Abstract
In earlier studies of the Iddm14 diabetes susceptibility locus in the rat, we identified an allele of the T-cell receptor (TCR) β-chain, Tcrb-V13S1A1, as a candidate gene. To establish its importance, we treated susceptible rats with a depleting anti-rat Vβ13 monoclonal antibody and then exposed them to either polyinosinic:polycytidylic acid or a diabetogenic virus to induce diabetes. The overall frequency of diabetes in the controls was 74% (n = 50), compared with 17% (n = 30) in the anti-Vβ13–treated animals, with minimal islet pathology in nondiabetic treated animals. T cells isolated from islets on day 5 after starting induction showed a greater proportion of Vβ13+ T cells than did peripheral lymph node T cells. Vβ13 transcripts recovered from day 5 islets revealed focused Jβ usage and less CDR3 diversity than did transcripts from peripheral Vβ13+ T cells. CDR3 usage was not skewed in control Vβ16 CDR3 transcripts. Anti-rat Vβ13 antibody also prevented spontaneous diabetes in BBDP rats. The Iddm14 gene is likely to be Tcrb-V13, indicating that TCR β-chain usage is a determinant of susceptibility to autoimmune diabetes in rats. It may be possible to prevent autoimmune diabetes by targeting a limited element of the T-cell repertoire.
Type 1 diabetes is a T-cell–mediated autoimmune disorder, the fundamental cause of which is unknown (1). Most strategies to prevent, arrest, or reverse it target T cells, either directly by altering number or function or indirectly via tolerizing antigens. To date, some interventions seem to preserve some β-cells after onset (2,3), but none safely and effectively prevent or reverse the disease.
A major need is for new, disease-specific modalities that avoid broad-spectrum targeting of immune-system components. This is difficult because multiple antigenic specificities participate in the diabetogenic response, and multiple alleles of immune-system genes contribute to the process. In addition, diverse antigenic specificities may be recognized by individuals with different major histocompatibility complex (MHC) haplotypes and may be presented in an atypical fashion (4). Treatments that target the whole T-cell population or cytokine receptors have the potential to increase the risk of infection and other complications.
To address these problems, we study rat models of the disease. Type 1–like autoimmune diabetes, both spontaneous and inducible, is relatively common among inbred rat strains that, like humans, express a high-risk class II MHC haplotype; in rats, it is designated RT1B/Du (5–7). We previously reported that Iddm14 (formerly designated Iddm4) is a dominant non-MHC susceptibility locus important for both spontaneous and induced autoimmune diabetes in multiple rat strains (8–14).
Studies of Iddm14 in eight RT1B/Du rat strains led to the identification of a susceptibility haplotype in the Tcrb-V locus (10). Single nucleotide polymorphism (SNP) haplotype mapping of this region of chromosome 4 encompassed Tcrb-V1–4, 5.1–5.2, 6, 7, 8.1–8.4, 9–13, and 15–20. These comprise the majority of the 24 Tcrb-V family members identified in previous studies and by our own bioinformatics (10). Our SNP haplotype mapping revealed that six rat strains susceptible to diabetes (KDP, BBDR, BBDP, LEW.1WR1, LEW.1AR1-iddm, and PVG-RT1u) all share one allele of β-chain variable region gene Tcrb-V13, designated Tcrb-V13S1A1 (15). Three rat strains that are resistant to, or confer resistance to, diabetes in genetic studies all express different alleles, either Tcrb-V13S1A2 (BN and WF rats) or Tcrb-V13S1A3P (F344 rats) (15). These polymorphisms are of interest because preferential usage of the Tcrb-V13S1A1 gene product, designated Vβ13a, by CD4+ but not CD8+ cells has been reported (15). Here, we report prevention of autoimmune diabetes by selective depletion of Vβ13a+ T cells in LEW.1WR1 and BBDP rats.
RESEARCH DESIGN AND METHODS
LEW.1WR1 and BBDP rats (RT1B/Du) were from BRM (Worcester, MA). BBDP rats become spontaneously diabetic at a rate of 60–90% between 50 and 120 days of age (5). LEW.1WR1 rats developed spontaneous diabetes at a rate of ~2.5% (16), but treatment with polyinosinic:polycytidylic acid (poly I:C) (16) or infection with viruses from several families (17) increases the frequency of diabetes to 30–100%. Animals were housed in viral antibody–free conditions, confirmed monthly to be serologically free of rat pathogens (11), and maintained in accordance with institutional and national guidelines (18).
Induction of diabetes.
“Triggered” diabetes in LEW.1WR1 rats of either sex at 21–30 days of age was induced with poly I:C (Sigma, St. Louis, MO), 1 μg/g i.p. three times weekly, as described (16). Typically, 80–100% of treated animals become diabetic within 12–18 days of the first injection (16). Treatment was for 40 days or until diabetes diagnosis. In one experiment, diabetes was induced by exposure to poly I:C (1 μg/g i.p.) on three consecutive days, followed 1 day later by injection of Kilham rat virus (KRV) (107 pfu), as described (17). Typically, 80–100% of animals become diabetic within 15–25 days. Poly I:C alone at this dose is not diabetogenic; KRV alone induces diabetes in ~40% of LEW.1WR1 rats (17). Animals were tested for glycosuria for 40 days or until the diagnosis of diabetes, defined as a plasma glucose >250 mg/dL (OneTouch; Johnson & Johnson, Milpitas, CA).
Diabetes prevention studies.
The hybridoma producing the 17D5 mouse anti-rat Vβ13 monoclonal antibody (mAb; IgG2a) recognizes the product of the Tcrb-V13S1A1 (Vβ13a) allele of the Tcrb-V13 (Vβ13) gene (15). The hybridoma producing the His42 mouse anti-rat Vβ16 (IgG2b) mAb (19) was a gift from Dr. Thomas Hünig. Both antibodies were prepared as ascites and purified by affinity chromatography. Mouse OKT8 anti-human CD8 mAb (IgG2a) was obtained from the American Type Culture Collection. In prevention studies, each mAb was administered intraperitoneally at a dose of 0.1 mg per rat in a volume of 0.5 mL. In studies in the LEW.1WR1 rat, mAb was injected three times weekly, and the first mAb injection was given 48 h before the first injection of poly I:C. BBDP rats were injected with mAb once weekly beginning at 45 days of age. Timing and total number of doses in each experiment is described in the results.
Measurement of T-cell depletion.
We quantified the effect of 17D5 and His42 on peripheral T-cell populations by measuring Vβ4, Vβ13, Vβ15, and Vβ16 mRNA transcripts by quantitative RT-PCR. This method was used because we lacked anti-Vβ13 and anti-β16 antibodies against a second epitope to allow us to distinguish if cells were depleted or only masked. Total RNA was isolated from spleens, mesenteric lymph nodes, and cervical lymph nodes (CLNs) at the onset of diabetes or at the end of the experiment. In brief, tissues were harvested and stored in RNAlater (Qiagen, Valencia, CA). RNA was prepared using Ultraspec (Biotecx, Houston, TX) and treated with Turbo DNA-free (Applied Biosystems, Carlsbad, CA) to prevent genomic contamination. cDNA was synthesized from 2 μg total RNA using the High Capacity RNA-to-cDNA Kit (Applied Biosystems).
The primers used for quantitative RT-PCR (qRT-PCR) were designed using Primer 3 (http://frodo.wi.mit.edu/primer3) and T-cell receptor (TCR)-Vβ gene sequences. Primers were selected to be of optimal size for real-time PCR, with the 5′ primer located in the leader sequence and the 3′ primer in a region of the gene that did not contain SNPs. All primers were synthesized by Integrated DNA Technologies (Coralville, IA). Primer sequences are given in the Supplementary Data. Quantitative PCR was performed with an ABI 7900HT Sequence Detector using SYBR Green PCR Mix (Applied Biosystems). Amplification data were collected and analyzed using software from ABI–SDS2.2.
Recovery and expansion of islet-infiltrating T cells.
To phenotype early islet-infiltrating T cells, we adapted the expansion method of Jarchum et al. (20). For each experiment, eight LEW.1WR1 rats were treated with poly I:C, as described above. Animals were killed 48 h after one dose of poly I:C (day 3 of diabetes pathogenesis) or 48 h after the second dose of poly I:C (day 5 of pathogenesis). Pancreatic islets were isolated as described (21,22). Handpicked isolated islets were cultured for 7 days in 24-well tissue culture plates at a density of 50 islets/mL/well, as described (20), to expand infiltrating T-cell populations. Culture medium consisted of RMPI-1640 supplemented with 10% FBS (Hyclone, Logan, UT), 1 mmol/L Na pyruvate, nonessential amino acids, 28 μmol/L β-mercaptoethanol, and 50 units/mL recombinant rat interleukin-2 (PeproTech, Rocky Hill, NJ). Cells were cultured in 5% CO2 95% air at 37°C. On day 7, islets and infiltrating cells were collected and passed through a 40-micron strainer to retain the islets. Infiltrating cells were analyzed by flow cytometry.
Flow cytometry.
Antibodies to the αβ TCR (clone R73), CD25 (clone OX-39), CD4 (clone OX-35), CD8α chain (clone OX-8), and Vβ13 TCR (clone 18B1) were from BD Pharmingen, and FoxP3 antibody (clone FJK-16a) was from eBiosciences. Isotype control antibodies (mouse IgG1, IgG2a, and IgG2b) and phycoethrin- or allophycocyanin-conjugated streptavidin were from Pharmingen (San Diego, CA). Antibodies either were directly conjugated with fluorochromes (fluorescein isothiocyanate, peridinin-chlorophyll-protein complex (PerCP), allophycocyanin, or Pacific Blue) or were used as biotin conjugates followed by streptavidin. For analyses of FoxP3, cells were permeabilized using the FoxP3 Staining Buffer Set (eBiosciences), per manufacturer’s instructions. Data were acquired using an LSR II instrument (BD Biosciences) and analyzed with FloJo software. A minimum of 100,000 viable cells in each sample were analyzed. The lymphocyte fraction was gated according to forward and side scatter.
Analyses of Tcrb-V13 transcripts.
To analyze combinatorial variants in the CDR3 region of Vβ13+ T cells in islets early in the course of diabetes, three cohorts of eight LEW.1WR1 rats were treated with intraperitoneal poly I:C, 1 μg/g on days 0 and +3, and killed on day +5, when spleens and pancreata were harvested. Islets were isolated as described (21,22), and hand-picked islets were immediately immersed in RNALater (Applied Systems-Ambion, Austin, TX) or Trizol (Invitrogen, Carlsbad, CA). Total RNA was isolated according to the manufacturers’ protocols and reverse transcribed into cDNA (High Capacity cDNA Reverse Transcription Kit; Applied Biosystems). Tcrb-Vβ13 transcripts were amplified with a Tcrb-Vβ13–specific forward primer and a Cβ1/Cβ2 reverse primer (Supplementary Materials) using platinum high-fidelity Taq DNA polymerase (Invitrogen). Transcripts were cloned directly into the pCR4-TOPO vector with the TOPO TA cloning kit (Invitrogen). Transformants were plated on LB-agar-ampicillin plates. Single clones were grown in LB-ampicillin overnight, and plasmids were isolated with the QIAprep spin mini-prep kit (Qiagen, Valencia, CA). Tcrb-V13S1A1 plasmid inserts were sequenced by GeneWiz (Plainfield, NJ) with the T3 primer. Sequences were aligned and compared with the Multiple Sequence Alignment program by Clustal (http://www.genome.jp/tools/clustalw/). To exclude artifacts of PCR amplification or bacterial clone expansion on the relative abundance of individual CDR3 regions, in particular to exclude multiple amplicons of any one transcript, a correlation plot was constructed with the relative abundance of Jβ isotypes, as determined by sequencing versus relative abundance of Jβ isotypes quantified by qRT-PCR (see below).
Analysis of Jβ relative abundance.
To analyze diversity and Jβ frequency in T cells, we developed a qRT-PCR assay for TCR-Jβ region abundance. The assay was applied to Tcrb-V13 and Tcrb-V16 transcripts from islets and spleens of poly I:C LEW.1WR1 rats and untreated LEW.1WR1 rats. Forward primers were designed for Tcrb-V13 and Tcrb-V16, and PCR was performed with reverse primers in each of 12 TCR-Jβ regions (Supplementary Data). The relative proportions of Jβ isotypes were calculated after normalization to total Tcrb-V13 and Tcrb-V16 transcript abundance, respectively, using the comparative Ct method for quantification of results, as described (http://www.ambion.com/techlib/basics/rtpcr/index.html).
Histology.
Pancreata were removed at diabetes onset or on day 40 and fixed in 10% buffered formalin. Paraffin-embedded sections were sectioned at 4-μm intervals and stained with hematoxylin and eosin. Intensity of insulitis was scored as follows: 0, no inflammatory mononuclear cell (MNC) infiltration; 1+, small numbers of infiltrating MNCs with preservation of islet architecture; 2+, moderate infiltrating MNCs with preservation of architecture; 3+, many MNCs with most islets affected and distortion of islet architecture; and 4+, florid infiltration and distorted islet architecture or end-stage islets with or without residual inflammation. Some sections were processed immunohistochemically for the presence of insulin and glucagon and scored as follows: 0, no evidence of hormone being present; 1+ hormonal staining detectable but in reduced number of islets and/or reduced number of cells per islet; and 2+ staining consistent with normal islet cell number.
Statistics.
Diabetes-free survival was analyzed using Kaplan-Meier methodology; equality of survival distributions was tested by log-rank statistic (23). Parametric data are given as arithmetic means ± 1 SD or ±SE, as indicated in the figure legends. Fisher exact statistic was used for analyzing 2 × 2 tables and the χ2 test for larger tables. Comparisons of three of more means used one-way and two-way ANOVAs and the either the Bonferroni or least significant differences procedure for a posteriori contrasts (23). Two-tailed P values <0.05 were considered statistically significant.
RESULTS
Depletion of Vβ13+ T cells prevents poly I:C-triggered autoimmune diabetes.
We first determined that a single injection of 17D5 mAb was associated with a reduction of ~60% in the number of Vβ13+ T cells in the spleen of LEW.1WR1 rats (data not shown). We then determined the protective effect of anti-Vβ13 mAb treatment on diabetes penetrance in LEW.1WR1 rats treated with poly I:C three times weekly.
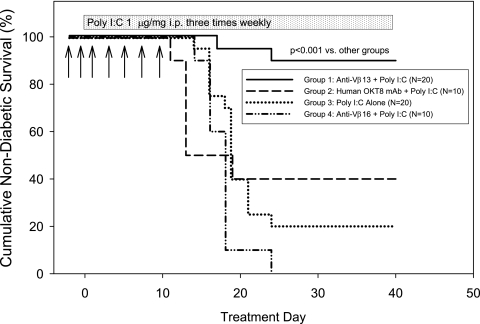
Trial 1 compared diabetes frequency in rats treated with either 17D5 anti-Vβ13 mAb or mouse anti-human OKT8 as a control. Trial 2 compared diabetes frequency in rats treated with 17D5 anti-Vβ13 mAb, His42 anti-Vβ16 mAb, or vehicle. The combined results of the two trials are shown in Fig. 1. Diabetes frequency in rats given poly I:C and 17D5 mAb was 10% (2 of 20). In contrast, diabetes frequency in controls averaged 85% (34 of 40; P < 0.001).
FIG. 1.
Frequency of diabetes in LEW.1WR1 rats treated with poly I:C given three times weekly (shaded bar). Compared with controls given an irrelevant mouse anti-human mAb (group 2) or the His42-depleting anti-rat Vβ16 mouse mAb (group 4) or no treatment (group 3), only anti-Vβ13 (group 1) protected against type 1 diabetes. Antibody injections (0.1 mg/dose) are indicated by arrows (overall log-rank statistic = 23.89; df = 3; P < 0.001). Diabetes frequency in groups 2, 3, and 4 was statistically similar (overall log-rank statistic = 0.30; df = 2; P = NS). The ratio of males to females in all treatment groups averaged 1:1.
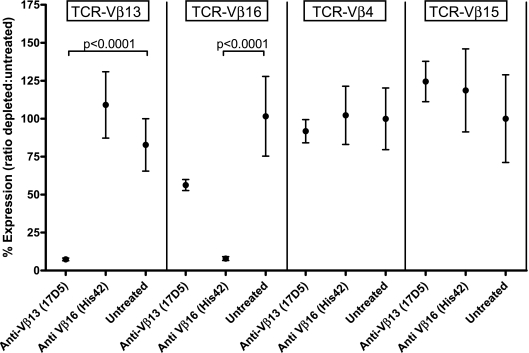
As shown in Fig. 2, treatment with 17D5 mAb was associated with a specific reduction in Tcrb-V13 mRNA transcripts, having no significant effect on Tcrb-V4, -15, or -16 transcripts; His42 mAb treatment was associated with a reduction of similar magnitude only in Tcrb-V16 transcripts.
FIG. 2.
To determine the extent and specificity of T-cell depletion resulting from treatment with the 17D5 and His42 mAbs, we measured relative transcript abundance for four Tcrb-V isotypes in T cells isolated from the spleens of rats that were either untreated, treated with 17D5 anti-rat Vβ13 mAb, or treated with His42 anti-rat Vβ16 mAb, as described in research design and methods. The control reference isotypes (Vβ4 and Vβ15) for this experiment were selected at random. The data show that Vβ13+ T cells are statistically significantly depleted by the 17D5 reagent and that Vβ16+ T cells are statistically significantly depleted by the His42 reagent; no other paired comparisons were statistically significant. Expression of each β-chain variable region transcript was adjusted for relative Cβ transcript abundance and is presented as the ratio of abundance in treated animals relative to untreated controls. Each data point represents the means ± SE of relative transcript abundance from 9 or 10 individual rats.
Depletion of Vβ13+ T cells prevents insulitis and preserves islet architecture.
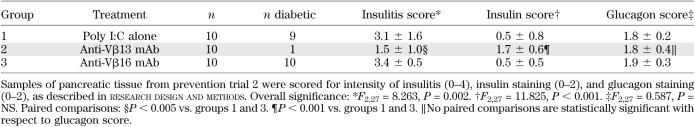
Pancreata from animals in trial 2 were analyzed histologically. As shown in Table 1, there was significantly less insulitis in animals treated with anti-Vβ13 mAb than in rats in either control group; 9 of 10 of these animals were nondiabetic. In one Vβ13 mAb–treated rat that became diabetic, there was extensive insulitis (not shown). Likewise, there was significantly more insulin detectable in the anti-Vβ13 mAb–treated group. In contrast, glucagon immunohistochemistry revealed no detectable differences among groups. Fig. 3 shows histological samples from a diabetic rat that had received anti-Vβ16 mAb and a nondiabetic rat treated with anti-Vβ13 mAb. The diabetic animal had intense insulitis (Fig. 3D), no detectable insulin (Fig. 3E), and glucagon that was readily detectable but suggestive of collapse of the central core of β-cells (Fig. 3F). In contrast, the anti-Vβ13–treated rat displays little or no insulitis (Fig. 3A), abundant insulin staining (Fig. 3B), and glucagon staining in a normal annular pattern (Fig. 3C).
TABLE 1.
Quantitative comparison of islet histology in treated and untreated rats
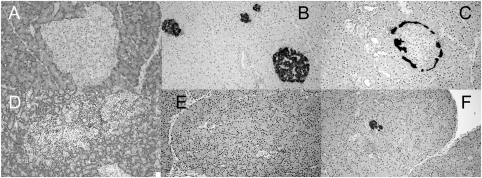
FIG. 3.
Representative histological sections from animals in trial 1 (see RESULTS). A–C: A nondiabetic rat treated with both poly I:C and anti-Vβ13 mAb. D–F: A diabetic animal treated with both poly I:C and anti-Vβ16 mAb. A and D: A normal-appearing islet and a distorted islet with intense insulitis, respectively. B and E: Abundant vs. absent insulin immunostaining, respectively. C: Normal immunostaining for glucagon in an annular pattern. F: Residual staining for glucagon with loss of normal architecture, which together with E is suggestive of selective loss of islet β-cells. Overall statistical analysis of the histological data is given in Table 1.
Depletion of Vβ13+ T cells prevents virus-triggered autoimmune diabetes.
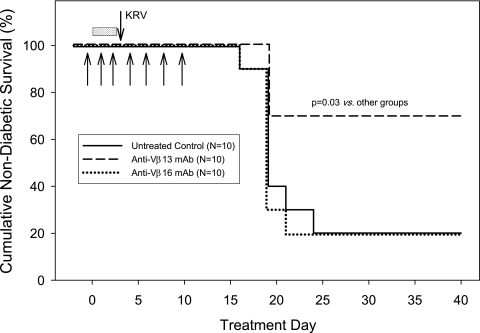
We also tested a second, possibly more physiologic model of induced diabetes, one triggered by virus. Trial 3 examined the protective efficacy of 17D5 anti-Vβ13 mAb in rats exposed to a small priming dose of poly I:C followed by infection with KRV. The priming dose of poly I:C is by itself nondiabetogenic but increases the penetrance of virus-triggered diabetes (from ~40 to ~100%) (17). As shown in Fig. 4, diabetes frequency in 17D5-treated rats was 30% (3 of 10) compared with 80% (8 of 10; P = 0.03) in both His42 mAb–treated animals and otherwise-untreated controls.
FIG. 4.
Frequency of diabetes in LEW.1WR1 rats treated with a small priming dose of poly I:C (shaded bar) and then inoculated with KRV (downward arrow), as described in research design and methods. Compared with controls given the His42-depleting anti-rat Vβ16 mouse mAb or no additional treatment, only anti-Vβ13 protected against type 1 diabetes. Antibody injections (0.1 mg/dose) are indicated by upward arrows (overall log-rank statistic = 6.95; df = 2; P = 0.03).
CD4+Vβ13+ T cells are abundant in islets early in the course of diabetes onset.
Because the above data indicate that cells expressing the gene product of Tcrb-V13S1A1 are important in diabetes pathogenesis, we hypothesized that, as a fraction of total T cells, the percentage of Vβ13a+ T cells in the islets of susceptible rats early in pathogenesis should be greater than the percentage in peripheral lymphoid tissues. Islets were isolated 48 h after one or two injections of poly I:C (i.e., on day 3 or day 5 of diabetes progression) and cultured for 7 days; the infiltrating T-cell phenotype was analyzed as described (20).
We first analyzed pooled islets harvested from eight individual rats on day 3 after injection with poly I:C on day 0. The percentage of TCR+Vβ13+ T cells recovered from isolated islets was 4.1%, which was indistinguishable from the percentage (5.3%) recovered from pooled fresh CLNs from eight untreated animals.
In two additional experiments, we analyzed islets harvested on day 5 after injection with poly I:C on days 0 and +3. The percentages of TCR+Vβ13+ T cells recovered from isolated islets were higher than from day 3 islets (5.2 and 9.6%), whereas the percentages in T cells from fresh CLNs were about the same (3.5 and 3.3%). In these two trials, the percentage of CD4+ islet–derived T cells that were Vβ13+ averaged 19.5%, whereas in CLNs the percentage of CD4+Vβ13+ T cells averaged 3.5%. In contrast, there were no apparent differences in the percentage of CD8+ cells that were Vβ13+ recovered from day 5 islets (average 2.5%) compared with CLN T cells (2.0%). Finally, we observed that only a minority of CD4+CD25+Vβ13+ recovered islet T cells were FoxP3+ (9.6%). The percentage of TCR+CD4+CD25+FoxP3+ T cells that were Vβ13+ was 5.9% in the islets and 3.8% in CLNs.
Vβ13/Jβ mRNA transcripts in prediabetic islets are skewed.
The gene products of Tcrb-V13S1A1 and Tcrb-V13S1A2 encode different amino acid sequences for both the CDR1 and CDR2 regions of the β-chain (15). This polymorphism distinguishes WF and other type 1 diabetes–resistant strains from BBDR, BBDP, KDP, and LEW.1WR1 type 1 diabetes–susceptible strains, all of which share the same class II MHC (10). CDR1 and CDR2 sequences are encoded within each Tcrb-V allele and are not altered by the combinatorial processes that create the CDR3 regions of the TCR.
To investigate the molecular mechanism by which Tcrb-V13S1A1 might confer susceptibility to autoimmunity, we performed a comparative analysis of Vβ13 mRNA transcripts from islets versus peripheral lymphoid tissues. These were cloned as cDNAs into plasmid vectors and sequenced. T cells were obtained from islets or spleens from rats on the fifth day of progression to diabetes or from the spleens of age-matched untreated control rats. The day 5 point was selected based on the flow cytometry data above.
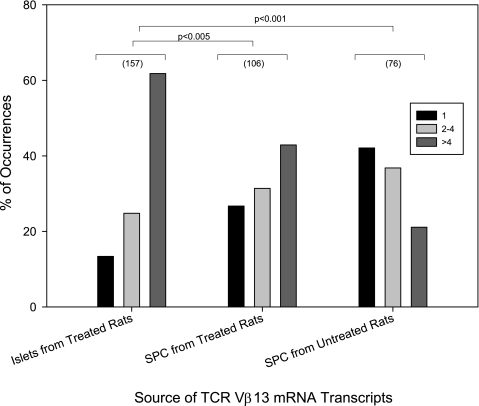
The number of times that an individual CDR3 sequence was present in Vβ13 transcripts was quantified. As shown in Fig. 5, relatively few CDR3 sequences (13%) were found only once in the pools of islet mRNA transcripts; more (25%) were detected two to four times, and nearly two-thirds (62%) were detected more than four times (n = 157). In contrast, relatively more CDR3 sequences (27%) were found only once in the pools of splenocyte mRNA transcripts; 31% were detected two to four times, and fewer than one-half (43%) were detected more than four times (n = 107; χ2=11.3; df = 2; P < 0.005 vs. islet-derived transcripts). Among spleen cells from untreated rats, the effect was more pronounced; 42% of CDR3 sequences were found only once in the pools of islet mRNA transcripts, 37% were found two to four times, and only 21% were found more than four times (n = 76; χ2=38.7; df = 2; P < 0.001 vs. islet-derived transcripts). The correlational analysis of the relative abundance of Jβ isotypes, as determined by sequencing versus relative abundance of Jβ isotypes (see research design and methods), showed significant positive correlation and demonstrates that uniqueness/diversity represented by CDR3 direct sequencing is an accurate measure of TCR-Vβ13 CDR3 diversity.
FIG. 5.
Frequency distribution for unique Vβ13a-CDR3 sequences. To assess the likelihood that Vβ13+ TCRs present in prediabetic islets might be involved in the recognition of a specific antigen or antigens, the number of times that a specific CDR3 sequence was present in a Vβ13 transcript was quantified as described in research design and methods. The total number of transcripts analyzed for each tissue is given in parentheses. Relatively few Vβ13a-CDR3 sequences were found only once in the pools of mRNA transcripts obtained from islets of poly I:C–treated rats (21 of 157); more were found two to four times (39 of 157) and nearly two-thirds were found more than four times (97of 157). In contrast, relatively more Vβ13a-CDR3 sequences were found only once in the mRNA transcripts from spleens of poly I:C–treated rats (28 of 106); a few more were found two to four times (33 of 106) and fewer than one-half were found more than four times (45 of 106). Among Vβ13a-CDR3 transcripts obtained from spleen cells of untreated rats, 32 of 76 were observed only once, 28 of 76 two to four times, and 16 of 76 more than four times (overall χ2 = 39.5; df = 4; P < 0.0001). SPC, spleen cells.
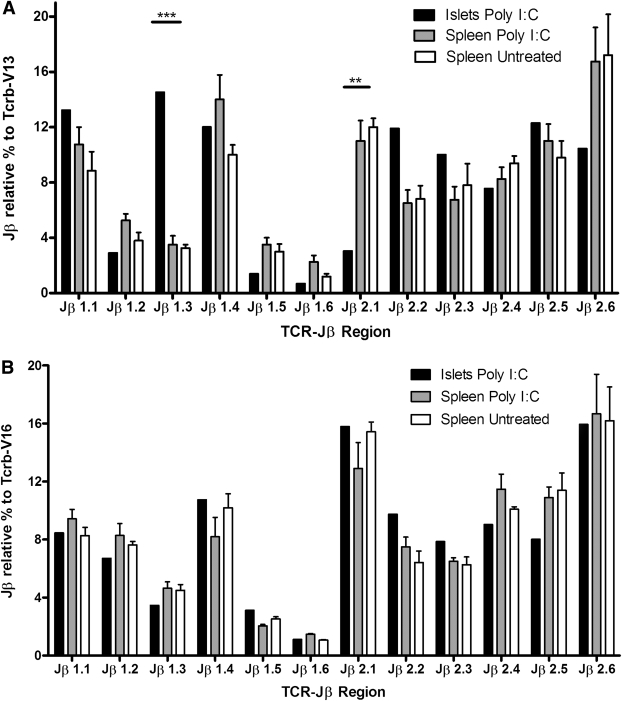
These data suggested that antigen-specific expansion of Vβ13+ T cells has occurred in the islets of prediabetic rats, but the possibility of unequal amplification of subsets of targets, artificially skewing the observed diversity, cannot be entirely excluded. To address this issue and validate further our inference regarding the relative skewing of islet versus peripheral Tcrb-V13 transcripts, we developed a qRT-PCR assay to quantify relative utilization of the 12 Tcrb-J regions expressed by Vβ13+ (diabetes-associated) versus Vβ16+ (control) T cells. We observed differential expression of Tcrb-J regions between islet-homing versus peripheral T cells in the Vβ13+ samples that, as expected, significantly correlated with Jβ abundance determined by direct sequencing (Supplementary Fig. 1), As shown in Fig. 6A, we observed skewed Tcrb-J usage in islet-infiltrating Vβ13+ T cells with overrepresentation of Jβ1.3 and underrepresentation of Jβ2.1. Spleen Vβ13+ T cells from poly I:C–treated and untreated rats did not differ with respect to the frequency of individual Jβ usage. In contrast, the representation of different Jβ segments in Vβ16+ T-cell transcripts was not skewed in the islets in comparison with peripheral Vβ16 transcripts. (Fig. 6B), further supporting a role for Vβ13+ T cells in the early recognition of antigen in islets.
FIG. 6.
Skewed usage of Jβ elements in Vβ13+ but not in Vβ16+ islet-infiltrating T cells. cDNA was prepared from mRNA isolated from islets and spleen cells of prediabetic LEW.1WR1 rats and from spleen cells of untreated control LEW.1WR1 rats as described in research design and methods. To generate the islet data points, mRNA from eight treated rats was pooled in each of two separate experiments; data are shown as the mean. Spleens from four or five treated and untreated rats were analyzed individually, and data are shown as means ± SE. A: Jβ element distribution among the Vβ13 transcripts obtained from each of these three sources. The data are presented as the percentage of Vβ13+ transcripts that used each of the indicated Jβ regions (see research design and methods). Two-way ANOVA revealed a significant overall main effect for Jβ usage by both Vβ13 (F = 18.46; df = 11; P < 0.001) and Vβ16 (F = 29.92; df = 11; P < 0.001) transcripts irrespective of the source of the mRNA. There were no statistically significant main effects for RNA source by either Vβ13 or Vβ16 transcripts. However, as shown in A, there was a highly significant interaction between Jβ usage and mRNA source (F = 3.17; df = 22; P < 0.001) for Vβ13 mRNA transcripts. Furthermore, this was specifically localized to overrepresentation of Jβ1.3 (***P < 0.001) and underrepresentation of Jβ2.1 (**P < 0.01). This is indicative of nonrandom usage of Jβ segments by Vβ13+ T cells present in the islets of prediabetic rats. In contrast, as shown in B, there was no interaction between Jβ usage and mRNA source in Vβ16+ transcripts, indicative of random Jβ isotype usage irrespective of tissue of origin or diabetes status.
Depletion of Vβ13+ T cells prevents spontaneous autoimmune diabetes.
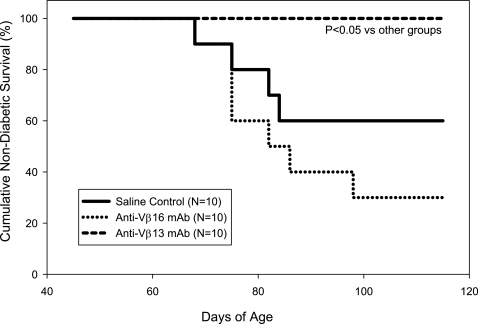
In a final experiment, we sought to exclude the possibility that the protective effect of anti-Vβ13 mAb was restricted to the LEW.1WR1 strain and/or autoimmune diabetes that had been induced by a trigger. To do so, we treated cohorts of spontaneously diabetic BBDP rats with vehicle, anti-Vβ13 mAb, or anti-Vβ16 mAb. As shown in Fig. 7, treatment with anti-Vβ13 mAb through 115 days of age completely prevented diabetes, whereas diabetes occurred in 40%, or vehicle-injected, and 70%, or anti-Vβ16 mAb–treated, rats (P < 0.01). Among the rats that were nondiabetic at the end of the experiment, underlying insulitis was more severe among vehicle-treated controls (mean score 2.5 ± 0.8; n = 6) than among rats treated with anti-Vβ16 mAb (1.0 ± 1.7; n = 3) or anti-Vβ13 mAb (1.3 ± 0.7; n = 10; F2,16 = 4.01; P < 0.05).
FIG. 7.
Frequency of spontaneous diabetes in BBDP rats that were either treated with saline, treated with 17D5 anti-rat Vβ13 mAb, or treated with His42 anti-rat Vβ16 mAb, as described in research design and methods. Injections were given once weekly between 45 and 100 days of age. The anti-Vβ13 mAb–treated group differed from both other groups (overall log-rank statistic = 10.10; df = 2; P < 0.01; untreated control vs. anti-Vβ13 mAb: log rank = 4.75; df = 1; P < 0.05).
DISCUSSION
These results demonstrate that depletion of T cells expressing a single β-chain variable region allele, Vβ13a, can prevent autoimmune diabetes in multiple rat models. Depletion of Vβ16+ T cells had no protective effect. The treatment was effective in animals in which diabetes was triggered by innate immune activation (poly I:C), triggered by parvoviral infection, or spontaneous. These functional results confirm and extend our hypothesis that Tcrb-V13S1A1 confers a high degree of susceptibility to rat autoimmune diabetes and is the Iddm14 gene.
The flow cytometry results suggest involvement of Vβ13+ T cells during early stages of disease progression. The limited data available do not permit identification of an autoreactive effector phenotype, but they do suggest that CD4+CD25+Vβ13+FoxP3− cells play a role in the day 5 inflammatory lesion. This is consistent with previous studies of diabetes in the LEW.1AR1-iddm rat (24) and the BB rat (25), showing that CD4+ T cells are capable of adoptively transferring diabetes. Of interest, we have reported a CD4 bias for rat T cells expressing the Vβ13S1A1 variant β-chain; this skews the TCR repertoire and substantially alters the CD4+:CD8+ ratio among Vβ13+ T cells (15). The importance of Vβ13+ early in disease is highlighted by our finding that weekly anti-Vβ13 mAb started immediately after onset does not reverse hyperglycemia (J.P.M., unpublished data).
Sequencing of Vβ13+ TCR β-chains present in islets on day 5 of disease progression showed that a relatively limited number of “focused” CDR3 specificities from Vβ13+ T cells are clonally expanded. These specificities may mediate islet-antigen recognition in the prediabetic LEW.1WR1 rat. Oligoclonality was not observed in control tissues or in T cells bearing Vβ16 TCR β-chains. Taken together, the genetic, prevention, and phenotyping datasets imply that an immunologic synapse comprising rat class II B/Du on antigen-presenting cells and Vβ13a on T cells confers the geometry required for an unknown antigenic determinant to initiate diabetes. We speculate that the interaction of CDR1 and CDR2 with peptide MHC (pMHC) could affect thymic selection (15) and that these differences account for the specificity of Tcrb-V13S1A1 in conferring susceptibility to diabetes in the rat. Unusual binding geometry of TCR with pMHC recently has been implicated in the generation of autoreactivity at the crystallographic level (26,26). Our data are consistent with recent literature suggesting that allelic and isotypic TCR α- and β-chain usage, independent of CDR3 usage, contributes to differential immunoreactivity in vitro and in vivo (27,28). Finally, we note that a small number, typically 2–5%, of WF rats that do not express Vβ13a can become induced to become diabetic (13), suggesting that non-Vβ13a+ TCRs can mediate autoimmune diabetes but with much lower efficiency.
Our results suggest that, when present in the context of a high-risk MHC class II haplotype (RT1B/Du), Tcrb-V13S1A1 is a major determinant of disease probability in the rat. We recognize that, based on human and mouse data, it has been assumed that redundancy among cognate rat TCRs would preclude reliance on any one allele of TCR α- or β-chains for disease susceptibility. Our data and data of others strongly suggest that this assumption may be faulty.
In the NOD mouse, no TCR chain family has been linked genetically to type 1 diabetes, but a limited TCRβ repertoire could nonetheless be important. Several analyses of T-cell repertoire, especially in early prediabetic stages, implicate an oligoclonal T-cell response by NOD mice (29,30). NOD and C57BL/6 mice share alleles at most Tcrb loci (http://phenome.jax.org), and thus Idd-congenic strains based on differences between NOD and C57BL/6 would unlikely be to show linkage to the Tcrb region. The one linkage test of a role for Tcrb-V used a parental strain (SJL) that has a major deletion of Tcrb-V and proved only that murine Tcrb-V5, -8, -9, -11, -12, and -13 are not required for insulitis or spontaneous diabetes (31). However, NOD8.3 TCR transgenic mice, in which the majority of T cells are skewed toward expression of an IGRP-specific Vβ8+ TCR, develop disease with rapid onset (32,33). In addition, the prediabetic NOD mouse has an oligoclonal T-cell response (29,30,34–36), and preferential TCR usage has been observed. In one case, it is a dramatic TCR α-chain restriction (predominantly AV13S3) (37). In another, preferential TCR β-chain usage (TRBV-15) has been reported (38).
Human datasets that fail to implicate the TCR in autoimmune diabetes are based on either classical gene mapping or SNP haplotyping. Neither method is ideal for detecting recessive protective alleles because “resistant” individuals would have to be homozygous for the loss of Tcrb-V elements, as we propose is important in the rat. Furthermore, stratification by four-digit MHC haplotype may be required to detect TCR effects in human type 1 diabetes, something that has to date not been reported.
We recognize that studies of anti-Vβ reagents in NOD mice have given conflicting results. Empirical treatment with an anti-Vβ8 mAb reportedly prevented disease in NOD/Wehi mice with cyclophosphamide-accelerated diabetes (39) and in islet isografted diabetic NOD mice (40). However, genetic analyses showed that NOD mice that cannot express Vβ8 nonetheless become diabetic. We would point out, however, that the selection of Vβ8 as a target was not based on genetic susceptibility analyses, as in our studies of rat Vβ13, and NOD mouse diabetes is notoriously easy to prevent with many reagents, including more than 10 different mAbs, among them IgG2a isotype control (41).
In conclusion, TCR β-chain usage may contribute to the penetrance of both spontaneous and induced autoimmune diabetes in genetically susceptible rats of multiple strains. The presence of abundant Vβ13+ T cells in prediabetic islets suggests that Vβ13a is a marker of the disease. Rats prone to autoimmune diabetes may offer the opportunity to determine how Tcrb-V chain variation contributes to autoimmunity and in detail how pMHC/TCR interaction initiates the development of β-cell–specific autoimmunity. Studies to knock down rat Tcrb-V13 expression and to clone diabetogenic Vβ13+ T cells are underway in our laboratories.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grants 7-11-BS-102, 7-08-RA-106 (to J.P.M.), and 7-06-BS-18 (to E.P.B.) from the American Diabetes Association and R21-AI-088480 (to E.P.B.), R43-DK-085910 (to J.P.M.), AI-46629, AI-73871, DK-32520 (to D.L.G.), and Diabetes Endocrinology Research Center Grant DK-32520 from the National Institutes of Health. This research made use of the Center Morphology Core.
B.Y. is an employee of BRM, Inc., the vendor of the LEW.1WR1 and BBDP rats. No other potential conflicts of interest relevant to this article were reported.
Z.L., R.E., and T.H. researched data, contributed to the discussion, and reviewed and edited the manuscript. L.C., J.H.L., and B.Y. researched data and contributed to the discussion. D.L.G. contributed to the discussion and reviewed and edited the manuscript. E.P.B. and J.P.M. researched data, contributed to the discussion, and wrote, reviewed, and edited the manuscript. J.P.M. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0867/-/DC1.
Z.L. is currently affiliated with the Department of Medical Microbiology, Weifang Medical University, Shandong, China.
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
See accompanying commentary, p. 976.
REFERENCES
- 1.Morran MP, Omenn GS, Pietropaolo M. Immunology and genetics of type 1 diabetes. Mt Sinai J Med 2008;75:314–327 [DOI] [PubMed] [Google Scholar]
- 2.Keymeulen B, Walter M, Mathieu C, et al. Four-year metabolic outcome of a randomised controlled CD3-antibody trial in recent-onset type 1 diabetic patients depends on their age and baseline residual beta cell mass. Diabetologia 2010;53:614–623 [DOI] [PubMed] [Google Scholar]
- 3.Pescovitz MD, Greenbaum CJ, Krause-Steinrauf H, et al. Type 1 Diabetes TrialNet Anti-CD20 Study Group Rituximab, B-lymphocyte depletion, and preservation of beta-cell function. N Engl J Med 2009;361:2143–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadinski BD, Zhang L, Crawford F, Marrack P, Eisenbarth GS, Kappler JW. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc Natl Acad Sci USA 2010;107:10978–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mordes JP, Poussier P, Rossini AA, Blankenhorn EP, Greiner DL. Rat models of type 1 diabetes: genetics, environment, and autoimmunity. In Animal Models of Diabetes: Frontiers in Research. 2nd ed Shafrir E, Ed. Boca Raton, FL, CRC Press, 2007, p. 1–39 [Google Scholar]
- 6.Ellerman KE, Like AA. Susceptibility to diabetes is widely distributed in normal class IIu haplotype rats. Diabetologia 2000;43:890–898 [DOI] [PubMed] [Google Scholar]
- 7.Awata T, Guberski DL, Like AA. Genetics of the BB rat: association of autoimmune disorders (diabetes, insulitis, and thyroiditis) with lymphopenia and major histocompatibility complex class II. Endocrinology 1995;136:5731–5735 [DOI] [PubMed] [Google Scholar]
- 8.Martin A-M, Maxson MN, Leif J, Mordes JP, Greiner DL, Blankenhorn EP. Diabetes-prone and diabetes-resistant BB rats share a common major diabetes susceptibility locus, iddm4: additional evidence for a “universal autoimmunity locus” on rat chromosome 4. Diabetes 1999;48:2138–2144 [DOI] [PubMed] [Google Scholar]
- 9.Blankenhorn EP, Rodemich L, Martin-Fernandez C, Leif J, Greiner DL, Mordes JP. The rat diabetes susceptibility locus Iddm4 and at least one additional gene are required for autoimmune diabetes induced by viral infection. Diabetes 2005;54:1233–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mordes JP, Cort L, Norowski E, et al. Analysis of the rat Iddm14 diabetes susceptibility locus in multiple rat strains: identification of a susceptibility haplotype in the Tcrb-V locus. Mamm Genome 2009;20:162–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mordes JP, Leif J, Novak S, DeScipio C, Greiner DL, Blankenhorn EP. The iddm4 locus segregates with diabetes susceptibility in congenic WF.iddm4 rats. Diabetes 2002;51:3254–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hornum L, DeScipio C, Markholst H, et al. Comparative mapping of rat Iddm4 to segments on HSA7 and MMU6. Mamm Genome 2004;15:53–61 [DOI] [PubMed] [Google Scholar]
- 13.Blankenhorn EP, Descipio C, Rodemich L, et al. Refinement of the Iddm4 diabetes susceptibility locus reveals TCRVbeta4 as a candidate gene. Ann NY Acad Sci 2007;1103:128–131 [DOI] [PubMed] [Google Scholar]
- 14.Fuller JM, Bogdani M, Tupling TD, et al. Genetic dissection reveals diabetes loci proximal to the gimap5 lymphopenia gene. Physiol Genomics 2009;38:89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stienekemeier M, Hofmann K, Gold R, Herrmann T. A polymorphism of the rat T-cell receptor beta-chain variable gene 13 (BV13S1) correlates with the frequency of BV13S1-positive CD4 cells. Immunogenetics 2000;51:296–305 [DOI] [PubMed] [Google Scholar]
- 16.Mordes JP, Guberski DL, Leif JH, et al. LEW.1WR1 rats develop autoimmune diabetes spontaneously and in response to environmental perturbation. Diabetes 2005;54:2727–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tirabassi RS, Guberski DL, Blankenhorn EP, et al. Infection with viruses from several families triggers autoimmune diabetes in LEW*1WR1 rats: prevention of diabetes by maternal immunization. Diabetes 2010;59:110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Institute of Laboratory Animal Resources Commission on Life Sciences, National Research Council: Guide for the Care and Use of Laboratory Animals. Washington, DC, National Academy Press, 1996 [Google Scholar]
- 19.Kampinga J, Kroese FG, Pol GH, et al. A monoclonal antibody to a determinant of the rat T cell antigen receptor expressed by a minor subset of T cells. Int Immunol 1989;1:289–295 [DOI] [PubMed] [Google Scholar]
- 20.Jarchum I, Takaki T, DiLorenzo TP. Efficient culture of CD8(+) T cells from the islets of NOD mice and their use for the study of autoreactive specificities. J Immunol Methods 2008;339:66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parker DC, Greiner DL, Phillips NE, et al. Survival of mouse pancreatic islet allografts in recipients treated with allogeneic small lymphocytes and antibody to CD40 ligand. Proc Natl Acad Sci USA 1995;92:9560–9564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacy PE, Kostianovsky M. Method for the isolation of intact islets of Langerhans from the rat pancreas. Diabetes 1967;16:35–39 [DOI] [PubMed] [Google Scholar]
- 23.Bewick V, Cheek L, Ball J. Statistics review 12: survival analysis. Crit Care 2004;8:389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arndt T, Wedekind D, Weiss H, et al. Prevention of spontaneous immune-mediated diabetes development in the LEW.1AR1-iddm rat by selective CD8+ T cell transfer is associated with a cytokine shift in the pancreas-draining lymph nodes. Diabetologia 2009;52:1381–1390 [DOI] [PubMed] [Google Scholar]
- 25.Métroz-Dayer M-D, Mouland A, Brideau C, Duhamel D, Poussier P. Adoptive transfer of diabetes in BB rats induced by CD4 T lymphocytes. Diabetes 1990;39:928–932 [DOI] [PubMed] [Google Scholar]
- 26.Sethi DK, Schubert DA, Anders AK, et al. A highly tilted binding mode by a self-reactive T cell receptor results in altered engagement of peptide and MHC. J Exp Med 2011;208:91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gras S, Chen Z, Miles JJ, et al. Allelic polymorphism in the T cell receptor and its impact on immune responses. J Exp Med 2010;207:1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burrows SR, Chen Z, Archbold JK, et al. Hard wiring of T cell receptor specificity for the major histocompatibility complex is underpinned by TCR adaptability. Proc Natl Acad Sci USA 2010;107:10608–10613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinn A, McInerney M, Huffman D, et al. T cells to a dominant epitope of GAD65 express a public CDR3 motif. Int Immunol 2006;18:967–979 [DOI] [PubMed] [Google Scholar]
- 30.Haskins K. Pathogenic T-cell clones in autoimmune diabetes: more lessons from the NOD mouse. Adv Immunol 2005;87:123–162 [DOI] [PubMed] [Google Scholar]
- 31.Shizuru JA, Taylor-Edwards C, Livingstone A, Fathman CG. Genetic dissection of T cell receptor V β gene requirements for spontaneous murine diabetes. J Exp Med 1991;174:633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verdaguer J, Yoon JW, Anderson B, et al. Acceleration of spontaneous diabetes in TCR-β-transgenic nonobese diabetic mice by β-cell cytotoxic CD8+ T cells expressing identical endogenous TCR-α chains. J Immunol 1996;157:4726–4735 [PubMed] [Google Scholar]
- 33.Dudek NL, Thomas HE, Mariana L, et al. Cytotoxic T-cells from T-cell receptor transgenic NOD8.3 mice destroy β-cells via the perforin and Fas pathways. Diabetes 2006;55:2412–2418 [DOI] [PubMed] [Google Scholar]
- 34.Drexler K, Burtles S, Hurtenbach U. Limited heterogeneity of T-cell receptor V β gene expression in the early stage of insulitis in NOD mice. Immunol Lett 1993;37:187–196 [DOI] [PubMed] [Google Scholar]
- 35.Baker FJ, Lee M, Chien YH, Davis MM. Restricted islet-cell reactive T cell repertoire of early pancreatic islet infiltrates in NOD mice. Proc Natl Acad Sci USA 2002;99:9374–9379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galley KA, Danska JS. Peri-islet infiltrates of young non-obese diabetic mice display restricted TCR β-chain diversity. J Immunol 1995;154:2969–2982 [PubMed] [Google Scholar]
- 37.Abiru N, Wegmann D, Kawasaki E, Gottlieb P, Simone E, Eisenbarth GS. Dual overlapping peptides recognized by insulin peptide B:9-23 T cell receptor AV13S3 T cell clones of the NOD mouse. J Autoimmun 2000;14:231–237 [DOI] [PubMed] [Google Scholar]
- 38.Li L, He Q, Garland A, et al. Beta cell-specific CD4+ T cell clonotypes in peripheral blood and the pancreatic islets are distinct. J Immunol 2009;183:7585–7591 [DOI] [PubMed] [Google Scholar]
- 39.Bacelj A, Charlton B, Mandel TE. Prevention of cyclophosphamide-induced diabetes by anti-V β 8 T-lymphocyte-receptor monoclonal antibody therapy in NOD/Wehi mice. Diabetes 1989;38:1492–1495 [DOI] [PubMed] [Google Scholar]
- 40.Bacelj A, Mandel TE, Charlton B. Anti-V β 8 antibody therapy prevents disease recurrence in fetal pancreas isografts in spontaneously diabetic nonobese diabetic mice. Transplant Proc 1992;24:220–221 [PubMed] [Google Scholar]
- 41.Atkinson MA, Leiter EH. The NOD mouse model of type 1 diabetes: as good as it gets? Nat Med 1999;5:601–604 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.