Abstract
Increased adiponectin levels have been shown to be associated with a lower risk of type 2 diabetes. To understand the relations between genetic variation at the adiponectin-encoding gene, ADIPOQ, and adiponectin levels, and subsequently its role in disease, we conducted a deep resequencing experiment of ADIPOQ in 14,002 subjects, including 12,514 Europeans, 594 African Americans, and 567 Indian Asians. We identified 296 single nucleotide polymorphisms (SNPs), including 30 amino acid changes, and carried out association analyses in a subset of 3,665 subjects from two independent studies. We confirmed multiple genome-wide association study findings and identified a novel association between a low-frequency SNP (rs17366653) and adiponectin levels (P = 2.2E–17). We show that seven SNPs exert independent effects on adiponectin levels. Together, they explained 6% of adiponectin variation in our samples. We subsequently assessed association between these SNPs and type 2 diabetes in the Genetics of Diabetes Audit and Research in Tayside Scotland (GO-DARTS) study, comprised of 5,145 case and 6,374 control subjects. No evidence of association with type 2 diabetes was found, but we were also unable to exclude the possibility of substantial effects (e.g., odds ratio 95% CI for rs7366653 [0.91–1.58]). Further investigation by large-scale and well-powered Mendelian randomization studies is warranted.
Adiponectin is an anti-inflammatory adipokine secreted by adipocytes and is inversely associated with the risk of type 2 diabetes (1); however, whether adiponectin is causal or merely a marker of prediabetes is not yet known. Use of genetics through Mendelian randomization (2,3) is one approach to investigate causality; thus the identification of genetic variation affecting adiponectin levels has drawn much attention. Through linkage and association studies, adiponectin levels have been linked to the ADIPOQ locus on chromosome 3q27 (4–8). The majority of adiponectin genetic investigations to date have been limited to common variants, but with the advent of massively parallel sequencing, we can now explore low-frequency variation within this gene as well. Here, we describe results from a deep resequencing experiment of the exons and flanking regions of ADIPOQ in 14,002 individuals. We describe the genetic variations observed and report genetic associations with adiponectin levels in a subset of 3,665 individuals with adiponectin measurements. For variants independently associated with adiponectin levels, we further evaluated their impact on type 2 diabetes susceptibility in a cohort of 5,145 type 2 diabetic and 6,374 control subjects.
RESEARCH DESIGN AND METHODS
We sequenced ADIPOQ in 14,002 individuals, including 12,514 Europeans, 594 African Americans, and 567 Indian Asians. Adiponectin levels were measured in a subset of 3,665 subjects of European origin from two studies: 1,579 from the Genetic Epidemiology of the Metabolic Syndrome (GEMS) study (9) and 2,086 from the Cohorte Lausannoise (CoLaus) study (10). The GEMS study is a large multinational study designed to explore the genetic basis of the metabolic syndrome. Subjects in our resequencing study were selected based on DNA availability and consisted of 787 dyslipidemic subjects with an elevated plasma triglyceride and a low serum HDL cholesterol and 792 normolipidemic control subjects having the combination of an elevated plasma triglyceride, a low serum HDL cholesterol, and a BMI >25 kg/m2. The CoLaus study is a single-center, population-based study to assess the prevalence of cardiovascular risk factors in the population of Lausanne, Switzerland. We included 2,086 subjects in this experiment based on availability of DNA and phenotype assessment. Genotyping was conducted in the Genetics of Diabetes Audit and Research in Tayside Scotland (GO-DARTS) study (11), which includes a total of 12,348 individuals, 5,145 type 2 diabetic subjects and 6,374 normoglycemic, population-based control subjects, all of European U.K. origin.
DNA sequencing and genotyping.
All three exons of ADIPOQ (NM_004797) plus 50 bases of flanking sequence (NCBI build 36.3) were selected for capture using a custom Roche NimbleGen (Madison, WI) HD2.1M sequence capture array. Paired-end sequencing was conducted for each 48-sample indexed pool. Variants were called using SOAPsnp (12) at a minimum depth of 7 and a minimum consensus quality of 20. Genotyping in the GO-DARTS study was performed using a Kaspar assay (http://www.kbioscience.co.uk/).
Adiponectin measurement.
Plasma adiponectin levels were measured using the ELISA assay (R&D Systems, Minneapolis, MN).
Statistical methods.
Linear regression analyses were carried out in the GEMS and CoLaus studies separately under an additive genetic model adjusted for significant covariates (P < 0.05) in each study, including dyslipidemia status, age, sex, collection site, waist and hip circumference in GEMS, and age, sex, waist and hip circumference, BMI, smoking, and alcohol usage in CoLaus. Adiponectin levels were log transformed and the extreme outliers were set to the 99.9 percentile of the distribution. Single nucleotide polymorphisms (SNPs) with at least 10 copies of the minor allele were analyzed individually, whereas nonsynonymous SNPs with <10 copies were aggregated (13). SNPs and subjects with >20% missing data were excluded from analysis. Multiple testing corrections were made by adjusting for the total number of tests performed in each study. Meta-analysis was performed using the inverse-variance method (14).
To identify the number of independent SNPs in and around ADIPOQ, we conducted variable selection by both frequentist and Bayes methods. In the frequentist approach, both criteria for a SNP to enter or leave the stepwise regression model were set as P < 0.005. Bayes variable selection analysis was conducted using the BTAS WinBUGs toolkit (15,16). In both frequentist and Bayes variable selection, missing genotype data were imputed by BEAGLE (17). We further conducted haplotype analysis using Haplo Stats (18) for the set of independent SNPs identified from the aforementioned variable selection analyses to determine whether any of the independent SNPs resided on the same shared haplotype.
In the GO-DARTS study, we tested for association under an additive model using logistic regression for the seven SNPs individually and also in conjunction by analyzing the linear combination of allele dosage data weighted by the effect estimates obtained from linear regression analysis.
Bioinformatics analysis.
We used PolyPhen2 (19) and SIFT (20) software to predict impact of nonsynonymous SNPs on the protein function, and PhyloP scores to measure conservation of a base pair across many species (21). Our most strongly associated SNP (rs17366653) is intronic, located 24 nucleotides upstream of the beginning of exon 3. The sequence flanking this position was submitted to a number of splice site or splice branch point bioinformatics prediction tools, including NetGene2 (22), MaxEnt (23), and the Alternative Splicing Desktop (ASD) (http://www.ebi.ac.uk/asd-srv/wb.cgi?method=6).
RESULTS
Observed variants.
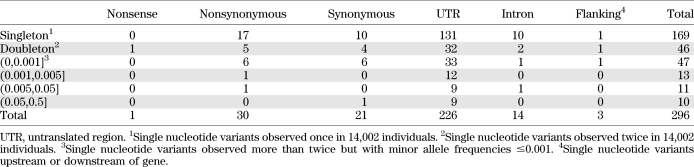
Resequencing of ADIPOQ in 14,002 subjects achieved an average depth of 27 reads and an average quality score of 84 (Supplementary Fig. 1). A total of 296 SNPs were observed (Supplementary Table 1). Among them, 52 (18%) were within the coding regions, consisting of 1 nonsense, 30 nonsynonymous, and 21 synonymous variants. Most SNPs were rare, including 169 (57%) singletons and 46 (16%) doubletons, each observed once or twice in 14,002 individuals (Table 1). There were no common nonsynonymous SNPs [minor allele frequency (MAF) >5%] and only two had MAF > 0.1%. The only nonsense variant detected was observed in two European individuals for whom adiponectin measurements were not available.
TABLE 1.
Single nucleotide variants observed in ADIPOQ in 14,002 individuals by frequency and type
We compared the frequencies of nonsynonymous SNPs observed in 12,518 European Caucasians, 588 African Americans, and 574 Indian Asians (Supplementary Table 1). Of the 30 nonsynonymous SNPs, 13 were unique to Europeans, 2 to African Americans, and 6 to Indian Asians. Only two, R55C and Y111H, were observed in all three populations. Overall, these variants were extremely rare, and a majority of them were private to each of the three populations included in our study.
Genetic association analyses.
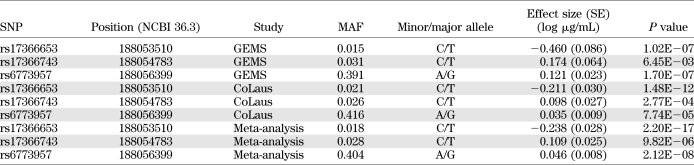
We identified three SNPs significantly associated with adiponectin levels. Details of the three SNPs and their effect on adiponectin levels are shown in Table 2. Two of them were previously described, Y111H (rs17366743) and a 3′ untranslated-region SNP (rs6773957). The third (rs17366653), which is the most significantly associated SNP, is previously unreported (P = 1.02E−07, MAF = 0.015 in GEMS; P = 1.48E−12, MAF = 0.020 in CoLaus; meta-P = 2.20E−17). The minor allele was estimated to decrease adiponectin levels by 0.24 μg/mL in the combined samples. Conditional analyses on rs17366743 and rs6773957 yielded highly significant results for rs17366653 (P < 1.1E−06 in GEMS and P < 2.0E−09 in CoLaus), indicating its effect is independent of the other two significant associations in the gene. Four rare nonsynonymous SNPs were found in GEMS and eight in CoLaus. Aggregation analysis gave no interesting results (P > 0.5) in GEMS and yielded a modest association in CoLaus (P = 0.02) (Supplementary Figs. 2 and 3).
TABLE 2.
Characteristics of three significant variants identified in analysis of resequencing data
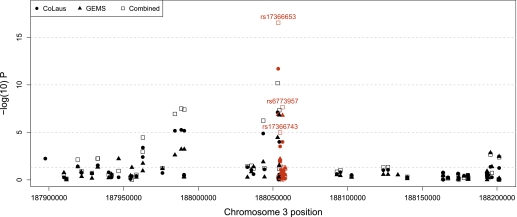
Recently, genome-wide association studies (GWAS) have identified associations between SNPs in and around ADIPOQ with adiponectin levels in European populations (7,8,24). For subjects included in our resequencing study with adiponectin measurements, Affymetrix 500 K genotype data were also available. We subsequently analyzed the 353 GWAS SNPs within 2 megabases (Mb) of ADIPOQ together with the 15 resequence SNPs with at least 10 copies of the minor allele. Results corresponding to analyses in GEMS, CoLaus, and meta-analysis are shown in Fig. 1. The novel association remained the most statistically significant. The effect of the variant was the largest among all the SNPs being evaluated. Conditional analyses on all other SNPs in this region showed that it was an independent signal from GWAS findings. Furthermore, imputation in a 2-Mb region based on different GWAS panels and data from the 1000 Genomes Project revealed this SNP could not be well imputed (r2 < 0.3). We thus conclude this novel association was previously missed by GWAS and its discovery is due to resequencing a large number of samples.
FIG. 1.
Association analysis results based on joint analysis of GWAS and resequence SNPs. The red and black symbols represent resequence and GWAS SNPs, respectively.
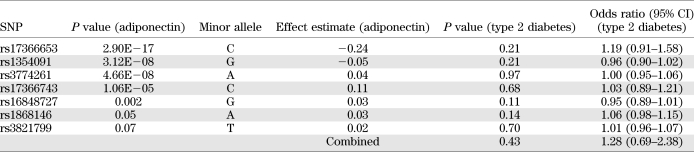
Seven independent associations (rs17366653, rs17366743, rs1354091, rs3774261, rs3821799, rs16848727, and rs1868146) were identified from the stepwise regression analysis in the GEMS and CoLaus combined dataset. Together, they explained 6% of the adiponectin variation. MAFs of these SNPs ranged from 0.02 to 0.46 (Supplementary Table 2). Bayes variable selection gave very similar results to the frequentist analysis above, with a posterior for the number of independent signals concentrated around 7 and 8 (Supplementary Fig. 4). Haplotype-specific tests corresponded well with individual SNP tests. We thus conclude that these SNPs exert independent effect on adiponectin levels. To examine whether any of the independent SNPs associated with adiponectin levels were contributing to type 2 diabetes risk, we carried out association analyses for the seven SNPs in the GO-DARTS study including 5,145 type 2 diabetic and 6,374 control subjects. None of the tests was statistically significant (P > 0.1) (Table 3). However, based on the effect estimate of adiponectin level change on type 2 diabetes risk (1), our statistical power to detect association between these SNPs and type 2 diabetes risk was estimated to range from 6 to 17% for an individual SNP and was at 42% for all seven SNPs combined (Supplementary Table 3).
TABLE 3.
Analysis of the seven independent SNPs associated with adiponectin levels in the GO-DARTS study
Testing for association between the seven independent SNPs associated with adiponectin levels and a host of metabolic and cardiovascular-related traits did not yield any significant findings in GEMS and CoLaus (Supplementary Table 4).
Bioinformatics analysis.
ASD, MaxEnt, and marginally NetGene2 predicted the minor allele of our most strongly associated SNP (rs17366653) to weaken the scores for splicing at the intron/exon 3 junction. This could lead to aberrant splicing at this site, resulting in an alternative transcript. Further experimental validation is required to validate this hypothesis.
DISCUSSION
By conducting a deep resequencing experiment, we identified a novel association between a low-frequency SNP in ADIPOQ and adiponectin levels in 3,665 individuals from two independent studies, and confirmed several associations previously identified via GWAS. However, all seven independent SNPs identified only explained about 6% of adiponectin variation in our samples. The unexplained phenotypic variation could be due to structural variations, infrequent noncoding variants, and environmental factors.
Our study provides the most complete assessment of genetic variation in ADIPOQ to date. Despite the large number of sequenced subjects, the vast majority of putatively functional variants detected were extremely rare (MAF <0.02%) and likely to be population specific. Recently, a rare (1.1%) nonsynonymous variant G45R of large effect was found in 1,240 Cuban Hispanics (25). This variant is not observed in our samples. The frequency of our most strongly associated SNP was ∼1.5% in Europeans, but only 0.3 and 0.6% in African Americans and Indian Asians, respectively. These observations illustrate the limited distribution of such low-frequency variants in different populations and underscore the importance of ethnicity in genetic association studies. An additional challenge of studying very rare variants is the difficulty of replication at the variant level. In CoLaus, we observed a nonsynonymous variant (P91R) of large effect, but only once in 14,002 sequenced subjects.
The inverse association between adiponectin levels and type 2 diabetes risk has been well established in epidemiology studies. By focusing on the adiponectin-encoding gene ADIPOQ, we were able to rule out concerns over pleiotropic effect of genetic variants associated with other traits. However, all the independent SNPs in ADIPOQ only explained 6% of the adiponectin variation in our samples. The estimated statistical power to detect a causal effect of adiponectin on the risk of type 2 diabetes through ADIPOQ was only 42% based on our study of 5,145 type 2 diabetic and 6,374 control subjects. Thus our negative finding is inconclusive to tease out the causal relationship between adiponectin and type 2 diabetes. Further investigation by large-scale and well-powered Mendelian randomization studies is warranted.
Supplementary Material
ACKNOWLEDGMENTS
The CoLaus study was supported by research grants from GlaxoSmithKline and from the Faculty of Biology and Medicine of Lausanne, Switzerland. Funding for the GEMS study was provided by GlaxoSmithKline. Funding for the GO-DARTS study was supported by the Wellcome Trust and genotyping and analysis by Wellcome Trust grant 083270/ Z/07/Z. J.R.P. is a Sir Henry Wellcome Postdoctoral Research Fellow (092447/Z/10/Z).
L.L.W., L.L., M.R.N., M.G.E., J.S., D.J.F., J.L.A., K.L.N., A.J.S., P.M.W., M.D.H., S.D.T., X.Y., L.R.C., S.L.C., V.M., J.C.W., and D.M.W. are employees of GlaxoSmithKline. No other potential conflicts of interest relevant to this article were reported.
L.L.W. researched data and wrote the manuscript. L.L., J.S., D.J.F., J.L.A., K.L.N., A.J.S., and X.Y. researched data. M.R.N., M.G.E., S.D.T., S.L.C., T.M.F., J.C.W., and D.M.W. contributed to discussion and reviewed and edited the manuscript. P.M.W., M.D.H., L.R.C., V.M., A.D.M., C.N.A.P., and J.R.P. reviewed and edited the manuscript. L.L.W. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank the co-primary investigators of the CoLaus study, Gerard Waeber (Centre Hospitalier Universitaire Vaudois, Lausanne, Switzerland) and Peter Vollenweider (Centre Hospitalier Universitaire Vaudois), and the primary investigator of the PsyColaus Study, Martin Preisig (Centre Hospitalier Universitaire Vaudois). The authors thank GEMS investigators: Scott Grundy (UT Southwestern Medical Center, Dallas, Texas), Jonathan Cohen, (UT Southwestern Medical Center), Ruth McPherson (Ottawa Heart Institute, Ottawa, Canada), Antero Kesaniemi (University of Oulu, Oulu, Finland), Robert Mahley (University of California, San Francisco, CA), Tom Bersot (University of California, San Francisco, CA), Philip Barter (The University of Adelaide, Adelaide, Australia), and Gerard Waeber (Centre Hospitalier Universitaire Vaudois). The authors are grateful to John Novembre and Daniel Wegmann (University of California, Los Angeles, CA) for helpful discussions. The authors thank study participants who provided written informed consent for the use of their DNA in genetic studies.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0985/-/DC1.
REFERENCES
- 1.Li S, Shin HJ, Ding EL, van Dam RM. Adiponectin levels and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA 2009;302:179–188 [DOI] [PubMed] [Google Scholar]
- 2.Ebrahim S, Davey Smith G. Mendelian randomization: can genetic epidemiology help redress the failures of observational epidemiology? Hum Genet 2008;123:15–33 [DOI] [PubMed] [Google Scholar]
- 3.Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol 2004;33:30–42 [DOI] [PubMed] [Google Scholar]
- 4.Hivert MF, Manning AK, McAteer JB, et al. Common variants in the adiponectin gene (ADIPOQ) associated with plasma adiponectin levels, type 2 diabetes, and diabetes-related quantitative traits: the Framingham Offspring Study. Diabetes 2008;57:3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jee SH, Sull JW, Lee JE, et al. Adiponectin concentrations: a genome-wide association study. Am J Hum Genet 2010;87:545–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindsay RS, Funahashi T, Krakoff J, et al. Genome-wide linkage analysis of serum adiponectin in the Pima Indian population. Diabetes 2003;52:2419–2425 [DOI] [PubMed] [Google Scholar]
- 7.Ling H, Waterworth DM, Stirnadel HA, et al. Genome-wide linkage and association analyses to identify genes influencing adiponectin levels: the GEMS study. Obesity (Silver Spring) 2009;17:737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richards JB, Waterworth D, O’Rahilly S, et al. GIANT Consortium A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet 2009;5:e1000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stirnadel H, Lin X, Ling H, et al. Genetic and phenotypic architecture of metabolic syndrome-associated components in dyslipidemic and normolipidemic subjects: the GEMS study. Atherosclerosis 2008;197:868–876 [DOI] [PubMed] [Google Scholar]
- 10.Firmann M, Mayor V, Vidal PM, et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc Disord 2008;8:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeggini E, Scott LJ, Saxena R, et al. Wellcome Trust Case Control Consortium Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008;40:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li R, Li Y, Fang X, et al. SNP detection for massively parallel whole-genome resequencing. Genome Res 2009;19:1124–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet 2008;83:311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177–188 [DOI] [PubMed] [Google Scholar]
- 15.Lunn DJ, Whittaker JC, Best N. A Bayesian toolkit for genetic association studies. Genet Epidemiol 2006;30:231–247 [DOI] [PubMed] [Google Scholar]
- 16.Lunn D, Thomas A, Best N, Spiegelalter D. WinBUGS-a Bayesian modelling framework: concepts, structure, and extensibility. Stat Comput 2000;10:325–337 [Google Scholar]
- 17.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 2007;81:1084–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet 2002;70:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res 2002;30:3894–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res 2001;11:863–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siepel A, Bejerano G, Pedersen JS, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 2005;15:1034–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunak S, Engelbrecht J, Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol 1991;220:49–65 [DOI] [PubMed] [Google Scholar]
- 23.Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 2009;37:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heid IM, Wagner SA, Gohlke H, et al. Genetic architecture of the APM1 gene and its influence on adiponectin plasma levels and parameters of the metabolic syndrome in 1,727 healthy Caucasians. Diabetes 2006;55:375–384 [DOI] [PubMed] [Google Scholar]
- 25.Bowden DW, An SS, Palmer ND, et al. Molecular basis of a linkage peak: exome sequencing and family-based analysis identify a rare genetic variant in the ADIPOQ gene in the IRAS family study. Hum Mol Genet 2010;19:4112–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.