Abstract
In diabetic patients and susceptible mice, insulin is a targeted autoantigen. Insulin B chain 9-23 (B:9-23) autoreactive CD4 T cells are key for initiating autoimmune diabetes in NOD mice; however, little is known regarding their origin and function. To this end, B:9-23–specific, BDC12-4.1 T-cell receptor (TCR) transgenic (Tg) mice were studied, of which, despite expressing a single TCR on the recombination activating gene–deficient background, only a fraction develops diabetes in an asynchronous manner. BDC12-4.1 CD4 T cells convert into effector (Teff) and Foxp3+-expressing adaptive regulatory T cells (aTregs) soon after leaving the thymus as a result of antigen recognition and homeostatic proliferation. The generation of aTreg causes the heterogeneous diabetes onset, since crossing onto the scurfy (Foxp3) mutation, BDC12-4.1 TCR Tg mice develop accelerated and fully penetrant diabetes. Similarly, adoptive transfer and bone marrow transplantation experiments showed differential diabetes kinetics based on Foxp3+ aTreg’s presence in the BDC12-4.1 donors. A single-specificity, insulin-reactive TCR escapes thymic deletion and simultaneously converts into aTreg and Teff, establishing an equilibrium that determines diabetes penetrance. These results are of particular importance for understanding disease pathogenesis. They suggest that once central tolerance is bypassed, autoreactive cells arriving in the periphery do not by default follow solely a pathogenic fate upon activation.
Type 1 diabetes (T1D) is an autoimmune disease where antigen-specific T cells can mediate the destruction of insulin-producing β-cells in mice and are thought to substantially contribute to human diabetes pathogenesis. The NOD mouse has served to model T1D pathogenesis for decades, and insulin-specific T cells determine disease development in NOD mice. Approximately 50% of CD4+ T-cell clones isolated from pancreata of prediabetic NOD mice react to insulin (1). Of these insulin-reactive clones, 90% respond specifically to the insulin B chain epitope B:9-23 (2). NOD mice with a genetic deletion of both native insulin genes (Ins1 and Ins2) expressing a mutated nonimmunogenic but hormonally active proinsulin transgene (dKO/B16:A mice) do not develop T1D, demonstrating that immunologic recognition of insulin is crucial for the initiation of diabetes in this model (3).
Current evidence suggests that insulin epitopes bind weakly and with a fast dissociation rate to the NOD-specific class II major histocompatibility complex (MHC) allele I-Ag7, so that the limited supply of this peptide results in partial failure during negative selection (4). However, preproinsulin-2 expression is detected in thymic medullary epithelial cells, is regulated by autoimmune regulator, and can result in thymic deletion (5). Of evidence, Ins2 gene ablation significantly accelerates T1D progression in NOD mice (6,7), whereas Ins1-deficient mice are protected from diabetes (8).
Ins1 and Ins2 gene expression has been found in pancreatic draining lymph nodes (PDLNs) and also in islets (9). However, it is not known which insulin epitopes and by which mechanism they activate insulin-specific T cells that escape negative selection and what the functional outcome is. Moreover, it is not well understood whether both effector and regulatory T cells (Teffs and Tregs) specific to insulin can be generated simultaneously in vivo. In a recent study, two populations of B:9-23–reactive cells were described: type A that recognizes the 13-21 register on professional antigen-presenting cells and type B that recognizes the 12-20 register. This single amino acid shift distinguishes both sets of B:9-23–reactive cells that either get deleted in the thymus (type A) or get activated in the periphery by other mechanisms (type B) (10).
To determine the fate of insulin-reactive cells in more detail, we analyzed the T-cell receptor (TCR) transgenic (Tg) mouse line specific for B:9-23, namely BDC12-4.1. BDC12-4.1 TCR Tg mice develop spontaneous insulitis but no diabetes in F1 mice (FVB x NOD), whereas some diabetes manifests in NOD.recombination activating gene (RAG)KO (back-cross one generation). Disease progression is modified by a series of genetic factors such as H-2g7 haplotype and the presence of additional T-/B-cell receptor–rearranged genes (RAG+ versus RAGKO) (11). Approximately 40% of the H-2g7.BDC12-4.1.NOD.RAGKO (termed here BDC12-4.1.RAGKO) mice develop spontaneous T1D by 40 weeks of age. This incomplete diabetes penetrance is strikingly different from several other CD4 Tg TCR NOD mouse models, where diabetes development either occurs in all mice or none, for example in BDC2.5 and 2H6 TCR Tg NOD mice on the RAG- and severe combined immunodeficiency–deficient backgrounds, respectively (12,13).
Currently, the function of TCR (antigen) specificity in diabetes development is controversial. It is not clear at which level antigen can determine the fate of a T cell (14), although it is shown that islet specificity is required for homing in the pancreas (15). Foxp3 is the master transcription factor for bona fide thymic-derived, natural Tregs (nTregs) (16). In addition to thymic nTreg differentiation, extrathymic Foxp3+ Treg development can result in mice in particular differentiation niches that allow conversion (adaptive [aTreg]) (17). From earlier studies, it was shown that the thymic and peripheral Treg TCR repertoires are similar but diverse from the T-conventional repertoire (18,19), suggesting that minimal T conventional to aTreg conversion occurs. In Foxp3-deficient (scurfy) mice, the pathogenic TCR repertoire is more similar to the nTreg compartment of wild-type (wt) mice, suggesting that nTregs bear autoreactive TCRs (20). Generating TCR Tg mice by means of retroviral transduction showed that nTreg-derived TCR clones cannot instruct the nTreg fate (21). Thus, although some results highlight the instructive nature of the TCR, TCR specificity does not completely determine the fate of a T cell.
Most previous studies with TCR Tg mice revealed that TCR stimulation determined by the affinity for the cognate antigen and the presence or absence of endogenous TCR rearrangement dictates whether nTregs will develop (22,23). In the absence of TCR rearrangement (RAGKO), no nTregs develop unless their cognate is present and in sufficient levels (24). In this study, we found that BDC12-4.1 TCR Tg, B:9-23–reactive, Treg cells can be efficiently selected in the thymus in RAG+ mice but not in RAGKO mice. However, CD4+CD25+Foxp3+ Treg cells could be readily detected in the periphery, spleen, pancreas, and PDLNs of BDC12-4.1.RAGKO mice. De novo, Foxp3-expressing aTregs were generated in the periphery as a result of antigen recognition and homeostatic proliferation and were responsible for the protection from T1D in 60% of the Tg mice. As evidence, when these mice were crossed onto the C57BL/6.scurfy background, diabetes developed faster and affected 100% of the Tg mice. We conclude that pathogenic T cells (Teffs) and aTregs with identical B:9-23–specific TCR specificity can be induced in the periphery from the same TCR-bearing precursors, but diabetes develops because counter-regulation by the autoreactive (insulin-specific) aTreg is inadequate. Thus, we identified a TCR that can instruct both peripheral Treg and Teff differentiation by insulin recognition.
RESEARCH DESIGN AND METHODS
Mice.
BDC12-4.1 TCR Tg mice were previously described (11). Heterozygous Foxp3wt/mut C57BL/6 mice I-Ab.RAG+ females (strain 4088) were purchased from The Jackson Laboratory (Bar Harbor, ME). A minimum of two crosses with NOD.BDC12-4.1.RAGKO background was needed to generate I-Ag7/g7.BDC12-4.1.Foxp3mut.RAGKO animals. In all experiments, Tg littermates served as genetic controls. Mice were maintained in La Jolla Institute for Allergy and Immunology animal facility or at the University of Colorado Health Sciences Center for Laboratory Animal Care in Denver under specific pathogen-free conditions according to each institution’s animal care and use committee guidelines for the use and care of laboratory animals.
Genotyping.
Genotyping for the BDC12-4.1 Tg TCRα and TCRβ, RAG, and I-A typing were performed as described previously (11). The presence of the Foxp3mut allele could be detected by PCR (forward primer, TCA GGC CTC AAT GGA CAA AA; reverse primer, CAT CGG ATA AGG GTG GCA TA) with a band size near 1,000 base pairs (bp). Females were classified as either heterozygous Foxp3wt/mut or homozygous Foxp3mut/mut by microsatellite typing with 6-carboxyfluorescein (FAM)-conjugated forward primer, ATG AGA AGAAGG AAG ATC AGCG, and an unlabeled reverse primer, ACC TGG GAA GGA ACT ATT GC. The microsatellites were analyzed with a real-time PCR ABI 3100 Avant Analyzer (Foster City, CA). The mutant peaks appear at 344 bp and the wt peaks are at 342 bp.
Glucose monitoring.
Mice were followed for diabetes by weekly testing of tail blood using either the Freestyle glucometer and test strips from Abbott Laboratories (Abbott Park, IL) or the ReliOn Ultima system (Alameda, CA). Mice whose glucose values exceeded 250 mg/dL were tested the next day and were considered diabetic if the second reading also exceeded 250 mg/dL.
Flow cytometry.
Cells were washed in fluorescence-activated cell sorter (FACS) staining buffer (0.1% sodium azide, 1 mmol/L EDTA, and 2% FCS in PBS) and incubated with 10 μl/mL Fc block (2.4G2) (BD-Pharmingen, San Diego, CA) for 10 min. After the blocking step, cells were stained for CD4, CD8α, CD44, CD62L, CD69, CD25 and CD127, killer cell lectin-like receptor subfamily G (KLRG)-1, CD103, V β 2 (Vβ2) (BDC12-4.1), or V β 4 (Vβ4) (2H6) (eBioscience or BD-Pharmingen, San Diego, CA). For intracellular Foxp3, Helios, or Ki-67 detection, cells were fixed with Foxp3 Fix/Perm buffer and stained with the corresponding monoclonal antibodies (eBioscience). For cytokine detection, splenocytes were activated in vitro for 4 h with anti-CD3/28 (10/5 μg/mL, respectively) or phorbol myristic acid/ionomycin (50/1000 ng/ml, respectively) in the presence of brefeldin A (BfA, 10 μg/mL) (Sigma-Aldrich, St. Louis, MO). Cells were costained for Foxp3, interferon-γ (IFN-γ), and interleukin-10 (IL-10) after fixation with the Foxp3 Fix/Perm kit. All antibody incubations were performed at 4°C for 30 min (isotype controls were included). Cells were immediately acquired on a FACSCalibur or LSRII flow cytometer (BD-Pharmingen) and analyzed using FlowJo software (Treestar).
Immunofluorescence histological staining.
Pancreas sections (6-μm) were cut by cryostat and dried overnight. For positive control, spleen sections were included. Upon rehydration with TBS-PI (Tris-buffered saline containing protease inhibitor, mini EDTA-free tablets; Roche) and fixation (0.4% paraformaldehyde, 15 min at room temperature [RT]), sections were blocked in 5% BSA and 2% goat serum TBS-PI (30 min RT). To block endogenous biotin activity, the avidin/biotin blocking kit from Vector was used according to the manufacturer’s instructions. As primary antibodies, anti–CD4-FITC (clone RM4-5, 1/50 dilution; BD-Pharmingen), guinea pig anti-swine insulin (1/100 dilution; Dako), and biotinylated anti-Foxp3 (clone FJK-16S, 1/50 dilution; eBioscience) were used. Primary antibody incubations were carried for 1.5–2 h at RT in a damp chamber. For detecting CD4, the Alexa Fluor 488 signal amplification kit for fluorescein dyes (Invitrogen) was used according to the manufacturer’s instructions. For detecting Foxp3 and insulin, Alexa Fluor 647–coupled streptavidin (1/1,000 dilution; Invitrogen) and Alexa Fluor 594–labeled polyclonal goat anti–guinea pig IgG antibody (1/1,000 dilution; Molecular Probes) were added overnight at 4°C or for 1.5 h at RT, respectively. Finally, nuclei were stained with Hoechst 33258 dye (1/200 dilution, 20 min; Invitrogen). After washing, sections were mounted with SlowFade Gold mounting reagent (Invitrogen), and coverslips were sealed with nail polish. Images were recorded using FluoView FV10i confocal microscope (Olympus), and analysis was performed with Fluoview ASW2.1 (Olympus) and Image Pro Analyzer 7.0 (Media Cybernetics) software packages.
Enzyme-linked immunosorbent assay.
Purified, FACS-sorted CD4SP T cells from the thymi of BDC12-4.1.RAGKO mice were cultured in 96-well plates at 0.25 × 106 cell/well in the presence of 0.5 × 106/well of T cell–depleted splenocytes (TDS) as antigen-presenting cells and insulin B:9-23 peptide sequence 1 (PHLVEALYLVCGERG) or 2 (SHLVEALYLVCGERG) at 50 μg/mL. TDS consisted of spleens magnetically depleted of CD90.2+ T cells with the use of αCD90.2 antibody-coated microbeads (Miltenyi Biotech Inc., Auburn, CA) from NOD.Ins2KO or Ins1KO donor mice. Supernatants were collected 3 days after culture for IL-4, transforming growth factor-β1 (TGF-β1), IFN-γ, and IL-10 detection. Cytokines present in the supernatants were quantified by a sandwich enzyme-linked immunosorbent assay (ELISA) using standard commercially available kits (Pharmingen OptEIA mouse; BD-Pharmingen).
Splenocyte transfer.
Spleens were extracted under aseptic conditions and homogenized by grinding between two glass microscope slides or passing through cell strainers. Erythrocytes (red blood cells) were lysed using red blood cell lysis buffer (R7757) (Sigma-Aldrich) or ACK buffer. Splenocytes were washed in sterile PBS three times. Cells were resuspended in the appropriate volume of sterile PBS before transfer. Varying amounts of primary, unsorted splenocytes (1.5 × 105 to 6 × 106 cells) were injected intraperitoneally in 200 μL into 6–8-week-old I-Ag7.C57BL/6.RAGKO or NOD.RAGKO recipients.
Bone marrow transplantation.
Bone marrow (BM) was harvested under aseptic conditions from the tibias and femurs of 4–5-week-old donors in MACS buffer (PBS, 0.5% BSA, and 2 mmol/L EDTA) and washed twice. Mature B and T cells were depleted by negative selection with CD4, CD8, and CD45RB/B220 magnetic microbeads and the autoMACS Magnetic Cell Sorter (Miltenyi Biotech Inc.) according to the manufacturer’s instructions. Ten million BM cells (in 200 μL) were injected retro-orbitally into sublethally X ray–irradiated (200 rads), 6–8-week-old I-Ag7.C57BL/6.RAGKO or NOD.RAGKO recipients.
Statistical analysis.
The statistical significance of the difference between means was determined using the two-tailed Student t test. All statistical analyses, including Kaplan-Meier survival curve analyses, were performed using GraphPad Prism version 4.01 for Windows (GraphPad Software, San Diego, CA). P values were as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
RESULTS
BDC12-4.1.RAGKO mice have both B:9-23–specific Teffs and Foxp3+ Tregs in the periphery, but accumulate Teffs as they age.
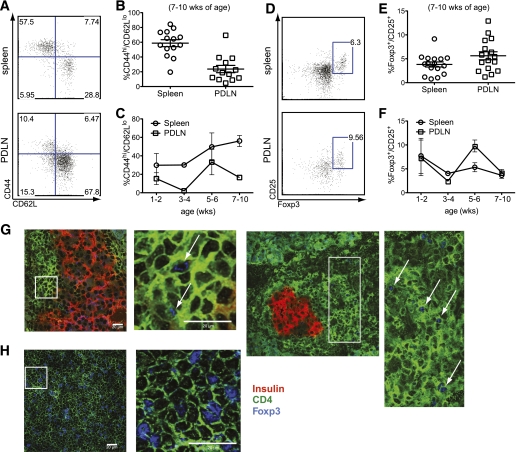
Earlier studies had shown that BDC12-4.1 mice only develop T1D on the RAGKO background with ∼40% penetrance by 40 weeks of age, whereas RAG+ mice develop insulitis but no overt diabetes (11). On the RAGKO background, no other TCRα (TCRα) chains were expressed, and only CD4 single-positive (SP) cells were able to mature in the thymus (Supplementary Fig. 1). To investigate whether Treg activity would contribute to diabetes resistance in BDC12-4.1.RAGKO mice, we performed detailed phenotypic analysis. A significant proportion (60–70% in the spleen and 20% in the PDLNs) of the remaining CD4+Vβ2+ cells displayed marked activation, exhibiting a typical Teff phenotype (CD44hiCD62Llow) by 7–10 weeks of age (Fig. 1A and B), whereas CD69 levels were more pronounced in the PDLNs (data not shown). Interestingly, when we searched for the presence of Tregs in BDC12-4.1.RAGKO mice, on average, ∼3% of CD4+Vβ2+CD127lo cells in the spleen and ∼5% in the PDLNs were CD25+Foxp3+ (Fig. 1D–F). Foxp3+ Tregs were also detected with immunofluorescence staining directly in the pancreatic infiltrate of BDC12-4.1.RAGKO mice (Fig. 1G and H). Thus, BDC12-4.1 T cells can develop into Teffs as well as aTregs likely after exiting the thymus in a naïve state (see next section).
FIG. 1.
BDC12-4.1.RAGKO mice have both B:9-23–specific Teffs and Foxp3+ Tregs in the periphery, but accumulate Teffs as they become older. A and B: Splenocytes and PDLN-derived cells from 7–10-week-old BDC12-4.1.RAGKO mice were analyzed by flow cytometry after gating on the CD4+Vβ2+ population (B:9-23–specific T cells) for the presence of CD44hi/CD62Llo Teffs. Mice showed downregulation of TCRβ expression in the spleen (∼30% of CD4+ cells were Vβ2high) (data not shown). Representative flow cytometric plots displaying the CD44/CD62 L profile in the spleen and PDLNs after gating on the CD4+Vβ2+ population (A), and cumulative data for more than ten 7–10-week-old mice are displayed (B), with each symbol representing individual mice. C: The frequency of Teffs was monitored over time (with age) in the spleen and PDLNs of BDC12-4.1.RAGKO mice. D–F: Tissues from the same mice were analyzed by flow cytometry after gating on the CD4+Vβ2+CD127lo population for the presence of CD25+Foxp3+ (Treg). It is interesting to note that Tregs and Teffs both appear to undergo cyclical changes in the pancreatic lymph node but not the spleen, possibly indicative of recurring changes in the Teff/Treg equilibrium. G: Pancreatic sections from 10-week-old BDC12-4.1.RAGKO mice were prepared and stained for insulin (red), CD4 (green), and Foxp3 (blue). Triple immunofluorescence labeling was performed as described in research design and methods. Higher magnification and capture was performed for the selected region, which is indicated with a white box. H: Spleen sections were stained similarly to serve as a positive control. White arrows indicate the presence of Foxp3+CD4+ T cells. (A high-quality digital representation of this figure is available in the online issue.)
Next, we monitored the Teff and aTreg frequencies over time and found that fewer CD44hiCD62Llow cells accumulated in the PDLNs compared with the spleen, correlating with the slightly increased aTreg frequency in that site (Fig. 1C and F). In the spleen, the number and activation status of Teffs consistently increased with age, whereas aTreg frequency remained relatively stable. In contrast, in the PDLNs, Teffs and aTregs both underwent cyclical increases and decreases.
Insulin-specific Foxp3+ Tregs do not originate in the thymus but are induced in the periphery.
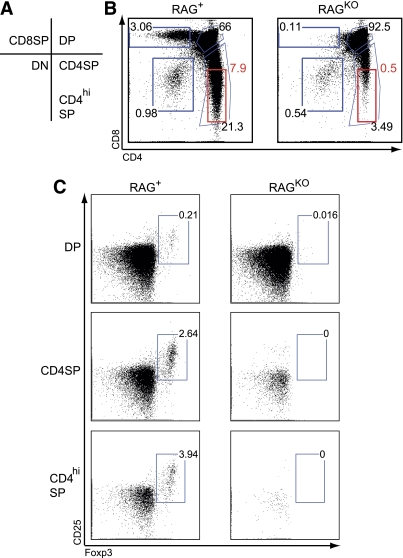
Foxp3+ Treg cells can be generated in the thymus as a result of agonist recognition during thymic selection. To define the precise origin of Foxp3+ cells in BDC12-4.1.RAGKO mice, we analyzed thymocytes by flow cytometry. The percentage of viable CD4/CD8 double-positive (DP) thymocytes was high, but they did not exhibit the typical characteristics of cognate antigen recognition; no CD4/CD8 coreceptor downregulation and marginal CD69 downregulation were observed (Supplementary Fig. 2). However, TCRβ levels were somewhat reduced, and a 90% reduction in mature CD4 SP cells was seen in RAGKO compared with RAG+ mice. This suggests that some degree of negative selection took place, most probably at the transition from the DP to SP stage (Supplementary Fig. 2). Had insulin-specific negative selection been complete, no CD4SP cells should be detected (25).
Interestingly, no Foxp3+ nTregs could be detected within the CD4SP population on the RAGKO background (Fig. 2), arguing strongly that the Foxp3+ Tregs detected in the periphery were in fact induced BDC12-4.1 aTregs. Our findings also suggest that the CD25+Foxp3+ cells detected in the thymus in RAG+ mice were the result of TCR rearrangement (pairing between the Tg Vβ2 chain with non-Tg Vα chains). These observations highlight the significance of de novo aTreg induction in enforcing tolerance. Autoreactive T cells convert into aTregs to keep Teffs of the same or other specificities under their direction.
FIG. 2.
Insulin-specific Foxp3+ Tregs do not originate in the thymus. Representative thymi from BDC12-4.1 TCR Tg mice on the RAG+ and RAGKO genetic backgrounds at 7–10 weeks of age were analyzed. A: The DP, CD4 SP, CD8SP, double-negative (DN), and CD4hiSP gating are shown. B: A representative dot blot is shown. After gating on the respective populations, analysis for the presence of nTregs (CD25+Foxp3+) was performed. On the RAGKO background, no nTregs could be identified in either DP, CD4SP, or CD4hiSP gated thymocyte populations (C). On the RAG+ background, nTreg development was readily detected as a result of TCR rearrangement (pairing of the Tg Vβ chain with non-Tg, endogenous Vα chains). (A high-quality color representation of this figure is available in the online issue.)
Insulin recognition in vitro can induce Foxp3 upregulation in BDC12-4.1 TCR Tg cells.
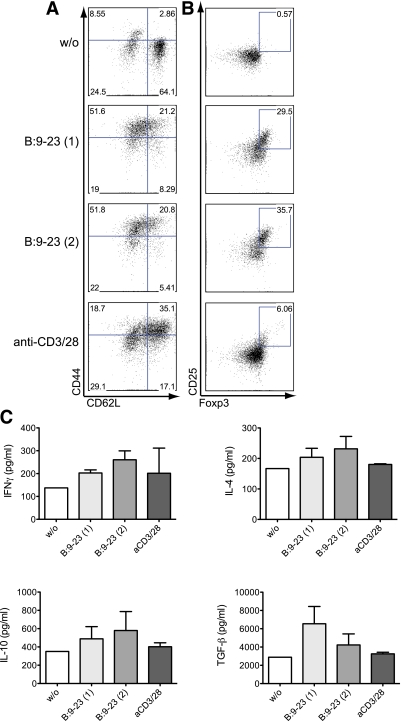
In contrast to humans, mice express two insulin genes, Ins1 and Ins2, and the B:9-23 peptides, to which BDC12-4.1 Tg cells react, differ in their amino acid sequence at position 9 (S for INS2 and P for INS1). ELIspot analysis showed that BDC12-4.1 Tg cells produce IFN-γ upon stimulation with both B:9-23 sequences, whereas they are not specific for B24-C36 or B16:A (Supplementary Research Design and Methods and Fig. 3). We hypothesized that one of the mechanisms involved in acquiring Foxp3 expression in the periphery would include insulin recognition. Therefore, we sorted CD4SP cells from the thymus of BDC12-4.1.RAGKO mice and placed them in culture with TDS from Ins2KO mice loaded with either B:9-23 peptide (INS1 or 2). After 48 h stimulation with either B:9-23 INS1 (1) or INS2 (2), upregulation of CD44 in conjunction with CD62 L downregulation occurred (Fig. 3A). Most importantly, B:9-23 stimulation also induced generation of CD25+Foxp3+ aTreg (Fig. 3B), suggesting that insulin recognition can induce BDC12-4.1 T cells to acquire both effector and regulatory properties. It is striking that a T cell with a fixed TCR can convert into a Treg upon antigen encounter in vitro in the absence of exogenously added TGF-β1. ELISA analysis of the cell culture supernatants revealed generation of IL-10, IFN-γ, and IL-4 and low amounts of TGF-β1 (Fig. 3C). This TGF-β1 expression could explain the Foxp3 induction and aTreg formation in the in vitro cultures. Altogether, activation of naïve, thymically derived CD4+ BDC12-4.1 TCR Tg cells (RAGKO background) in vitro resulted in the generation of effector and Foxp3-expressing T cells and the production of cytokines with proinflammatory and immunoregulatory properties.
FIG. 3.
BDC12-4.1 CD4SP cells convert into Teffs and Tregs upon antigen recognition in vitro. CD4+CD8− (CD4SP) thymocytes from BDC12-4.1.RAGKO mice were FACS-sorted and cultured in the presence of TDS from Ins2KO NOD mice. The stimulation lasted for 48 h in the absence (without [w/o]) or presence of 50 μg/mL B:9-23 peptide (Ins1 or Ins2 sequence), B:9-23(1) and B:9-23(2), respectively. For control, plate-bound anti-CD3 (1 μg/mL) plus anti-CD28 (0.5 μg/mL) were used to stimulate the cells. Cells were consequently stained and analyzed by flow cytometry after gating on CD4+CD8− for the expression of CD44 and CD62 L (A) or CD25 and Foxp3 (B). Cell supernatant was quantified with ELISA for the presence of four cytokines: IL-10, IL-4, IFN-γ, and TGF-β1 4 days after stimulation (C). (A high-quality color representation of this figure is available in the online issue.)
Peripheral Foxp3+ aTregs coproduce IL-10 and IFN-γ.
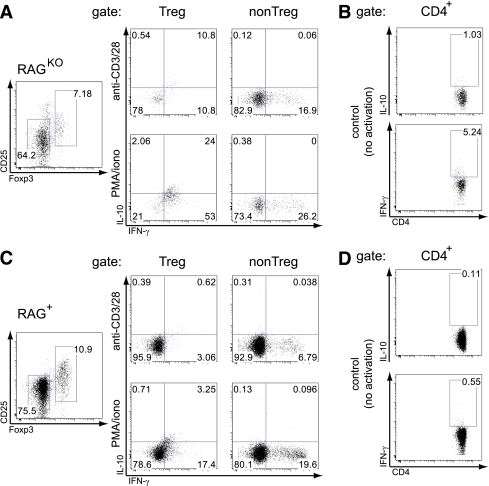
In order to address how the Foxp3-expressing CD4+ T cells in BDC12-4.1.RAGKO TCR Tg mice might exert their Treg function, we determined their cytokine profile. As shown in Fig. 4A, polyclonal activation of ex vivo splenocytes showed that non-Treg cells (CD25−Foxp3−) produced predominantly IFN-γ, whereas Treg cells (CD25+Foxp3+) coproduced IFN-γ and IL-10. Interestingly, the IFN-γ/IL-10 coproduction signature was specific for the Treg cells derived from BDC12-4.1.RAGKO and not BDC12-4.1.RAG+ mice (Fig. 4B). Thus, BDC12-4.1 mice generate Foxp3+ aTregs that coproduce IFN-γ and IL-10 in the periphery.
FIG. 4.
aTregs from BDC12-4.1.RAGKO mice coexpress IFN-γ and IL-10, whereas Teffs produce solely IFN-γ. Total splenocytes from BDC12-4.1.RAGKO (A and B) and BDC12-4.1.RAG+ (C and D) were incubated in vitro for 4 h with BfA (10 μg/mL) either alone (control, no activation) or in the presence of BfA and anti-CD3/28 (10/5 μg/mL, respectively) or phorbol myristic acid/ionomycin (50/1000 ng/ml, respectively). Cells were costained for Foxp3, IFN-γ, and IL-10 after fixation with the Foxp3 Fix/Perm kit. The cytokine profile is shown after gating on the Treg (CD4+Foxp3+) or non-Treg (CD4+Foxp3−) population. In the control (no activation), gating was performed on total CD4+ cells. One representative mouse (for each genotype) of at least three that were tested in two independent experiments is shown. (A high-quality color representation of this figure is available in the online issue.)
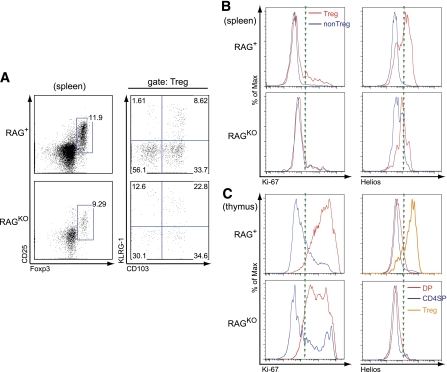
Since BDC12-4.1.RAGKO mice exhibit a strong degree of lymphopenia (11), we wondered whether a proportion of Foxp3+ cells were the result of homeostatic expansion. Since homeostatically expanded Foxp3+ cells express the CD103+/KLRG-1+ signature (26), we performed flow cytometry staining analysis for both markers after gating on the CD25+Foxp3+ (Treg) population. Indeed, as shown in Fig. 5A, >20% of the Tregs coexpressed CD103/KLRG-1 and therefore were likely derived from homeostatic expansion. We also found that Teff and Treg populations stained marginally positive for Ki-67 (Fig. 5B). In control BDC12-4.1.RAG+ animals, Treg renewal was more pronounced. To confirm the non–thymically derived lineage of Foxp3+ cells in BDC12-4.1.RAGKO mice, we performed additional staining for the nTreg-specific marker, Helios (27). Indeed, <5% of the Treg population in BDC12-4.1.RAGKO mice expressed Helios (Fig. 5B and C). In summary, Foxp3+ aTregs develop in the periphery of BCDC12-4.1.RAGKO mice as a result of insulin recognition and homeostatic expansion.
FIG. 5.
aTregs in BDC12-4.1.RAGKO mice do not express Helios, show marginal proliferation, and result from homeostatic expansion. A: Splenocyte-derived Treg from 7–10-week-old BDC12-4.1.RAG+ or RAGKO TCR Tg mice were examined for the presence of the CD103/KLRG-1 coexpression signature by flow cytometry. More than 20% of CD4+Vβ2+ Foxp3-expressing cells in the RAGKO background were likely the result of homeostatic expansion. B: Majority of Tregs in BDC12-4.1.RAGKO mice are not proliferating and do not express Helios compared with Tregs from BDC12-4.1.RAG+. Ki-67 and Helios histogram overlay was performed after gating on Treg (CD25+Foxp3+) (red line) and non-Treg (CD25−Foxp3−) (blue line) cells. C: For control, thymi from the same mice were analyzed. Histogram overlay depicts the expression of Ki-67 and Helios after gating on DP (red line), CD4SP (blue line), and Treg (CD4+CD25+Foxp3+, only for RAG+) populations (orange line).
Lack of Foxp3 in vivo results in 100% diabetes penetrance in BDC12-4.1 TCR Tg mice in an MHC-dependent manner.
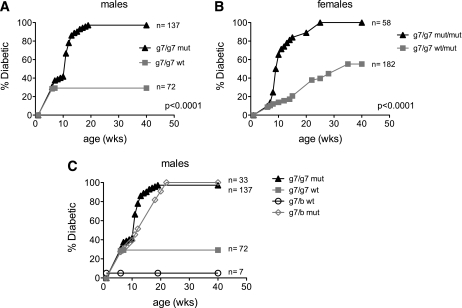
We found a strong correlation between the age and glucose intolerance in prediabetic BDC12-4.1.RAGKO mice (Supplementary Fig. 4). This suggested that despite the induction of aTregs, Teffs accumulated and eliminated more and more β-cells. However, only a proportion of mice eventually turn diabetic. In order to address whether Foxp3+ cells in the periphery were responsible for this T1D protection in 60% of BDC12-4.1.RAGKO mice, we back-crossed Foxp3 mutant (scurfy) C57BL/6 mice to Tg BDC12-4.1.RAGKO NOD mice for two generations (see figure legend for details). As depicted in Fig. 6A and B, spontaneous autoimmune diabetes was significantly accelerated with 100% penetrance when the scurfy Foxp3 mutation was introduced into BDC12-4.1.(I-Ag7/g7) RAGKO mice. At least one copy of I-Ag7 was needed for diabetes development, and one copy of the I-Ab gene somewhat delayed, but did not prevent, diabetes (Fig. 6C). However, despite the presence of a single I-Ab allele, all Foxp3-mutant (mut) BDC12-4.1.RAGKO mice still progressed to diabetes (Fig. 6C).
FIG. 6.
Lack of Foxp3 (scurfy mutation) significantly accelerates diabetes onset and increases penetrance in BDC12-4.1.RAGKO mice. I-Ag7.BDC12-4.1.RAGKO.Foxp3wt (g7/g7 wt) TCR Tg male mice were crossed with I-Ab.Foxp3wt/mut(scurfy) (C57BL/6 background) females (b/b wt/mut). The resulting back-cross generation 1 (BC1) I-Ag7/b.BDC12-4.1.RAG+/−.Foxp3wt/mut female and I-Ag7/b.BDC12-4.1.RAG+/−.Foxp3mut male mice were selected and intercrossed to generate all combinations. Of those, I-Ag7/g7.BDC12-4.1.RAGKO.Foxp3mut (g7/g7 mut) and I-Ag7/g7.BDC12-4.1.RAGKO.Foxp3wt (g7/g7 wt) male (A) or I-Ag7/g7.BDC12-4.1.RAGKO.Foxp3wt/mut (g7/g7 wt/mut) and I-Ag7/g7.BDC12-4.1.RAGKO.Foxp3mut/mut (g7/g7 mut/mut) female (B) offspring were monitored for diabetes development. In the absence of functional Foxp3, both male and female I-Ag7/g7.BDC12-4.1.RAGKO mice diabetes develops faster, and disease is 100% penetrant (P < 0.0001). C: Diabetes development in BDC12-4.1.RAGKO depends on both MHC I-Ag7/g7 (g7) and intact Foxp3. T1D development curve shown for the resulting BDC12-4.1.RAGKO TCR Tg male mice from the aforementioned intercross. Whereas diabetes incidence in Foxp3 wt mice depends on the presence of the protective MHC allele I-Ab (0% diabetes in g7/b wt vs. 20% in g7/g7 wt), mice bearing the scurfy mutation (mut) develop fast and fully invasive diabetes irrespective their MHC genotype (100% diabetes in both g7/b mut and g7/g7 mut); females show similar results (data not shown).
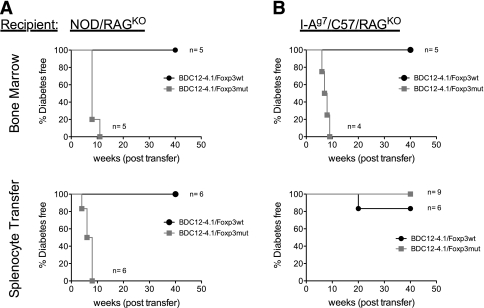
The above results conclusively confirmed our initial hypothesis that in BDC12-4.1.NOD.RAGKO mice, pathogenic as well as regulatory cells coemerge, and the generation of Tregs was crucial to curb diabetes development. A set of adoptive transfer experiments was performed to confirm pathogenic and regulatory roles of BDC12-4.1 CD4+ T cells. We reasoned that if the scurfy mutation truly eliminated any regulation by BDC12-4.1 T cells, then adoptive transfer of total splenocytes or BM transplantation (BMT) from BDC12-4.1.RAGKO.Foxp3mut mice into NOD.RAGKO or I-Ag7.C57BL/6.RAGKO (g7/B6.RAGKO) recipients would cause diabetes much faster than transfer of cells from BDC12-4.1.Foxp3wt donors. As shown in Fig. 7A, BMT from BDC12-4.1.RAGKO.Foxp3mut donors into both NOD and g7/B6.RAGKO recipients led to more diabetes development when the donors had the Foxp3 scurfy mutation (Foxp3mut) than when the donors did not (Foxp3wt). Similar observations were made when splenocytes were transferred (Fig. 7B). Interestingly, in this set of experiments, only when NOD.RAGKO, and not I-Ag7/57.RAGKO, were used as hosts did diabetes occur, underlining the importance of other insulin-dependent diabetes susceptibility genes for influencing disease progression. Taken together, these results provide strong evidence that the aTreg population observed in BDC12-4.1.RAGKO TCR Tg mice provide active peripheral tolerance and control pathogenic cells of the same specificity.
FIG. 7.
BMT and splenocyte adoptive transfers into RAGKO recipients from BDC12-4.1.RAGKO.Foxp3mut but not Foxp3wt donors resulted in diabetes development. A: BMT from BDC12-4.1.RAGKO donors into recipients that are I-Ag7.RAGKO either on the NOD or C57BL/6 background leads to significantly faster diabetes when the donors have the Foxp3 scurfy mutation (Foxp3mut) as compared with Foxp3wt (P = 0.0016 for NOD.RAGKO and P = 0.003 for I-Ag7.C57.RAGKO recipients). B: The same acceleration of diabetes is observed after splenocyte transfer into NOD.RAGKO hosts. In this experimental setting, homeostatic expansion is thought to have contributed to the pathogenicity of the cells. However, I-Ag7.C57.RAGKO recipients do not support the effector function of Foxp3mut B:9-23–specific (BDC12-4.1 TCR Tg) CD4+ T cells, suggesting that other insulin-dependent diabetes loci are important to support their function in this scenario (P = 0.003 for NOD.RAGKO and P = 0.25 for I-Ag7.C57.RAGKO recipients). In all cases, cells were derived from nondiabetic donors.
DISCUSSION
Here we show that the heterogeneous and much-reduced T1D development in mice uniformly expressing a TCR specific for a critical insulin B chain epitope (BDC12-4.1.RAGKO TCR Tg mice) was due to de novo generation of insulin-specific Tregs in the periphery. A significant proportion of insulin B:9-23–specific T cells escaped negative selection and then recognized insulin in the periphery, which led to their differentiation into Teffs and aTregs. This is the first time where a T cell with a fixed (autoreactive) TCR can follow both Teff and aTreg fates when exposed to similar differentiation niches in vivo.
In accordance, in vitro CD4SP–sorted thymocytes that responded to B:9-23 stimulation produced IL-10 and TGF-β1 and a fraction became Foxp3+. With increasing age, the number of Teffs increased in the spleen, whereas the number of Tregs remained relatively constant, which explains the development of T1D in a fraction of the BDC12-4.1.RAGKO mice. Accordingly, mice became more intolerant to glucose as they aged. Insulin-specific Teffs and Tregs underwent cyclical changes in the PDLNs, which could be a sign of a more active, iterative immunological process in that location; indeed, earlier studies had described similar cyclical alterations in islet-reactive CD8 responses in NOD mice. Alternatively, insulin-specific Teffs could have migrated to the pancreas, causing the decrease in cell counts in the PDLNs. In agreement, histological examination showed the presence of few Foxp3+ but far more Foxp3−CD4+ cells in the pancreas, especially in islets that were massively infiltrated.
In the mouse, Foxp3 is the most specific marker for CD4+ Treg cells (28), and accumulating evidence suggests that Foxp3+ Treg can be generated from peripheral naïve or already differentiated T cells in vivo under specific conditions (29). Such aTregs have been described to develop at lower levels of antigen availability under tolerogenic priming conditions (for example at mucosal surfaces or in the presence of TGF-β1) and are favored by homeostatic expansion (23). Usually, most Foxp3+ Treg originate in the thymus as a result of the regular T-cell selection process (25). Thymic differentiation of naturally occurring Tregs is regulated by TCR affinity, CD28 costimulation, common γ-c cytokine (IL-2) recognition, and TGF-β1 production (30,31). Previous studies with TCR Tg mouse models, where agonist peptides were expressed in the thymus, showed that self-peptide recognition not only induces negative selection but also nTreg differentiation, which suggests that the TCR repertoire of Treg cells overlaps somewhat with that of pathogenic T cells (23,32).
Since insulin is expressed in the thymus, we hypothesized that BDC12-4.1 Foxp3+ nTregs may also develop. However, only on the RAG+ background could thymic nTregs be observed, whereas on the RAGKO they were completely absent. We have evidence to suggest that in the BDC12-4.1.RAG+ background, the presence of additional TCR α chains enabled thymic Treg commitment. However, since nTregs did not develop in the thymus of RAGKO mice, either insulin B:9-23 expression is not adequate to drive their differentiation, medullary epithelial cells lack some important costimulatory signals, or the affinity/avidity of the trimolecular TCR/peptide/I-Ag7 complex is too low in NOD mice. In the future, it would be of interest to analyze thymi from BDC12-4.1 mice on diabetes-resistant genetic backgrounds, i.e., on BALB/c.
In the periphery, where the environment is largely lymphopenic, T cells start to proliferate. Perhaps these environmental cues together with stronger insulin recognition and presentation in the PDLNs permit autoreactive T-cell activation and differentiation into Teffs and aTregs. Despite the immediate effects of the immune system on the function and viability of insulin-producing β-cells, other factors, such as androgens, could have contributed to their decline (33). This is seen by the persistent gender discrepancy in several genetic scenarios (i.e., RAGKO, I-Ag7 homozygosity, etc.). However, in order to address how the insulin-specific aTreg modulated disease progression, we examined their cytokine production profile. aTregs in BDC12-4.1.RAGKO mice coproduce IFN-γ and IL-10, whereas Teffs produced solely IFN-γ. It is not clear whether some IFN-γ can be important for Treg function as we and others had shown previously (34,35). To corroborate this notion, T-bet expression (the master regulator of IFN-γ expression) in Treg cells is essential for their suppressive activity in the scurfy (Foxp3-deficient) model (36). The CD103/KLRG-1 coexpression signature (26) together with the absence of high amounts of folate receptor-4 and Helios, both distinctive markers for nTregs (37) (data not shown), strongly supports the adaptive origin.
The BDC12-4.1 TCR Tg mouse is the first single-Tg, RAG-deficient animal that exhibits spontaneous in vivo development of aTregs and Teffs with identical TCR sequence. Perhaps insulin is a unique autoantigen, and presentation in the context of I-Ag7 provides a permissive signal to instruct the acquisition of aTreg and Teff phenotype in cells bearing identical TCRs. It is important to note that the heterogeneous disease observed in BDC12-4.1.RAGKO animals, where the TCR repertoire is restricted to one insulin-specific Tg TCR, is usually not observed in other Tg or retrogenic CD4+ TCR models for T1D. For example, BDC2.5 mice recognizing the chromogranin autoantigen on immunodeficient backgrounds develop fulminant diabetes in 100% of the animals, suggesting that no aTreg development takes place (38,39). Of evidence, when BDC2.5.RAGKO mice are placed onto the scurfy background, no alteration in disease kinetics is seen.
The present findings support an intriguing TCR Tg model, where autoreactive cells escaping negative selection convert in the periphery into Teffs and aTregs, where their balance in the periphery determines the degree of islet destruction and consequently diabetes development. We conclude that it might be useful in further studies to assess therapies that might “tip the balance” toward antigen-specific Tregs rather than Teffs, and monitoring their frequency and the ratio could be an important biomarker to predict disease progression after certain types of therapeutic intervention.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a U01 National Institute of Allergy and Infectious Diseases prevention center grant to M.v.H., and the National Institutes of Health (DK-32083), Autoimmunity Prevention Center (AI050864), Diabetes Endocrine Research Center (P30-DK-57516), Juvenile Diabetes Research Foundation (4-2007-1056 and 11-2005-15), Children's Diabetes Foundation, and Brehm Coalition support to G.E.
No potential conflicts of interest relevant to this article were reported.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
G.F. and J.J. designed and conducted experiments, analyzed the data, and wrote the manuscript. A.D., T.J., and Y.C. performed experiments. M.N. performed experiments, provided important guidance, contributed to the discussion of the results, and critically improved the manuscript. P.P. performed immunofluorescence staining. F.L., G.S., M.C., and H.C. provided important guidance, contributed to the discussion of the results, and critically improved the manuscript. G.E. and M.v.H. supervised the study and wrote the manuscript. G.E. and M.v.H. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Malina Mc’Clure for excellent mouse husbandry, Natalie Amirian for assisting with histological preparation and staining, and Priscilla Colby for administrative assistance (all from La Jolla Insitute for Allergy and Immunology).
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db11-0671/-/DC1.
J.J. is currently affiliated with Partek, Inc., Chesterfield, Missouri.
REFERENCES
- 1.Wegmann DR, Norbury-Glaser M, Daniel D. Insulin-specific T cells are a predominant component of islet infiltrates in pre-diabetic NOD mice. Eur J Immunol 1994;24:1853–1857 [DOI] [PubMed] [Google Scholar]
- 2.Nakayama M, Abiru N, Moriyama H, et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature 2005;435:220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakayama M, Beilke JN, Jasinski JM, et al. Priming and effector dependence on insulin B:9-23 peptide in NOD islet autoimmunity. J Clin Invest 2007;117:1835–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadinski BD, Zhang L, Crawford F, Marrack P, Eisenbarth GS, Kappler JW. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc Natl Acad Sci USA 2010;107:10978–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kont V, Laan M, Kisand K, Merits A, Scott HS, Peterson P. Modulation of Aire regulates the expression of tissue-restricted antigens. Mol Immunol 2008;45:25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thébault-Baumont K, Dubois-Laforgue D, Krief P, et al. Acceleration of type 1 diabetes mellitus in proinsulin 2-deficient NOD mice. J Clin Invest 2003;111:851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan Y, Rudert WA, Grupillo M, He J, Sisino G, Trucco M. Thymus-specific deletion of insulin induces autoimmune diabetes. EMBO J 2009;28:2812–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moriyama H, Abiru N, Paronen J, et al. Evidence for a primary islet autoantigen (preproinsulin 1) for insulitis and diabetes in the nonobese diabetic mouse. Proc Natl Acad Sci USA 2003;100:10376–10381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leroux L, Desbois P, Lamotte L, et al. Compensatory responses in mice carrying a null mutation for Ins1 or Ins2. Diabetes 2001;50(Suppl. 1):S150–S153 [DOI] [PubMed] [Google Scholar]
- 10.Mohan JF, Petzold SJ, Unanue ER. Register shifting of an insulin peptide-MHC complex allows diabetogenic T cells to escape thymic deletion. J Exp Med 2011;208:2375–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jasinski JM, Yu L, Nakayama M, et al. Transgenic insulin (B:9-23) T-cell receptor mice develop autoimmune diabetes dependent upon RAG genotype, H-2g7 homozygosity, and insulin 2 gene knockout. Diabetes 2006;55:1978–1984 [DOI] [PubMed] [Google Scholar]
- 12.Du W, Wong FS, Li MO, et al. TGF-beta signaling is required for the function of insulin-reactive T regulatory cells. J Clin Invest 2006;116:1360–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You S, Slehoffer G, Barriot S, Bach JF, Chatenoud L. Unique role of CD4+CD62L+ regulatory T cells in the control of autoimmune diabetes in T cell receptor transgenic mice. Proc Natl Acad Sci USA 2004;101(Suppl. 2):14580–14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burton AR, Vincent E, Arnold PY, et al. On the pathogenicity of autoantigen-specific T-cell receptors. Diabetes 2008;57:1321–1330 [DOI] [PubMed] [Google Scholar]
- 15.Lennon GP, Bettini M, Burton AR, et al. T cell islet accumulation in type 1 diabetes is a tightly regulated, cell-autonomous event. Immunity 2009;31:643–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science 2003;299:1057–1061 [PubMed] [Google Scholar]
- 17.Curotto de Lafaille MA, Lafaille JJ. Natural and adaptive foxp3+ regulatory T cells: more of the same or a division of labor? Immunity 2009;30:626–635 [DOI] [PubMed] [Google Scholar]
- 18.Lathrop SK, Santacruz NA, Pham D, Luo J, Hsieh CS. Antigen-specific peripheral shaping of the natural regulatory T cell population. J Exp Med 2008;205:3105–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wong J, Mathis D, Benoist C. TCR-based lineage tracing: no evidence for conversion of conventional into regulatory T cells in response to a natural self-antigen in pancreatic islets. J Exp Med 2007;204:2039–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol 2006;7:401–410 [DOI] [PubMed] [Google Scholar]
- 21.Bautista JL, Lio CW, Lathrop SK, et al. Intraclonal competition limits the fate determination of regulatory T cells in the thymus. Nat Immunol 2009;10:610–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olivares-Villagómez D, Wang Y, Lafaille JJ. Regulatory CD4(+) T cells expressing endogenous T cell receptor chains protect myelin basic protein-specific transgenic mice from spontaneous autoimmune encephalomyelitis. J Exp Med 1998;188:1883–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apostolou I, Sarukhan A, Klein L, von Boehmer H. Origin of regulatory T cells with known specificity for antigen. Nat Immunol 2002;3:756–763 [DOI] [PubMed] [Google Scholar]
- 24.Jordan MS, Boesteanu A, Reed AJ, et al. Thymic selection of CD4+CD25+ regulatory T cells induced by an agonist self-peptide. Nat Immunol 2001;2:301–306 [DOI] [PubMed] [Google Scholar]
- 25.Hogquist KA, Baldwin TA, Jameson SC. Central tolerance: learning self-control in the thymus. Nat Rev Immunol 2005;5:772–782 [DOI] [PubMed] [Google Scholar]
- 26.Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc Natl Acad Sci USA 2010;107:5919–5924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thornton AM, Korty PE, Tran DQ, et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. J Immunol 2010;184:3433–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakaguchi S. Naturally arising Foxp3-expressing CD25+CD4+ regulatory T cells in immunological tolerance to self and non-self. Nat Immunol 2005;6:345–352 [DOI] [PubMed] [Google Scholar]
- 29.Wan YY, Flavell RA. The roles for cytokines in the generation and maintenance of regulatory T cells. Immunol Rev 2006;212:114–130 [DOI] [PubMed] [Google Scholar]
- 30.Lio CW, Hsieh CS. Becoming self-aware: the thymic education of regulatory T cells. Curr Opin Immunol 2011;23:213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ouyang W, Beckett O, Ma Q, Li MO. Transforming growth factor-beta signaling curbs thymic negative selection promoting regulatory T cell development. Immunity 2010;32:642–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawahata K, Misaki Y, Yamauchi M, et al. Generation of CD4(+)CD25(+) regulatory T cells from autoreactive T cells simultaneously with their negative selection in the thymus and from nonautoreactive T cells by endogenous TCR expression. J Immunol 2002;168:4399–4405 [DOI] [PubMed] [Google Scholar]
- 33.Rosmalen JG, Pigmans MJ, Kersseboom R, Drexhage HA, Leenen PJ, Homo-Delarche F. Sex steroids influence pancreatic islet hypertrophy and subsequent autoimmune infiltration in nonobese diabetic (NOD) and NODscid mice. Lab Invest 2001;81:231–239 [DOI] [PubMed] [Google Scholar]
- 34.Fousteri G, Dave A, Bot A, Juntti T, Omid S, von Herrath M. Subcutaneous insulin B:9-23/IFA immunisation induces Tregs that control late-stage prediabetes in NOD mice through IL-10 and IFNgamma. Diabetologia 2010;53:1958–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazarevic V, Glimcher LH. T-bet in disease. Nat Immunol 2011;12:597–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol 2009;10:595–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi T, Hirota K, Nagahama K, et al. Control of immune responses by antigen-specific regulatory T cells expressing the folate receptor. Immunity 2007;27:145–159 [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, Benoist C, Mathis D. How defects in central tolerance impinge on a deficiency in regulatory T cells. Proc Natl Acad Sci USA 2005;102:14735–14740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z, Herman AE, Matos M, Mathis D, Benoist C. Where CD4+CD25+ T reg cells impinge on autoimmune diabetes. J Exp Med 2005;202:1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.