Oppermann U, Kavanagh KL. The crystal structure of human GLRX5: iron-sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem. J. 2010;433:303–311. doi: 10.1042/BJ20101286. [DOI] [PubMed] [Google Scholar]
- 5.Muhlenhoff U, Gerber J, Richhardt N, Lill R. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 2003;22:4815–4825. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Manzaneque MT, Tamarit J, Belli G, Ros J, Herrero E. Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell. 2002;13:1109–1121. doi: 10.1091/mbc.01-10-0517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye H, Jeong SY, Ghosh MC, Kovtunovych G, Silvestri L, Ortillo D, Uchida N, Tisdale J, Camaschella C, Rouault TA. Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J. Clin. Invest. 2010;120:1749–1761. doi: 10.1172/JCI40372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ojeda L, Keller G, Muhlenhoff U, Rutherford JC, Lill R, Winge DR. Role of glutaredoxin-3 and glutaredoxin-4 in the iron regulation of the Aft1 transcriptional activator in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:17661–17669. doi: 10.1074/jbc.M602165200. [DOI] [PubMed] [Google Scholar]
- 9.Pujol-Carrion N, Belli G, Herrero E, Nogues A, de la Torre-Ruiz MA. Glutaredoxins Grx3 and Grx4 regulate nuclear localisation of Aft1 and the oxidative stress response in Saccharomyces cerevisiae. J. Cell Sci. 2006;119:4554–4564. doi: 10.1242/jcs.03229. [DOI] [PubMed] [Google Scholar]
- 10.Kumanovics A, Chen OS, Li L, Bagley D, Adkins EM, Lin H, Dingra NN, Outten CE, Keller G, Winge D, Ward DM, Kaplan J. Identification of FRA1 and FRA2 as genes involved in regulating the yeast iron regulon in response to decreased mitochondrial iron-sulfur cluster synthesis. J. Biol. Chem. 2008;283:10276–10286. doi: 10.1074/jbc.M801160200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huynen MA, Spronk CA, Gabaldon T, Snel B. Combining data from genomes, Y2H and 3D structure indicates that BolA is a reductase interacting with a glutaredoxin. FEBS Lett. 2005;579:591–596. doi: 10.1016/j.febslet.2004.11.111. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Mapolelo DT, Dingra NN, Keller G, Riggs-Gelasco PJ, Winge DR, Johnson MK, Outten CE. Histidine 103 in Fra2 is an iron-sulfur cluster ligand in the [2Fe-2S] Fra2-Grx3 complex and is required for in vivo iron signaling in yeast. J. Biol. Chem. 2011;286:867–876. doi: 10.1074/jbc.M110.184176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Witte S, Villalba M, Bi K, Liu Y, Isakov N, Altman A. Inhibition of the c-Jun N-terminal kinase/AP-1 and NF-kappaB pathways by PICOT, a novel protein kinase C-interacting protein with a thioredoxin homology domain. J. Biol. Chem. 2000;275:1902–1909. doi: 10.1074/jbc.275.3.1902. [DOI] [PubMed] [Google Scholar]
- 14.Cha H, Kim JM, Oh JG, Jeong MH, Park CS, Park J, Jeong HJ, Park BK, Lee YH, Jeong D, Yang DK, Bernecker OY, Kim do H, Hajjar RJ, Park WJ. PICOT is a critical regulator of cardiac hypertrophy and cardiomyocyte contractility. J. Mol. Cell. Cardiol. 2008;45:796–803. doi: 10.1016/j.yjmcc.2008.09.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng NH, Zhang W, Chen WQ, Jin J, Cui X, Butte NF, Chan L, Hirschi KD. A mammalian monothiol glutaredoxin, Grx3, is critical for cell cycle progression during embryogenesis. FEBS J. 2011;278:2525–2539. doi: 10.1111/j.1742-4658.2011.08178.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu Y, Wang J, Ray PS, Guo H, Huang J, Shin-Sim M, Bukoye BA, Liu B, Lee AV, Lin X, Huang P, Martens JW, Giuliano AE, Zhang N, Cheng NH, Cui X. Thioredoxin-like 2 regulates human cancer cell growth and metastasis via redox homeostasis and NF-kappaB signaling. J. Clin. Invest. 2011;121:212–225. doi: 10.1172/JCI43144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cha MK, Kim IH. Preferential overexpression of glutaredoxin3 in human colon and lung carcinoma. Cancer Epidemiol. 2009;33:281–287. doi: 10.1016/j.canep.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Haunhorst P, Berndt C, Eitner S, Godoy JR, Lillig CH. Characterization of the human monothiol glutaredoxin 3 (PICOT) as iron-sulfur protein. Biochem. Biophys. Res. Commun. 2010;394:372–376. doi: 10.1016/j.bbrc.2010.03.016. [DOI] [PubMed] [Google Scholar]
- 19.Gibson LM, Dingra NN, Outten CE, Lebioda L. Structure of the thioredoxin-like domain of yeast glutaredoxin 3. Acta Crystallogr. D Biol. Crystallogr. 2008;64:927–932. doi: 10.1107/S0907444908021641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann B, Uzarska MA, Berndt C, Godoy JR, Haunhorst P, Lillig CH, Lill R, Muhlenhoff U. The multidomain thioredoxin-monothiol glutaredoxins represent a distinct functional group. Antioxidants & redox signaling. 2011;15:19–30. doi: 10.1089/ars.2010.3811. [DOI] [PubMed] [Google Scholar]
- 21.Riemer J, Hoepken HH, Czerwinska H, Robinson SR, Dringen R. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal. Biochem. 2004;331:370–375. doi: 10.1016/j.ab.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 22.Beinert H. Semi-micro methods for analysis of labile sulfide and of labile sulfide plus sulfane sulfur in unusually stable iron-sulfur proteins. Anal. Biochem. 1983;131:373–378. doi: 10.1016/0003-2697(83)90186-0. [DOI] [PubMed] [Google Scholar]
- 23.Broderick JB, Henshaw TF, Cheek J, Wojtuszewski K, Smith SR, Trojan MR, McGhan RM, Kopf A, Kibbey M, Broderick WE. Pyruvate formate-lyase-activating enzyme: strictly anaerobic isolation yields active enzyme containing a [3Fe-4S](+) cluster. Biochem. Biophys. Res. Commun. 2000;269:451–456. doi: 10.1006/bbrc.2000.2313. [DOI] [PubMed] [Google Scholar]
- 24.Kasai T, Inoue M, Koshiba S, Yabuki T, Aoki M, Nunokawa E, Seki E, Matsuda T, Matsuda N, Tomo Y, Shirouzu M, Terada T, Obayashi N, Hamana H, Shinya N, Tatsuguchi A, Yasuda S, Yoshida M, Hirota H, Matsuo Y, Tani K, Suzuki H, Arakawa T, Carninci P, Kawai J, Hayashizaki Y, Kigawa T, Yokoyama S. Solution structure of a BolA-like protein from Mus musculus. Protein Sci. 2004;13:545–548. doi: 10.1110/ps.03401004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cameron JM, Janer A, Levandovskiy V, Mackay N, Rouault TA, Tong WH, Ogilvie I, Shoubridge EA, Robinson BH. Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am. J. Hum. Genet. 2011;89:486–495. doi: 10.1016/j.ajhg.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou YB, Cao JB, Wan BB, Wang XR, Ding GH, Zhu H, Yang HM, Wang KS, Zhang X, Han ZG. hBolA, novel non-classical secreted proteins, belonging to different BolA family with functional divergence. Mol. Cell. Biochem. 2008;317:61–68. doi: 10.1007/s11010-008-9809-2. [DOI] [PubMed] [Google Scholar]
- 27.Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Johnson MK, Rouhier N. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe-2S] clusters. EMBO J. 2008;27:1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Picciocchi A, Saguez C, Boussac A, Cassier-Chauvat C, Chauvat F. CGFS-type monothiol glutaredoxins from the cyanobacterium Synechocystis PCC6803 and other evolutionary distant model organisms possess a glutathione-ligated [2Fe-2S] cluster. Biochemistry. 2007;46:15018–15026. doi: 10.1021/bi7013272. [DOI] [PubMed] [Google Scholar]
- 29.Yeung N, Gold B, Liu NL, Prathapam R, Sterling HJ, Willams ER, Butland G. The E. coli Monothiol Glutaredoxin GrxD Forms Homodimeric and Heterodimeric FeS Cluster Containing Complexes. Biochemistry. 2011;50:8957–8969. doi: 10.1021/bi2008883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han S, Czernuszewicz RS, Kimura T, Adams MWW, Spiro TG. Fe2S2 protein resonance Raman-spectra revisited - Structural variations among adrenodoxin, ferredoxin, and red paramagnetic protein. J. Amer. Chem. Soc. 1989;111:3505–3511. [Google Scholar]
- 31.Kuila D, Schoonover JR, Dyer RB, Batie CJ, Ballou DP, Fee JA, Woodruff WH. Resonance Raman studies of Rieske-type proteins. Biochim. Biophys. Acta. 1992;1140:175–183. doi: 10.1016/0005-2728(92)90007-o. [DOI] [PubMed] [Google Scholar]
- 32.Moulis JM, Davasse V, Golinelli MP, Meyer J, Quinkal I. The coordination sphere of iron-sulfur clusters: Lessons from site-directed mutagenesis experiments. J. Biol. Inorg. Chem. 1996;1:2–14. [Google Scholar]
- 33.Iwasaki T, Kounosu A, Kolling DRJ, Crofts AR, Dikanov SA, Jin A, Imai T, Urushiyama A. Characterization of the pH-dependent resonance Raman transitions of archaeal and bacterial Rieske [2Fe-2S] proteins. J. Amer. Chem. Soc. 2004;126:4788–4789. doi: 10.1021/ja031976p. [DOI] [PubMed] [Google Scholar]
- 34.Rotsaert FAJ, Pikus JD, Fox BG, Markley JL, Sanders-Loehr J. N-isotope effects on the Raman spectra of Fe2S2 ferredoxin and Rieske ferredoxin: evidence for structural rigidity of metal sites. J. Biol. Inorg. Chem. 2003;8:318–326. doi: 10.1007/s00775-002-0417-4. [DOI] [PubMed] [Google Scholar]
- 35.Kounosu A, Li ZR, Cosper NJ, Shokes JE, Scott RA, Imai T, Urushiyama A, Iwasaki T. Engineering a three-cysteine, one-histidine ligand environment into a new hyperthermophilic archaeal Rieske-type [2Fe-2S] ferredoxin from Sulfolobus solfataricus. J. Biol. Chem. 2004;279:12519–12528. doi: 10.1074/jbc.M305923200. [DOI] [PubMed] [Google Scholar]
- 36.Tirrell TF, Paddock ML, Conlan AR, Smoll EJ, Jr, Nechushtai R, Jennings PA, Kim JE. Resonance Raman studies of the (His)(Cys)(3) 2Fe-2S cluster of MitoNEET: comparison to the (Cys)(4) mutant and implications of the effects of pH on the labile metal center. Biochemistry. 2009;48:4747–4752. doi: 10.1021/bi900028r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito Y, Shibayama H, Tanaka H, Tanimura A, Matsumura I, Kanakura Y. PICOT is a molecule which binds to anamorsin. Biochem. Biophys. Res. Commun. 2011;408:329–333. doi: 10.1016/j.bbrc.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 38.Netz DJ, Stumpfig M, Dore C, Muhlenhoff U, Pierik AJ, Lill R. Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 2010;6:758–765. doi: 10.1038/nchembio.432. [DOI] [PubMed] [Google Scholar]
- 39.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.