Abstract
Down-regulation of miR-138 (microRNA-138) has been frequently observed in various cancers, including HNSCC (head and neck squamous cell carcinoma). Our previous studies suggest that down-regulation of miR-138 is associated with mesenchymal-like cell morphology and enhanced cell migration and invasion. In the present study, we demonstrated that these miR-138-induced changes were accompanied by marked reduction in E-cad (E-cadherin) expression and enhanced Vim (vimentin) expression, characteristics of EMT (epithelial–mesenchymal transition). On the basis of a combined experimental and bioinformatics analysis, we identified a number of miR-138 target genes that are associated with EMT, including VIM, ZEB2 (zinc finger E-box-binding homeobox 2) and EZH2 (enhancer of zeste homologue 2). Direct targeting of miR-138 to specific sequences located in the mRNAs of the VIM, ZEB2 and EZH2 genes was confirmed using luciferase reporter gene assays. Our functional analyses (knock-in and knock-down) demonstrated that miR-138 regulates the EMT via three distinct pathways: (i) direct targeting of VIM mRNA and controlling the expression of VIM at a post-transcriptional level, (ii) targeting the transcriptional repressors (ZEB2) which in turn regulating the transcription activity of the E-cad gene, and (iii) targeting the epigenetic regulator EZH2 which in turn modulates its gene silencing effects on the downstream genes including E-cad. These results, together with our previously observed miR-138 effects on cell migration and invasion through targeting RhoC (Rho-related GTP-binding protein C) and ROCK2 (Rho-associated, coiled-coil-containing protein kinase 2) concurrently, suggest that miR-138 is a multi-functional molecular regulator and plays major roles in EMT and in HNSCC progression.
Keywords: enhancer of zeste homologue 2 (EZH2), epithelial–mesenchymal transition, microRNA-138 (miR-138), squamous cell carcinoma, vimentin (Vim), zinc finger E-box-binding homoeobox 2 (ZEB2)
INTRODUCTION
MicroRNAs are endogenous small non-coding RNAs that control the target gene expression at the post-transcriptional level. Several microRNAs have been functionally classified as protooncogenes or tumour suppressors and are aberrantly expressed in different cancer types [1,2]. Deregulation (e.g. overexpression or loss of expression) of these `cancerous' microRNAs can figure prominently in tumour initiation and progression by facilitating an inappropriate cellular programme that promotes uncontrolled proliferation, favours survival, induces EMT (epithelial–mesenchymal transition) and/or promotes invasive behaviour.
miR-138 (microRNA-138) has been thought to regulate a number of essential biological processes, including the development of mammary glands [3], regulating dendritic spine morphogenesis [4], modulating cardiac patterning during embryonic development [5] and thermotolerance acquisition [6]. The deregulation of miR-138 has been frequently observed in a number of cancer types, including thyroid cancer [7], lung cancer [8] and leukaemia [9]. The frequent down-regulation of miR-138 has also been observed in HNSCC (head and neck squamous cell carcinoma), including cases that originated from various anatomic sites such as the pharynx [10], the tongue [11–14] and other sites of the oral cavity [15]. Two miR-138 precursor genes, termed miR-138-1 and miR-138-2, were identified recently in the mouse genome [16], and their human homologues were mapped to chromosome 3p21.33 and 16q13 respectively. Interestingly, LOH (loss of heterozygosity) at both chromosome loci have been detected frequently in HNSCC [17–19].
Recent studies by us have demonstrated that the down-regulation of miR-138 in HNSCC cell lines enhances cell migration and invasion [10,12], and is associated with marked morphological changes (e.g. loss of polarity and cell–cell adhesion, and the acquisition of mesenchymal-like cell morphology) that are characteristics of EMT. However, the molecular mechanism(s) underlying the observed effect of miR-138 on EMT is poorly understood. The aim of the present study was to investigate the functional roles of miR-138 in EMT.
MATERIALS AND METHODS
Cell culture and transfection
The human HNSCC cell lines used in the present study were maintained in DMEM/F12 (where DMEM is Dulbecco's modified Eagle's medium) supplemented with 10% FBS (foetal bovine serum), 100 units/ml penicillin and 100 μg/ml streptomycin (Gibco) at 37°C in a humidified incubator containing 5% CO2. For functional analysis, miR-138 mimics and non-targeting miRNA mimics (Dharmacon), LNA (locked nucleic acid) knock-down probe specific to miR-138 (anti-miR-138 LNA) and negative control LNA (Exiqon), and gene-specific siRNAs (small interfering RNAs; On-TargetPlus SMARTpool, Dharmacon) were transfected into cells using DharmaFECT Transfection Reagent 1 as described previously [10,12]. For the induction of EMT, cells were treated with 10 ng/ml TGFβ (transforming growth factor β) 1 as described previously [20,21].
Fluorescence immunocytochemical analysis
Immunofluorescence analysis was performed as described previously [12]. In brief, cells were cultured on eight-chamber polystyrene vessel tissue-culture-treated glass slides (BD Biosciences) fixed with ice-cold methanol, permeabilized with 0.5% Triton X-100/PBS and blocked with 1% BSA in PBS. The slides were incubated with primary antibodies against E-cad (E-cadherin) (1:100), EZH2 (enhancer of zeste homologue 2; 1:100) (BD Biosciences), Vim (vimentin l) (1:200; Cell Signaling Technologies) or ZEB2 (zinc finger E-box-binding homeobox 2; 1:50; Sigma–Aldrich). The slides were then incubated with a FITC-conjugated anti-rabbit IgG antibody (1:50; Santa Cruz Biotechnology) and Alexa Fluor® 594-conjugated goat anti-mouse IgG antibody (1:400; Invitrogen). The slides were mounted with ProLong Gold antifade reagent containing DAPI (4′ ,6-diamidino-2-phenylindole; Invitrogen) following the manufacturer's protocol. The slides were then examined with a fluorescence microscope (Carl Zeiss).
Western blot analysis
Western blots were performed as described previously [12] using antibodies specific against E-cad, EZH2 (BD Biosciences), Vim, Snai2 (Snail homologue 2), Suz12 (suppressor of zeste 12; Cell Signaling Technologies), Eed (embryonic ectoderm development protein; Millipore), ZEB2 and β-actin (Sigma–Aldrich).
In vitro cell migration and invasion assays
Transwell assays were performed to assess cell migration and invasion using BD BioCoat Control Cell Culture Inserts (containing an 8.0 μm PET Membrane without matrix) or BD BioCoat BD Matrigel™ Invasion Chamber (containing a layer of BD Matrigel™ Basement Membrane Matrix) respectively. In brief, cells were treated with appropriate microRNA and/or siRNA reagents and then seeded in the upper Boyden chambers of the cell culture inserts. After 24 h of incubation (for migration) or 48 h of incubation (for invasion), cells remaining in the upper chamber or on the upper membrane were carefully removed. Cells adhering to the lower membrane were stained with Diff-Quik stain (Polyscience), imaged and counted using an inverted microscope equipped with a digital camera (Jenco). Experiments were performed in triplicate.
qRT-PCR (quantitative real-time PCR) analysis
To examine the expressional changes of the EMT-related genes, a RT2 Profiler PCR array for human EMT (Qiagen/SABiosciences) was used which consists of qRT-PCR assays for 84 EMT-related genes. Two additional qRT-PCR assays for RhoC (Rho-related GTP-binding protein C) and EZH2 (Ori-Gene) were also included in the experiments. The relative expression level was computed using the 2ΔΔCt analysis method, where actin was used as an internal reference [22]. For VIM, ZEB2 and EZH2, independent qRT-PCR assays were also performed using TaqMan Gene Expression Assays (Applied Biosystems). Experiments were performed in triplicate.
MicroRNA target prediction
The candidate targets of miR-138 were identified using miRGen [23], an integrated online database which contains a collection of five bioinformatics tools, including 4-way PicTar, 5-way PicTar, TargetScanS, miRanda at http://www.microrna.org and miRanda at miRBase. In addition, TargetScanHuman 5.1 [24] was also used for predicting the miR-138 targets. As such, the miR-138 targets are predicted by three different methods (PicTar, TargetScan and miRanda) with two different versions for each method. For the present study, genes that were predicted by at least one method were defined as potential miR-138 targets.
Dual-luciferase reporter assay
The luciferase reporter gene constructs for Vim (pGL-VimE1, pGL-VimE2 and pGL-VimE3) were created by cloning a 62-bp fragment and a 60-bp fragment from the coding region (position 518–579 and position 861′920, GenBank® accession number NM_003380, containing the miRNA-138-binding sites E1 and E2 respectively), and a 56-bp fragment from the 3′-UTR (untranslated region; position 1815′1870, containing the miRNA-138-binding site E3) of the Vim respectively, into the XbaI site of the pGL3-Control firefly luciferase reporter vector (Promega). The corresponding mutant constructs (pGL-VimE1m, pGL-VimE2m and pGL-VimE3m) were created by replacing the seed regions (position 2′8) of the miR-138-binding sites with 5′-TTTTTTT-3′. The luciferase reporter gene constructs for ZEB2 (pGL-ZEBE1, pGL-ZEBE2 and pGL-ZEBE3) were created by cloning a 60-bp fragment and a 70-bp fragment from the coding region (position 1751′1810 and position 3341′3410, GenBank® accession number NM_014795, containing the miRNA-138-binding sites E1 and E2 respectively), and a 70-bp fragment from the 3′-UTR (position 4891′4960, GenBank® accession number NM_014795, containing the identified miR-138-binding site E3) of the ZEB2 gene into the XbaI site of the pGL3 firefly luciferase reporter vector. The mutant constructs for ZEB2 (pGL-ZEBE1m, pGL-ZEBE2m and pGL-ZEBE3m) were created by mutating the seed region (position 2′8) of the miR-138-binding sites to 5′-TTTTTTT-3′. For EZH2, a 70-bp fragment from the coding region (position 1111′1180, GenBank® accession number NM_004456, containing the conserved miRNA-138-binding site Ec1) and a 60-bp fragment from the 3′-UTR of the EZH2 gene (position 2561′2620, GenBank® accession number NM_004456, containing the conserved miRNA-138-binding site Ec2) were cloned into the XbaI site of the pGL3 reporter vector. The mutant constructs for EZH2 (pGL-Ec1m and pGL-Ec2m) were created by replacing the seed region (position 2′8) of the miR-138-binding site with 5′-TTTTTTT-3′. An additional pair of constructs were also created for EZH2 which contained sequences of a previously described poorly conserved miR-138-binding site [6] from the human EZH2 gene (position 2388′2450, GenBank® accession number NM_004456, named pGL-Ep-hsa) and from the chicken EZH2 gene (position 2414′2476, GenBank® accession number XM_418879, named pGL-Ep-gga). The constructs were then verified by sequencing. Cells were transfected with the reporter constructs containing the targeting sequence from the three genes using Lipofectamine™ 2000 (Invitrogen). The pRL-TK vector (Promega) was co-transfected as an internal control for normalization of the transfection efficiency. The luciferase activities were then determined as described previously [12] using a Glomax 20/20 luminometer (Promega). Experiments were performed in quadruplicate.
Statistical analysis
Statistical analysis was performed using Student's t test. P < 0.05 was considered statistically significant.
RESULTS
miR-138 regulates EMT in HNSCC cell lines
The cell lines 1386Ln and 686Tu are previously established HNSCC cell lines with the miR-138 level in 1386Ln significantly lower than in 686Tu [10]. As shown with immunofluorescence analysis in Figures 1(A) and 1(B), ectopic transfection of the miR-138 mimic to the 1386Ln cells led to a dramatic decrease in Vim expression and a significant increase in E-cad expression. When the 686Tu cells were treated with anti-miR-138 LNA, a decrease in E-cad expression and an increase in Vim expression were observed (Figures 1C and 1D). These changes in E-cad and Vim expression were also confirmed by Western blot analysis (Figure 1E). Similar results were also observed in UM1 cells that were treated with the miR-138 mimic, and SCC9 and SCC15 cells treated with anti-miR-138 LNA (Supplementary Figure S1 at http://www.BiochemJ.org/bj/440/bj4400023add.htm). As shown in Figures 1(F) and 1(G), whereas the increased miR-138 level in 1386Ln resulted in reduced cell migration and cell invasion, the reduced miR-138 level in 686Tu led to enhanced cell migration and invasion. This finding is in agreement with our previous observation [12], and is in agreement with the notion that co-ordinated regulation of EMT is essential to cell motility, invasion and metastasis.
Figure 1. Effects of miR-138 on EMT in HNSCC cells.
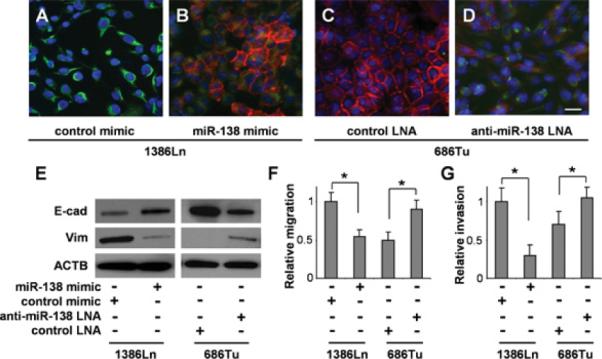
The 1386Ln cells were transfected with miR-138 mimic or negative control mimic. The 686Tu cells were treated with anti-miR-138 LNA or negative control LNA. The expressional changes in E-cad and Vim were measured in these cells by fluorescent immunocytochemical analysis (A–D; red, E-cad; green, Vim; blue, DAPI nuclear staining) and Western blotting (E). The miR-138-induced changes in the cell migration (F) and invasion (G) were measured in these cells as described in the Material and methods section. Representative images of × 100 magnification are shown. Results are representative of at least three independent experiments with similar results and the error bars represent S.D. *, P < 0.05. Scale bar, 10 μm.
As illustrated in Figure 2(A), treatment of 686Tu cells with TGF-β, a potent EMT inducer, down-regulated the expression of E-cad and enhanced the expression of Vim. TGF-β treatment also led to the down-regulation of miR-138 (Supplementary Figure S2 at http://www.BiochemJ.org/bj/440/bj4400023add.htm). The TGFβ-induced changes in E-cad and Vim expression were blocked by ectopic transfection of miR-138 mimic. As expected, TGFβ treatment led to enhanced cell migration and invasion (Figures 2B and 2C). The TGFβ-induced increase in cell migration and invasion were blocked by transfection of miR-138 mimic. As illustrated in Figure 2(D), ectopic transfection of the miR-138 mimic to the 1386Ln cells led to a reduced expression of Vim and enhanced expression of E-cad. These changes in E-cad and Vim expression were accompanied by reduced cell migration and invasion (Figures 2E and 2F). The miR-138-induced down-regulation of cell migration and invasion can be partially reversed when E-cad was knocked-down by siRNA.
Figure 2. Effect of miR-138 on cell migration and invasion in HNSCC cells.
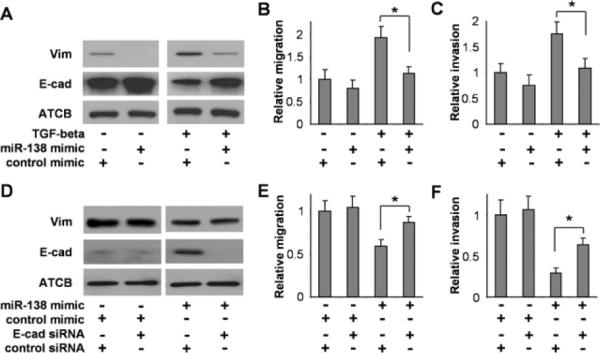
686Tu cells were incubated with 10 ng/ml TGFβ or vehicle for 48 h, and then transfected with either the miR-138 or negative control mimic for 24 h. The expression of Vim and E-cad (A), the cell migration (B) and invasion (C) were measured. 1386Ln cells that were treated with siRNA against E-cad or control siRNA, were co-treated with either miR-138 mimic or control mimic. The expression of Vim and E-cad (D), the cell migration (E) and invasion (F) were measured. Results are representative of at least three independent experiments with similar results and the error bars represent S.D.. *, P < 0.05.
To fully explore the roles of miR-138 in EMT, it is important to identify the functionally relevant targets (e.g. mRNA). We utilized an EMT-specific qRT-PCR array to determine the differential expression of 86 EMT-related genes on 1386Ln cells transfected with the miR-138 and negative control mimics. Of the 86 genes tested, 23 genes were altered after miR-138 treatment (>2-fold, P < 0.10). These include nine down-regulated genes and 14 up-regulated genes (Supplementary Table S1 at http://www.BiochemJ.org/bj/440/bj4400023add.htm). To identify the potential targets of miR-138, a bioinformatics-based analysis was carried out based on a combination of six different sequence-based microRNA target prediction algorithms. Our analysis revealed that the set of down-regulated genes was significantly enriched with the predicted targets of miR-138 (Supplementary Table S2 at http://www.BiochemJ.org/bj/440/bj4400023add.htm), whereas no significant enrichment of the predicted miR-138 targets was observed in the set of up-regulated transcripts. Of these nine down-regulated genes, four are potential direct targets of miR-138, including EZH2, ZEB2, RHOC and VIM (Table 1).
Table 1.
miR-138 targets identified by EMT-specific qRT-PCR array and bioinformatics prediction
| Transcripts down-regulated by miR-138 treatment (qRT-PCR) |
miR-138 targets predicted by bioinformatic tools |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene name | Fold difference (miR-138/control) | P value | PicTar 4-way | PicTar 5-way | TargetScanS | TargetScan Human 5.1 | miRanda (microrna.org) | miRanda (miRBase) |
| EZH2 | 0.24 | 0.001 | 1 | 1 | 1 | 1 | 1 | |
| SERPINE1 | 0.31 | 0.009 | ||||||
| PLEK2 | 0.32 | 0.065 | ||||||
| ZEB2 | 0.38 | 0.022 | 1 | |||||
| SNAI2 | 0.43 | 0.001 | ||||||
| MMP3 | 0.43 | 0.001 | ||||||
| RHOC | 0.44 | 0.008 | 1 | 1 | 1 | 1 | ||
| VIM | 0.48 | 0.005 | 1 | 1 | 1 | |||
| CDH2 | 0.49 | 0.078 | ||||||
The expressional changes for the 86 EMT-related genes we examined are shown in Supplementary Table S1 at at http://www.BiochemJ.org/bj/440/bj4400023add.htm. Genes that were predicted to be candidate targets of miR-138 were identified by the number 1 Gene names in bold have a fold difference < 0.5 (P < 0.10) and are bioinformatically predicted to be miR-138 targets. The down-regulation of EZH2 by miR-138 has recently been demonstrated in chicken model [6]. The down–regulation of the RHOC gene by miR-138 has been experimentally confirmed in our recent study [12]. CDH2, cadherin 2; MMP3, matrix metallopeptidase 3; PLEK2, pleckstrin 2.
miR-138 targets Vim in HNSCC cells
Based on the bioinformatics analysis, three miR-138-targeting sequences were identified in the VIM mRNA (Figure 3A). The first and second targeting sequences (E1 and E2) are located in the coding region and the third sequence (E3) is located in the 3′-UTR of the VIM mRNA. To confirm that miR-138 directly targets these sequences, dual-luciferase reporter assays were performed using the construct in which these targeting sites were cloned into the 3′-UTR of the reporter gene (pGL-VimE1, pGL-VimE2 and pGLVimE3). As illustrated in Figure 3(B), when cells were transfected with the miR-138 mimic, the luciferase activities were reduced significantly for both reporter constructs as compared with the cells transfected with negative control. When the seed regions of the targeting sites were mutated (pGL-VimE1m, pGL-VimE2m and pGL-VimE3m), the miR-138 effects on the luciferase activity were abolished. Furthermore, as shown by qRT-PCR (Figure 3C), ectopic transfection of miR-138 in 1386Ln cells led to significant down-regulation of VIM expression, and knock-down of miR-138 in 686Tu cells enhanced VIM expression. Similar results were observed in additional cell lines (Supplementary Figure S1). The miR-138-induced change in Vim expression was also confirmed by immunofluorescence analysis (Figures 3D–3K). When 1386Ln cells were treated with miR-138 mimic, a marked reduction in Vim staining was observed (Figures 3D and 3E). The reduction in Vim was accompanied by an increase in E-cad staining (Figures 3H and 3I). In contrast, when 686Tu cells were treated with anti-miR-138 LNA, an apparent increase in Vim staining was observed (Figures 3F and 3G). This increase in Vim expression was accompanied by a decrease in E-cad staining (Figures 3J and 3K).
Figure 3. miR-138 regulates EMT by targeting Vim.
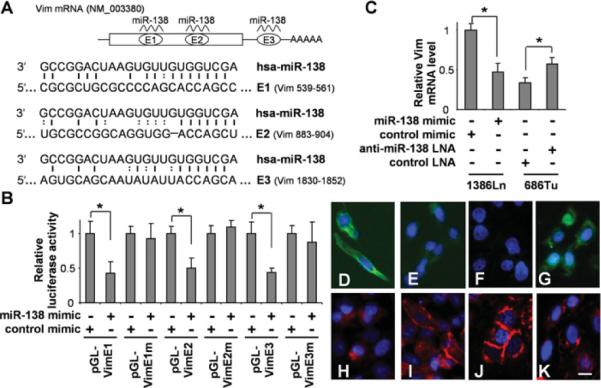
(A) Two predicted miR-138-targeting sequences were located in the coding region (E1 and E2) and 3′-UTR (E3) of the VIM mRNA. (B) Dual-luciferase reporter assays were performed to test the interactions of miR-138 and its targeting sequences in theVIM mRNA using constructs containing the predicted targeting sequences (pGL-VimE1, pGL-VimE2 and pGL-VimE3) and the corresponding mutated targeting sequences (pGL-VimE1m, pGL-VimE2m and pGL-VimE3m) cloned into the 3′-UTR of the reporter gene. The 1386Ln cells were transfected with miR-138 mimic or negative control mimic. The 686Tu cells were treated with anti-miR-138 LNA or negative control LNA. The expressional changes of the VIM gene were measured in these cells with qRT-PCR (C). Fluorescence immunocytochemical analysis was also performed to determine the effect of miR-138 on the expression of Vim and E-cad. Immunofluorescence analyses were carried out on these cells for Vim (D–G) and E-cad (H–K). (D and H) 1386Ln cells were transfected with negative control mimic. (E and I) 1386Ln cells were transfected with miR-138 mimic. (F and J) 686Tu cells were treated with negative control LNA. (G and K) 686Tu cells were treated with anti-miR-138 LNA. Red, E-cad; green, Vim; blue, DAPI nuclear staining. Representative images of ×200 magnification are shown. Results are representative of at least three independent experiments with similar results and the error bars represent S.D. *, P < 0.05. Scale bar, 5 μm.
miR-138 down-regulates transcriptional repressors (ZEB2 and Snai2) and promotes E-cad expression in HNSCC cells
Three miR-138-binding sites were predicted in the ZEB2 mRNA, two located in the coding region and one located in the 3′-UTR (Figure 4A). As shown in Figure 4(B), dual-luciferase reporter assays were performed using constructs in which these targeting sites were cloned into the 3′-UTR of the reporter gene (pGLZEBE1, pGL-ZEBE2 and pGL-ZEBE3). A dramatic reduction in luciferase activity was observed for pGL-ZEBE3, when the cells were transfected with miR-138 as compared with those of the cells transfected with negative control. Relatively small, but statistically significant, reductions in luciferase activity were observed for pGL-ZEBE1 and pG–ZEBE2 in cells transfected with miR-138 as compared with those transfected with negative control. When the seed regions of these targeting sites were mutated (pGL-ZEBE1m, pGL-ZEBE2m and pGL-ZEBE3m), the miR-138 effect on luciferase was abolished. Furthermore, as shown by Western blot analysis (Figure 4C), ectopic transfection of miR-138 or siRNA against ZEB2 in 1386Ln cells led to significant down-regulation of ZEB2 expression, and knock-down of miR-138 in 686Tu cells enhanced the ZEB2 expression. The changes in ZEB2 expression were inversely correlated with changes in E-cad expression. The miR-138-induced change in ZEB2 expression was also confirmed by qRT-PCR (Figure 4D). Similar results were observed in additional cell lines (Supplementary Figure S1). As shown in Figures 4(E) and 4(F), when 1386Ln cells were treated with the miR-138 mimic, a marked reduction in ZEB2 nuclear staining was observed. The reduction in ZEB2 staining was accompanied by an increase in E-cad staining (Figures 4G and 4H). In contrast, when 686Tu cells were treated with anti-miR-138 LNA, an apparent increase in ZEB2 staining was observed (Figures 4I and 4J). This increase in ZEB2 expression was accompanied by a decrease in E-cad staining (Figures 4K and 4L).
Figure 4. miR-138 regulates EMT by targeting ZEB2.
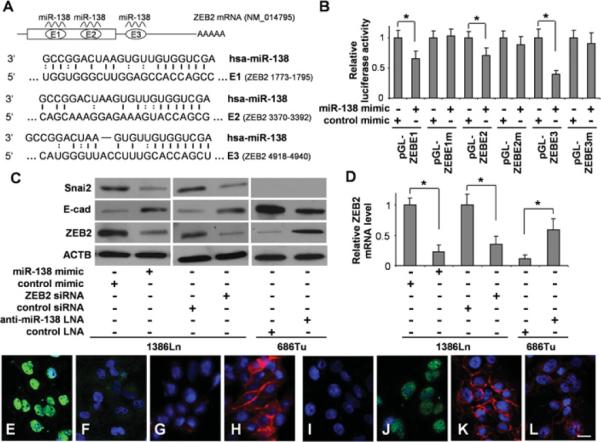
(A) The predicted highly conserved miR-138-targeting sequences in the coding region (E1 and E2) and 3′-UTR (E3) of ZEB2 mRNA. (B) Dual-luciferase reporter assays were performed to test the interactions of miR-138 and its targeting sequences in ZEB2 mRNA using constructs containing the predicted targeting sequences (pGL-ZEBE1, pGL-ZEBE2 and pGL-ZEBE3) and mutated targeting sequences (pGL-ZEBE1m, pGL-ZEBE2m and pGL-ZEBE3m) cloned into the 3′-UTR of the reporter gene. (C) Western blot analyses were performed to examine the effects of miR-138 on ZEB2, E-cad and SNAI2 gene expression at the protein level on 1386Ln cells treated with either miR-138 mimic, negative control mimic or siRNA against ZEB2 or control siRNA, and 686Tu cells treated with either anti-miR-138 LNA or negative control LNA. (D) qRT-PCR assays were also performed to examine the effects of miR-138 on ZEB2 gene expression in these cells at the mRNA level. Fluorescence immunocytochemical analysis was also performed to determine the effect of miR-138 on the expression of ZEB2 and E-cad. Immunofluorescence analyses were carried out on these cells for ZEB2 (E, F, I and J) and E-cad (G, H, K and L). (E and G) 1386Ln cells were transfected with negative control mimic. (F and H) 1386Ln cells were transfected with miR-138 mimic. (I and K) 686Tu cells were treated with negative control LNA. (J and L) 686Tu cells were treated with anti-miR-138 LNA. Red, E-cad; green, ZEB2; Blue, DAPI nuclear staining. Representative images of ×200 magnification are shown. Results are representative of at least three independent experiments with similar results and the error bars represent S.D. *, P < 0.05.
It is noteworthy that we also observed a significant reduction in Snai2 expression at both the mRNA level (Table 1) and the protein level (Figure 4C) in 1386Ln cells treated with the miR-138 mimic. Reduced Snai2 expression was also observed in UM1 cells that were treated with the miR-138 mimic (Supplementary Figure S1). Although no apparent expression of Snai2 was observed in 686Tu cells (Figure 4C), enhanced Snai2 expression was observed in SCC9 and SCC15 cells that were treated with anti-miR-138 LNA (Supplementary Figure S1). However, we failed to identify any miR-138-targeting site in the SNAI2 mRNA.
miR-138 targets epigenetic regulator EZH2 and regulates E-cad expression in HNSCC cells
PcG (polycomb group) proteins such as EZH2 are important epigenetic regulators. The EZH2-mediated repression of E-cad expression is associated with EMT in several cancer types [25–27]. Our bioinformatics analysis identified two conserved miR-138 targeting sequences located in the coding region (Ec1) and in the 3′ -UTR (Ec2) of the EZH2 mRNA (Figure 5A). In addition, a poorly conserved targeting site (Ep) that overlaps with the junction of the coding region and 3′ -UTR has been reported previously in chickens [6]. However, in humans, two bases in the seed region (positions 4 and 6) of this Ep site are replaced with non-consensus bases. As shown in Figure 5(B), the luciferase activities of the reporter constructs containing the conserved sites (pGL-Ec1 and pGL-Ec2) were reduced significantly when cells were transfected with miR-138 mimic in comparison with those transfected with negative control. No apparent change was observed in the luciferase activities of the reporter constructs containing the mutated sites (pGL-Ec1m and pGL-Ec2m) when cells were treated with either the miR-138 or control mimic. For the reporter construct containing the chicken Ep site (pGL-Ep-gga), the luciferase activity was reduced significantly when the cells were transfected with the miR-138 mimic as compared with the cells transfected with negative control. No apparent change was observed in luciferase activity of the reporter construct containing the human Ep site (pGL-Ep-hsa) when cells were treated with either the miR-138 or control mimic. Furthermore, as shown by Western blot analysis (Figure 5C), ectopic transfection of miR-138 or siRNA specific against EZH2 in 1386Ln cells led to significant down-regulation of EZH2 expression. Knock-down of miR-138 in 686Tu cells enhanced the EZH2 expression. The changes in EZH2 expression were inversely correlated with changes in E-cad expression. Similar results were observed in additional cell lines (see Supplementary Figure S1). We also examined the expression of Eed and Suz12, two additional core components of the PRC2 (polycomb repressive complex 2). Although no change in Eed expression was observed, a slight decrease and an apparent increase in Suz12 levels were observed in 1386Ln cells treated with EZH2 siRNA and in 686Tu cells treated with anti-miR-138 LNA. The miR-138-induced change in EZH2 expression was also confirmed by qRT-PCR (Figure 5D) and fluorescence immunocytochemical analysis (Figures 5E–5L). As shown in Figures 5(E) and 5(F), when 1386Ln cells were treated with the miR-138 mimic, a marked reduction in EZH2 nuclear staining was observed. The reduction in EZH2 staining was accompanied by an increase in E-cad staining (Figures 5G and 5H). In contrast, when 686Tu cells were treated with anti-miR-138 LNA, an apparent increase in EZH2 staining was observed (Figures 5I and 5J). This increase in EZH2 expression was accompanied by a decrease in E-cad staining (Figures 5K and 5L).
Figure 5. miR-138 regulates EMT by targeting EZH2.
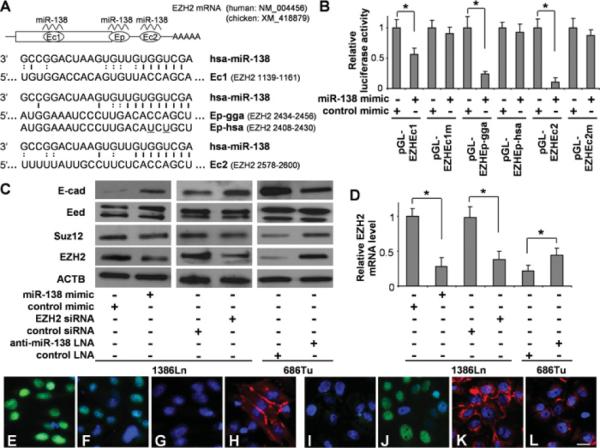
(A) Two highly conserved miR-138-targeting sequences (Ec1 and Ec2) and a poorly conserved miR-138 target sequence (Ep) were identified in the EZH2 mRNA. For Ep, both human (Ep-hsa) and chicken (Ep-gga) sequences are shown. (B) Dual-luciferase reporter assays were performed to test the interactions of miR-138 and the identified targeting sequences in the EZH2 gene using constructs containing the predicted targeting sequences (pGL-Ec1, pGL-Ep-gga, pGL-Ep-hsa and pGL-Ec2) and mutated Ec-targeting sequences (pGL-Ec1m and pGL-Ec2m) cloned into the 3′-UTR of the reporter gene. (C) Western blot analyses were performed to examine the effects of miR-138 on the expression of PRC2 proteins (EZH2, Suz12 and Eed) and E-cad on 1386Ln cells treated with either miR-138 mimic or negative control mimic, or siRNA against ZEB2 or control siRNA, and 686Tu cells treated with either anti-miR-138 LNA or negative control LNA. (D) qRT-PCR assays were also performed to examine the effects of miR-138 on EZH2 expression in these cells at the mRNA level. Fluorescence immunocytochemical analysis was also performed to determine the effect of miR-138 on the expression of EZH2 and E-cad. Immunofluorescence analyses were carried out on these cells for EZH2 (E, F, I and J) and E-cad (G, H, K and L). (E and G) 1386Ln cells were transfected with negative control mimic. (F and H) 1386Ln cells were transfected with miR-138 mimic. (I and K) 686Tu cells were treated with negative control LNA. (J and L) 686Tu cells were treated with anti-miR-138 LNA. Red, E-cad; green, ZEB2; blue, DAPI nuclear staining. Representative images of ×200 magnification are shown. Results are representative of at least three independent experiments with similar results and the error bars represent S.D. *, P < 0.05. Scale bar, 5 μm.
DISCUSSION
Cancer cells can de-differentiate through activation of specific biological pathways associated with EMT, thereby gaining the ability to migrate and invade. Previous studies have suggested that deregulation of microRNAs (including miR-200 family members) is associated with EMT and the progression of a number of different cancers [28–34]. However, the involvement of microRNA in EMT of the HNSCC cells has not been investigated fully. The down-regulation of miR-138 is a frequent event in HNSCC and has been consistently observed by multiple laboratories [10–15]. Our preliminary results show that miR-138 regulates cell migration and invasion by targeting RhoC and ROCK2 (Rho-associated, coiled-coil-interacting protein kinase 2) concurrently [12]. These miR-138-induced changes in cell migration and invasion are accompanied by marked morphological changes (e.g. loss of polarity and cell–cell adhesion, increased motility, and the acquisition of mesenchymal phenotype) which are characteristics of EMT. In the present study, we test the effects of miR-138 on the expression of 86 EMT-related genes using a qRT-PCR array. We found nine down-regulated genes and 14 up-regulated genes. Our bioinformatics analysis revealed that four of these nine down-regulated genes are potential direct targets of miR-138, including EZH2, ZEB2, RHOC and VIM. It is noteworthy that our qRT-PCR-based experiments measure the differential expression at the mRNA level, and are only sensitive to the targets that are regulated by microRNA-mediated degradation, but not to the targets that are regulated by microRNA-mediated translational inhibition. We anticipate that a portion of true miR-138 targets will not be detected by our approach. Nevertheless, the present study identified a panel of EMT-related genes that are regulated by miR-138, including the experimentally confirmed miR-138 target gene RHOC [12]. In the present paper we demonstrated that miR-138 regulates EMT via three novel and distinct pathways (Figure 6): (i) direct targeting of VIM mRNA and controlling the expression of VIM at the post-transcriptional level, (ii) regulating the transcriptional repressors (ZEB2 and Snai2) which in turn regulate the transcription activity of the E-cad gene, (iii) targeting the epigenetic regulator EZH2 which in turn modulates its gene-silencing effects on the downstream genes including E-cad.
Figure 6.
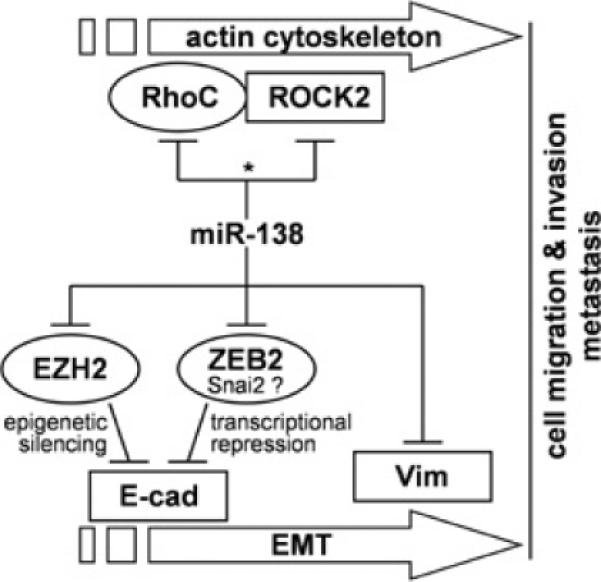
Potential roles of miR-138 in EMT and cancer cell metastasis
Vim is an essential structural cytoskeletal protein constituting IFs (intermediate filaments) of mesenchymal cells. During EMT, the cellular IF status changes from a keratin-rich network (with connections to adherens junctions and hemidesmosomes) to a Vim-rich network connecting to focal adhesions. The results of the present study demonstrated that Vim is a functional target of miR-138 and that the down-regulation of miR-138 in HNSCC is associated with enhanced Vim expression. It is noteworthy that miR-138 has also been thought to play important roles in developmental processes [3–6]. Given the apparent involvement of Vim in EMT, it is possible that the miR-138-mediated regulation of Vim may have functional relevance to developmental processes. Further studies are required to define the precise mechanisms through whichmiR-138 andVim regulate these diverse biological processes.
ZEB2 is a member of the zinc finger E-box-binding homeobox family of proteins that function as transcriptional repressors and interact with activated SMADs, the transducers of TGFβ signalling. ZEB2 is a well-established inducer of EMT and a potent repressor of E-cad expression [35]. The results of the present paper demonstrate a direct interaction between miR-138 and ZEB2 mRNA which leads to the post-transcriptional suppression of ZEB2 expression, which in turn regulates the expression of E-cad and EMT. It is noteworthy that the ZEB family repressors (both ZEB1 and ZEB2) are targeted by a number of different microRNAs, including the miR-200 family [36]. Interestingly, ZEB1 has been showed to suppress the expression of miR-200 family members indicating that miR-200 members and ZEB factors reciprocally control each other in a negative-feedback loop [32,37]. It is not clear whether miR-138 is also regulated by ZEB factors (or its other target genes) by similar feedback mechanism(s). Further studies are needed to explore this potential regulatory mechanism. In addition to ZEB2, our data suggested that miR-138 also down-regulates Snai2, a member of Snail family of zinc finger transcriptional repressors that play an important role in the regulation of E-cad expression and EMT [35]. However, we failed to identify any miR-138-targeting site in the SNAI2 mRNA. It is possible that miR-138 indirectly regulates SNAI2 by targeting factor(s) that control SNAI2 gene expression. Alternatively, Snai2 may be regulated by miR-138 through a non-canonical targeting site.
A third EMT-related gene that targeted by miR-138 is EZH2. EZH2 is a critical component of the PRC2 complex that includes non-catalytic subunits Suz12 and Eed. It catalyses trimethylation on Lys27 of histone 3 protein (H3K27Me3), which in turn leads to chromatin condensation and epigenetic silencing of the downstream genes [38]. One of the well-established downstream target genes of EZH2 is E-cad, and the EZH2-mediated repression of E-cad is associated with EMT in several cancer types [25–27]. A previous study reported that overexpression of EZH2 in cancer cells down-regulates the expression of E-cad through histone H3K27 trimethylation at the promoter of the gene [25]. Knockdown of EZH2 in vitro has been shown to restore E-cad expression [39,40]. We confirmed that EZH2 is required for the suppression of E-cad in our cell types. We further demonstrated that the miR-138 down-regulates the expression of the EZH2 gene by binding to a conserved targeting site located in the 3′ -UTR of the EZH2 mRNA. This miR-138-mediated EZH2 down-regulation is reversely correlated with E-cad expression and EMT in HNSCC cells. Interestingly, miR-138 has been showed to target EZH2 in chickens [6]. Two miR-138-targeting sites were identified in the chicken EZH2 mRNA: an evolutionarily conserved site located in the 3′ -UTR which is also present in the 3′ -UTR of the human EZH2 mRNA, and a poorly conserved site that is over-lapped with the translational stop codon in the chicken EZH2. In humans, the seed region of this poorly conserved site has two base substitutions (at positions 4 and 6), which makes it non-functional. Nevertheless, the fact that EZH2 expression is down-regulated by miR-138 in both chickens and humans suggested that the miR-138-mediated suppression of EZH2 is an evolutionarily conserved molecular event. We also observed an apparent increase in Suz12 levels in 686Tu cells treated with anti-miR-138 LNA. However, no miR-138-targeting site was identified in the SUZ12 mRNA, and currently, the biological significance of these observed changes in SUZ12 expression is not clear. It is noteworthy that miR-200 has been shown to directly target Suz12 and control the E-cad expression by regulating the PRC2 complex [41]. Additional studies will be needed to fully explore the potential concordant effect(s) of anti-EMT microRNAs (e.g. miR-138 and miR-200) on PRC2-mediated repression of E-cad.
In summary, the results of the present study demonstrated that miR-138 regulates EMT in HNSCC cells. This, together with our previous observation that miR-138 regulates cell migration and invasion by concurrently targeting RhoC and ROCK2 [12], suggested that miR-138 is a multi-functional molecular regulator and plays major roles in EMT. Further studies are required to explore its potential as a novel therapeutic target for cancer patients at risk of metastasis.
Supplementary Material
ACKNOWLEDGEMENT
We thank Ms Katherine Long for her editorial assistance prior to submission.
FUNDING This work was supported, in part, by the National Institutes for Health Public Health Service grants [grant numbers CA135992, CA139596 and DE014847] and supplementary funding from the University of Illinois at Chicago Center for Clinical and Translational Science [grant number UL1RR029879]. Y. J. is supported by a National Institutes for Health T32 training grant [grant number DE018381].
Abbreviations used
- DAPI
4′,6-diamidino-2-phenylindole
- E-cad
E-cadherin
- Eed
embryonic ectoderm development protein
- EMT
epithelial–mesenchymal transition
- EZH2
enhancer of zeste homologue 2
- HNSCC
head and neck squamous cell carcinoma
- IF
intermediate filament
- LNA
locked nucleic acid
- miR-138
microRNA-138
- PRC2
polycomb repressive complex 2
- qRT-PCR
quantitative real-time PCR
- RhoC
Rho-related GTP-binding protein C
- ROCK2
Rho-associated, coiled-coil-containing protein kinase 2
- siRNA
small interfering RNA
- Snai2
Snail homologue 2
- Suz12
suppressor of zeste 12
- TGFβ
transforming growth factor β
- UTR
untranslated region
- Vim
vimentin
- ZEB2
zinc finger E-box-binding homeobox 2
Footnotes
AUTHOR CONTRIBUTION Antonia Kolokythas, Yang Dai and Xiaofeng Zhou conceived the project. Xiqiang Liu, Cheng Wang, Zujian Chen, Yi Jin and Yun Wang performed the laboratory analyses. Yang Dai conducted the statistical analyses. Xiqiang Liu, Cheng Wang, Antonia Kolokythas, Yang Dai and Xiaofeng Zhou drafted the paper. All authors read and approved the final paper.
REFERENCES
- 1.Esquela-Kerscher A, Slack FJ. Oncomirs: microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 2.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 3.Wang C, Li Q. Identification of differentially expressed microRNAs during the development of Chinese murine mammary gland. J. Genet. Genomics. 2007;34:966–973. doi: 10.1016/S1673-8527(07)60109-X. [DOI] [PubMed] [Google Scholar]
- 4.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, et al. A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morton SU, Scherz PJ, Cordes KR, Ivey KN, Stainier DY, Srivastava D. microRNA-138 modulates cardiac patterning during embryonic development. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17830–17835. doi: 10.1073/pnas.0804673105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kisliouk T, Yosefi S, Meiri N. miR-138 inhibits EZH2 methyltransferase expression and methylation of histone H3 at lysine 27, and affects thermotolerance acquisition. Eur. J. Neurosci. 2011;33:224–235. doi: 10.1111/j.1460-9568.2010.07493.x. [DOI] [PubMed] [Google Scholar]
- 7.Mitomo S, Maesawa C, Ogasawara S, Iwaya T, Shibazaki M, Yashima-Abo A, Kotani K, Oikawa H, Sakurai E, Izutsu N, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99:280–286. doi: 10.1111/j.1349-7006.2007.00666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, et al. miR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12085–12090. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao X, Yang L, Hu J, Ruan J. miR-138 might reverse multidrug resistance of leukemia cells. Leuk. Res. 2010;34:1078–1082. doi: 10.1016/j.leukres.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Jiang L, Wang A, Yu J, Shi F, Zhou X. MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett. 2009;286:217–222. doi: 10.1016/j.canlet.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang L, Dai Y, Liu X, Wang C, Wang A, Chen Z, Heidbreder CE, Kolokythas A, Zhou X. Identification and experimental validation of G protein α inhibiting activity polypeptide 2 (GNAI2) as a microRNA-138 target in tongue squamous cell carcinoma. Hum. Genet. 2011;129:189–197. doi: 10.1007/s00439-010-0915-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang L, Liu X, Kolokythas A, Yu J, Wang A, Heidbreder CE, Shi F, Zhou X. Down-regulation of the Rho GTPase signaling pathway is involved in the microRNA-138 mediated inhibition of cell migration and invasion in tongue squamous cell carcinoma. Int. J. Cancer. 2010;127:505–512. doi: 10.1002/ijc.25320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong TS, Liu XB, Chung-Wai Ho A, Po-Wing Yuen A, Wai-Man Ng R, Ignace Wei W. Identification of pyruvate kinase type M2 as potential oncoprotein in squamous cell carcinoma of tongue through microRNA profiling. Int. J. Cancer. 2008;123:251–257. doi: 10.1002/ijc.23583. [DOI] [PubMed] [Google Scholar]
- 14.Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin. Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- 15.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 16.Obernosterer G, Leuschner PJ, Alenius M, Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piccinin S, Gasparotto D, Vukosavljevic T, Barzan L, Sulfaro S, Maestro R, Boiocchi M. Microsatellite instability in squamous cell carcinomas of the head and neck related to field cancerization phenomena. Br. J. Cancer. 1998;78:1147–1151. doi: 10.1038/bjc.1998.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogg RP, Honorio S, Martinez A, Agathanggelou A, Dallol A, Fullwood P, Weichselbaum R, Kuo MJ, Maher ER, Latif F. Frequent 3p allele loss and epigenetic inactivation of the RASSF1A tumour suppressor gene from region 3p21.3 in head and neck squamous cell carcinoma. Eur. J. Cancer. 2002;38:1585–1592. doi: 10.1016/s0959-8049(01)00422-1. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Gleich L, Pavelic ZP, Li YQ, Gale N, Hunt S, Gluckman JL, Stambrook PJ. Cervical metastases of head and neck squamous cell carcinoma correlate with loss of heterozygosity on chromosome 16q. Int. J. Oncol. 1999;14:557–561. doi: 10.3892/ijo.14.3.557. [DOI] [PubMed] [Google Scholar]
- 20.Sun L, Diamond ME, Ottaviano AJ, Joseph MJ, Ananthanarayan V, Munshi HG. Transforming growth factor-β 1 promotes matrix metalloproteinase-9-mediated oral cancer invasion through snail expression. Mol. Cancer. Res. 2008;6:10–20. doi: 10.1158/1541-7786.MCR-07-0208. [DOI] [PubMed] [Google Scholar]
- 21.Diamond ME, Sun L, Ottaviano AJ, Joseph MJ, Munshi HG. Differential growth factor regulation of N-cadherin expression and motility in normal and malignant oral epithelium. J. Cell Sci. 2008;121:2197–2207. doi: 10.1242/jcs.021782. [DOI] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Megraw M, Sethupathy P, Corda B, Hatzigeorgiou AG. miRGen: a database for the study of animal microRNA genomic organization and function. Nucleic Acids Res. 2007;35:D149–D155. doi: 10.1093/nar/gkl904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani RS, Tomlins SA, Mehra R, Laxman B, Cao X, Yu J, et al. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene. 2008;27:7274–7284. doi: 10.1038/onc.2008.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herranz N, Pasini D, Diaz VM, Franci C, Gutierrez A, Dave N, Escriva M, Hernandez-Munoz I, Di Croce L, Helin K, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol. Cell. Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang WY, Yang PM, Chang YF, Marquez VE, Chen CC. Methotrexate induces apoptosis through p53/p21-dependent pathway and increases E-cadherin expression through downregulation of HDAC/EZH2. Biochem. Pharmacol. 2011;81:510–517. doi: 10.1016/j.bcp.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 28.Mongroo PS, Rustgi AK. The role of the miR-200 family in epithelial-mesenchymal transition. Cancer Biol. Ther. 2010;10:219–222. doi: 10.4161/cbt.10.6312548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–119. doi: 10.4161/rna.5.3.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 32.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braun J, Hoang-Vu C, Dralle H, Huttelmaier S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. 2010;29:4237–4244. doi: 10.1038/onc.2010.169. [DOI] [PubMed] [Google Scholar]
- 34.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 36.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–3118. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 37.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res. 2008;68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 38.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 39.Fujii S, Ochiai A. Enhancer of zeste homolog 2 downregulates E-cadherin by mediating histone H3 methylation in gastric cancer cells. Cancer Sci. 2008;99:738–746. doi: 10.1111/j.1349-7006.2008.00743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao ZY, Cai MY, Yang GF, He LR, Mai SJ, Hua WF, Liao YJ, Deng HX, Chen YC, Guan XY, et al. EZH2 supports ovarian carcinoma cell invasion and/or metastasis via regulation of TGF-β1 and is a predictor of outcome in ovarian carcinoma patients. Carcinogenesis. 2010;31:1576–1583. doi: 10.1093/carcin/bgq150. [DOI] [PubMed] [Google Scholar]
- 41.Iliopoulos D, Lindahl-Allen M, Polytarchou C, Hirsch HA, Tsichlis PN, Struhl K. Loss of miR-200 inhibition of Suz12 leads to polycomb-mediated repression required for the formation and maintenance of cancer stem cells. Mol. Cell. 2010;39:761–772. doi: 10.1016/j.molcel.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


