Abstract
The activity of 6-phosphofructo-1-kinase is strictly controlled by fructose-2,6-bisphosphate, the level of which is regulated by another enzyme, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFK2/FBP2). PFK2/FBP2 is a bifunctional enzyme, having kinase and phosphatase activities, and regulates both glycolysis and gluconeogenesis. Here, we examined the hormonal regulation of the PFK2/FBP2 gene in vitro using the reporter assay, the electromobility shift assay (EMSA), and the chromatin immunoprecipitation (ChIP) assay in HuH7 cells and also using the mouse liver in vivo. We found that the transcriptional activity of the PFK2/FBP2 gene was stimulated by insulin and inhibited by cAMP and glucocorticoid. Liver X receptor (LXR) α showed a potent and specific stimulatory effect on PFK2/FBP2 gene transcription. Deletion and mutagenesis analyses identified the LXR response element (LXRE) in the 5′-promoter region of the PFK2/FBP2 gene. Binding of LXRα was confirmed by the EMSA and ChIP assay. Endogenous PFK2/FBP2 mRNA in the mouse liver was increased in the fasting/refeeding state compared with the fasting state. Altogether, PFK2/FBP2 gene transcription is found to be regulated in a way that is more similar to other glycolytic enzyme genes than to gluconeogenic genes. Furthermore, our data strongly suggest that LXRα is one of the key regulators of PFK2/FBP2 gene transcription.
6-Phosphofructo-1-kinase (PFK1) plays a pivotal role in the regulation of glycolysis through the Embden-Meyerhof pathway. The activity of PFK1 is known to be regulated by the adenosine 5’-monophosphate–to–adenosine 5’-triphosphate ratio and citrate, but, in addition, it also is known to be regulated by the intracellular level of fructose 2,6-bisphosphate (F2,6BP), which is a product of another enzyme, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFK2/FBP2). Of interest, PFK2/FBP2 is a bifunctional enzyme harboring kinase and phosphatase activities, with its NH2-terminal region acting as a kinase and the COOH-terminal region as a bisphosphatase. Thus, PFK2/FBP2 catalyzes the synthesis and degradation of F2,6BP, and, as a result, the enzyme is involved in both glycolysis and gluconeogenesis (1–4). There are four isoenzyme genes of PFK2/FBP2 (PFKFB1, PFKFB2, PFKFB3, and PFKFB4) that are differentially expressed, among which PFKFB1 is the dominant isoform in the liver (5).
The kinase and phosphatase activities of the liver isoform of PFK2/FBP2 are regulated by phosphorylation/dephosphrylation at Ser-32 by protein kinase and phosphatase. Indeed, during the fed state, insulin and carbohydrate dephosphorylate PFK2/FBP2 in the liver, and this makes the enzyme kinase dominant. Subsequently, fructose-6-phosphate is converted to F2,6BP, which activates PFK1 and inhibits fructose-1,6-bisphosphatase and finally stimulates glycolysis (Fig. 1A). On the other hand, during the fasting state, glucagon phosphorylates PFK2/FBP2 via cAMP/protein kinase A and makes the enzyme phosphotase dominant. As a result, decreased F2,6BP activates fructose-1,6-bisphosphatase and inhibits PFK1 and finally stimulates gluconeogenesis. By this mechanism, glycolysis and gluconeogenesis do not occur simultaneously (4).
FIG. 1.
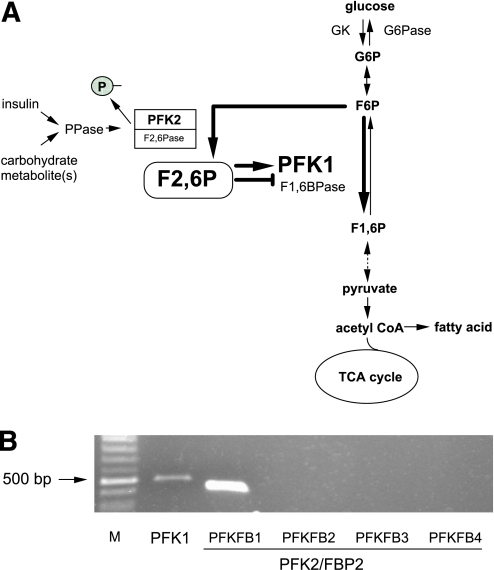
A: Schematic representation of the role of PFK2/FBP2 in the F2,6P-mediated regulation of PFK1 in the glycolytic pathway in the liver. G6P, glucose-6-phosphate; TCA, tricarboxylic acid cycle; GK, glucokinase; G6Pase, glucose-6-phosphatase; F6P, fructose-6-phosphate; F2,6P, fructose-2,6-bisphosphate; PPase, protein phosphatase. B: Endogenous expression of PFK1 (PFKL; liver type) and each isoform of PFK2/FBP2 (PFKFB1 through PFKFB4) mRNAs in HuH7 cells analyzed by RT-PCR. The figure shows photographs of the ethidium bromide–stained products using agarose gel electrophoresis. Complementary DNA produced from the reverse transcriptase reaction, using total RNA from the cells, was amplified by PCR with pairs of oligonucleotide primers specific for each mRNA. M, molecular size marker (100-bp ladder). (A high-quality color representation of this figure is available in the online issue.)
Glucose metabolism is regulated not only at the enzyme activity level but also at the gene expression level (6,7). In general, transcription of the glycolytic enzyme gene is stimulated by insulin/glucose, whereas that of the gluconeogenic enzyme gene is enhanced by glucagon/cAMP. Glucocorticoid also is involved in the regulation of a variety of glycolytic/gluconeogenic enzyme genes. In this context, it is interesting to note how the expression of the bifunctional PFK2/FBP2 gene is regulated by these hormones and factors mentioned above. Previous studies have shown that the amount of rat PFK2/FBP2 mRNA was increased by fasting/refeeding and by insulin in vivo (8), whereas the inhibitory effect of glucagon/cAMP was observed in adult rat hepatocytes in vitro (9). In addition, glucocorticoid increased PFK2/FBP2 mRNA in rat hepatoma cell lines (10,11). These findings suggest that hormones/factors involved in glucose metabolism have profound effects not only on PFK2/FBP2 activity but also on its mRNA expression. However, the molecular mechanism of PFK2/FBP2 gene expression is not yet characterized, and the transcription factor(s) involved is unidentified. Thus, in this study, we first examined the transcriptional regulation of the PFK2/FBP2 gene both in vitro and in vivo. We also tried to identify the transcription factor(s) and its cis-acting element(s) responsible for the hormonal/nutritional regulation of PFK2/FBP2 gene expression.
RESEARCH DESIGN AND METHODS
Reagents and plasmids.
Insulin, dexamethasone, and forskolin were obtained from Sigma (St. Louis, MO), and the specific liver X receptor (LXR) α agonist TO-901317 was from Calbiochem (San Diego, CA). The 5′-promoter regions of the human PFK2/FBP2 gene (−1197/+33 bp; +1 designates the transcription start site) and the human LXRα gene (−2191/+77 bp) were cloned by PCR and incorporated into a pA3Luc luciferase reporter plasmid (PFK2/FBP2-Luc and LXRα-Luc, respectively). A variety of deletion mutants of the PFK2/FBP2 promoter-reporter plasmids (−698, −448, and −282/+33 bp), a heterologous promoter construct containing only the LXR response element (LXRE) 2 of the PFK2/FBP2 gene (−191/−165 bp) and the thymidine kinase (TK) minimal promoter, and expression plasmids for the human glucocorticoid receptor, insulin receptor, LXRα, LXRβ, retinoid X receptor α (RXRα, sterol response element–binding protein 1c (SREBP1c) (12), carbohydrate response element–binding protein (ChREBP), and max-like protein X (Mlx) (13) also were constructed by standard molecular cloning techniques.
Cell culture.
The human hepatocyte–derived HuH7 cells were grown in DMEM (Sigma) supplemented with 10% fetal bovine serum. Cultured cells were maintained at 37°C in a humid atmosphere with 5% CO2.
Experiments.
In each experiment, cells were plated and cultured in 24-well plates with 50% confluency. Twenty-four hours after plating, the cells were transfected with the test plasmids (reporter plasmid with/without expression plasmid[s] when necessary) using the FuGene 6 transfection reagent (Roche, Indianapolis, IN). When different amounts of an expression plasmid(s) were used in an experiment, a complementary amount of the same plasmid without complementary DNA was used so that the net amount of the plasmids transfected was equal among the groups. On the next day, the medium was changed to serum-free medium with 0.1% BSA.
To see the effects of the hormones involved in glucose metabolism on PFK2/FBP2 gene 5′-promoter activity, each factor/reagent, in 1,000× concentration, was added directly into the culture medium, and the cells were incubated for a defined time interval. Dexamethasone was used to see the effect of pure glucocorticoid. Forskolin, an activator of adenylate cyclase, was used as a surrogate of glucagon/cAMP. At the end of incubation, the culture medium was removed, and the cells were harvested for the determination of promoter activity.
To see the effects of the coexpression of various transcription factors on PFK2/FBP2 gene 5′-promoter activity, both the PFK2/FBP2-Luc reporter plasmid and the test expression plasmid(s) were cotransfected as above. The cells were harvested 24–48 h after the end of transfection, followed by the determination of promoter activity.
Luciferase assay.
The luciferase assay was performed as previously described (14), and light output was measured for 20 s at room temperature using a luminometer (Berthold Lumat LB9507; Berthold, Bad Wildbad, Germany).
RT-PCR in vitro.
Endogenous expression of PFK1 (PFKL; liver type) and each isoform of PFK2/FBP2 mRNAs in HuH7 cells was examined by RT-PCR analysis. The cells were plated in 3.5-cm-diameter dishes, and total RNA was extracted using the RNeasy RNA extraction kit (Qiagen, Hilden, Germany). One microgram each was then applied for the reverse transcription reaction using Superscript III reverse transcriptase (Invitrogen, San Diego, CA), followed by PCR reaction (25–30 cycles; 94°C, 30 s; 60°C, 30 s; and 72°C 2 min) using Blend Taq DNA polymerase (Toyobo, Tokyo, Japan). The following were the primer sets used: for PFK1 sense, 5′-GGAGAACTTCATGTGGTGTGAGAG-3′, antisense, 5′-GAGAAGTTAGACTTCTCCTTGG-3′; PFKFB1 sense, 5′-CCAACACTACCAGAGAACGAC-3′, antisense, 5′-CTCTGATGTTGAGTTCACTCTC-3′; PFKFB2 sense, 5′-CTAACACGCTACCTCAACTGG-3′, antisense, 5′-GACTATCTTGCTCTGGATGTAG-3′; PFKFB3 sense, 5′-GTGAAGCAGTACAGCTCCTAC-3′, antisense, 5′-GGATGTTCATCAGGTAGTACAC-3′; and PFKFB4 sense, 5′-CTGACTCGATACCTGAACTGG-3′, antisense, 5′-CCTATCCAGGTCCTCATCTAG-3′. The sequences for both sense and antisense primers in each mRNA were obtained from a different exon to distinguish the mRNA-derived PCR product from the genomic DNA–derived product.
Real-time PCR in vivo.
Endogenous expression of PFK2/FBP2 mRNA in the liver in vivo was examined by quantitative real-time PCR analysis. C57/BL6 mice were divided into two groups (fasting [F] and fasting/refeeding [FR]; n = 6 in each group). Following the 24-h fast, mice in the FR group were fed for 3.5 h with standard laboratory chow and then killed. Mice from the F group were killed without refeeding. Trunk blood was obtained for the measurements of blood glucose and plasma insulin, and liver tissue was isolated, followed by the extraction of total RNA for the determination of endogenous PFK2/FBP2 mRNA. Total RNA was extracted using the RNEasy RNA extraction kit (Qiagen).
For real-time RT-PCR, reagents, software, and equipment were from Applied Biosystems (Foster City, CA). TaqMan reactions were performed using the TaqMan Universal PCR Master Mix and the StepOnePlus Real-Time PCR System, and data analysis was performed using sequence detection system software. TaqMan probes for the mouse PFK2/FBP2 mRNA (Applied Biosystems), and those for the mouse glyceraldehyde-3-phosphate dehydrogenase mRNA (Applied biosystems) as an internal control, were used. The quantity of the PFK2/FBP2 mRNA in an unknown sample was determined from the Ct value using the standard curve. A control without a template was included in each experiment. Nontemplate controls, standard dilutions, and samples were assayed in duplicate.
Electromobility shift assay.
Electromobility shift assay (EMSA) analysis was carried out to see the binding of LXRα/RXRα on the putative LXRE identified. Except for the supershift experiment (see below), we used a commercially available non–RI EMSA kit (LightShift Chemiluminescent EMSA kit; Pierce, Rockford, IL), according to the manufacturer’s instructions. In brief, the nuclear extract of the HuH7 cells transfected with LXRα/RXRα expression plasmids was prepared using NE-PER nuclear and cytoplasmic extraction reagents (Pierce, Rockford, IL) and then incubated for 20 min with the double-stranded 3′-end-biotinylated oligonucleotide probes (50 fmol) encompassing the LXR binding sites. The probes used are shown in Fig. 6A. The mixture was subjected to 6% nondenaturing polyacrylamide gel (160 V for 4 h). Finally, the biotinylated DNA was transferred to a nylon membrane and cross-linked, and then the biotin-labeled DNA was detected with a digital-imaging apparatus (LightCapture; Atto, Tokyo, Japan).
FIG. 6.
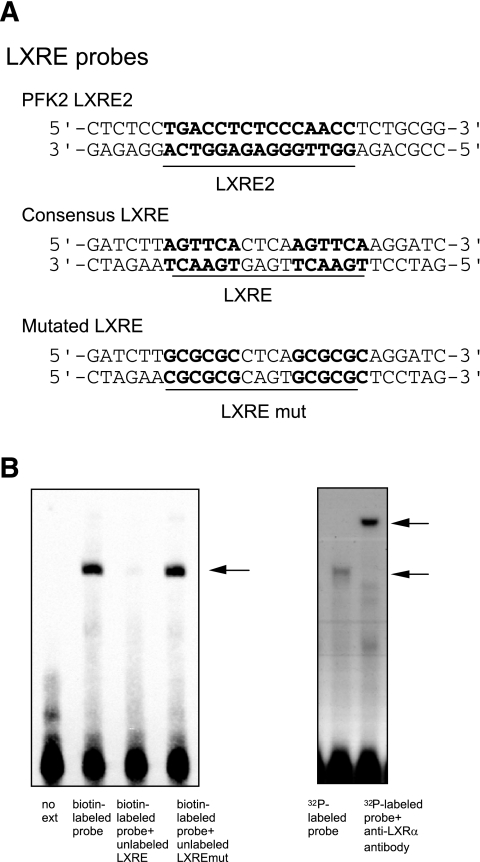
EMSA analysis. A: The nucleotide sequences of the probes used in the analysis. B. HuH7 cells were transfected with LXRα/RXRα expression plasmids, and the nuclear extract was applied for the EMSA. A specific band with a biotin-labeled probe containing the LXRE2 element of the PFK2/FBP2 promoter was recognized and was completely competed with an excess amount of unlabeled probe containing consensus LXRE but not with unlabeled probe containing mutated LXRE (left panel). For supershift analysis, mouse LXRα and human RXRα recombinant proteins were synthesized and used instead of the nuclear extract, and 32P-labeled probe was used (see research design and methods). A clear supershifted band was observed when the proteins were pretreated with anti-LXRα antibody (right panel).
To confirm the specificity of the band identified, a supershift experiment was performed, as described previously (15). Mouse LXRα and human RXRα recombinant proteins were synthesized from constructs in the pSG5 expression plasmid (16), using the TNT T7 Quick Coupled Transcription/Translation System (Promega). Binding reactions contained 20 mmol/L HEPES (pH 7.6), 50 mmol/L KCl, 12% glycerol, 1 mmol/L dithiothreitol, 1 μg poly(dI-dC)(dI-dC), and 4 μL of each of the synthesized nuclear receptors or unprogrammed reticulocyte lysates. Double-stranded oligonucleotides (PFK2 LXRE2 region: 5′-AGCTCTCCTGACCTCTGCCCAACCTCTC-3′) were labeled with [α-32P] deoxy-cytidine triphosphate by a fill-in reaction using a Klenow fragment of DNA polymerase. Binding reactions were performed at room temperature for 30 min. Then, 1 μL of anti-human LXRα ligand–binding domain mouse monoclonal antibody (Perseus Proteomics, Tokyo, Japan) was added as indicated in the figure, and the mixture was incubated for an additional 30 min at room temperature. The protein-DNA complexes were resolved on a 5% polyacrylamide gel in 0.5× Tris-base EDTA (TBE, 45 mmol/L).
Chromatin immunoprecipitation assay.
Chromatin immunoprecipitation (ChIP) assay was carried out using the ChIP-IT Express chromatin immunoprecipitation kit (Active Motif, Carlsbad, CA), according to the manufacturer’s instructions, to further confirm the binding of LXRα/RXRα on the putative LXRE in vivo. In brief, HuH7 cells transfected with LXRα/RXRα expression plasmids were homogenized by a Dounce homogenizer and centrifuged, and the shared chromatin in the supernatant was incubated with protein G–coated magnetic beads, protease inhibitor cocktail, and anti-LXRα antibody (Perseus Proteomics) for 4 h at 4°C. The complex then was washed twice, treated with the reverse cross-liking buffer, and then incubated with protease K for 1 h at 37°C. Finally, the reaction solution was applied for PCR. The primer sets used are shown in Fig. 7A. Primer sets for negative control also were used following the manufacturer’s instructions.
FIG. 7.
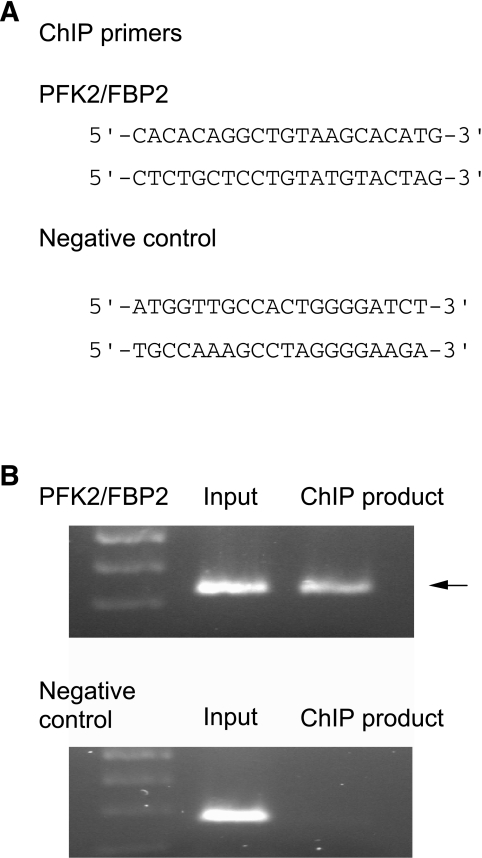
ChIP assay. A: The nucleotide sequences of the primers for PCR used in the assay. Primers recommended by the manufacturers’ instruction of the ChIP-IT Express chromatin immunoprecipitation kit was used as a negative control (see research design and methods). B: HuH7 cells were transfected with LXRα/RXRα expression plasmids, and the cells were homogenized with Dounce-type homogenizer and then treated with anti-LXRα antibody. The protein-DNA complex was separated with protein G–coated beads, treated with protease K, and then applied for PCR amplification (for details, see research design and methods).
Insulin and glucose assays.
Blood glucose was measured by the glucose oxidase method. Plasma insulin was measured by the mouse insulin enzyme-linked immunosorbent assay kit (Morinaga Institute of Biological Science, Tokyo, Japan).
Statistical analysis.
Samples in each group of the experiments were in triplicate or quadruplicate. All data were expressed as means ± SEM. When the statistical analyses were performed, data were compared by one-way ANOVA with the Fisher's protected least significant difference test, and P values <0.05 were considered significant.
RESULTS
Endogenous expression of PFK1 and PFK2/FBP2 genes in HuH7 cells.
There are four PFK2/FBP2 isoenzymes in humans (PFKFB1, PFKFB2, PFKFB3, and PFKFB4) (4) and at least three PFK1 isoenzymes (PFKL, PFKM, and PFKP/PFKC) (17–19). Among them, PFKL and PFKFB1 are known to be expressed in the liver in vivo. Indeed, we found endogenous expression of PFKL and PFKFB1 mRNA in HuH7 cells analyzed by RT-PCR (Fig. 1B), indicating that this cell line is appropriate for the transcriptional regulation of the PFKFB1 isoform of the PFK2/FBP2 gene in vitro.
Effects of glucoregulatory hormones on the transcriptional activity of the PFK2/FBP2 gene.
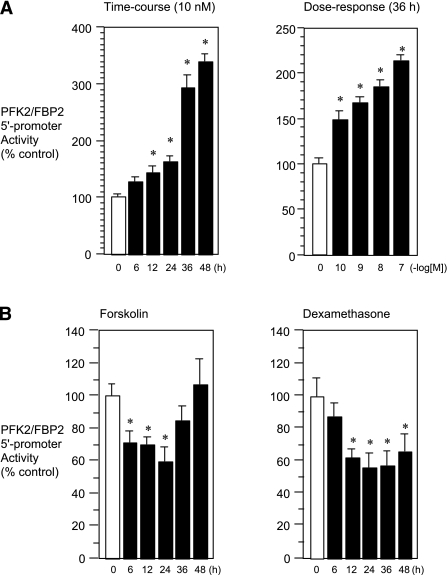
We first examined the effects of hormones related to glucose metabolism, such as insulin, glucagon/cAMP, and glucocorticoid, on the transcriptional activity of the PFK2/FBP2 gene. We found that insulin significantly stimulated the transcription of the PFK2/FBP2 gene in time- and dose-dependent manners (Fig. 2A). In contrast, forskolin/cAMP, a surrogate of glucagon, and dexamethasone showed significant inhibitory effects (Fig. 2B). Considering the fact that insulin is an anabolic/glycolytic hormone, whereas glucagon/cAMP is a gluconeogenic hormone, our results suggest that the regulation of the PFK2/FBP2 gene is similar to that of other glycolytic rather than gluconeogenic genes, at the transcriptional level.
FIG. 2.
Effects of glucoregulatory hormones/factors on the 5′-promoter activity of the PFK2/FBP2 gene. A: Time-course and dose-response effects of insulin. HuH7 cells were transfected transiently with the PFK2/FBP2-Luc plasmid and insulin receptor expression plasmid and then treated with insulin (10 nmol/L) for various time intervals (6–48 h) or treated with various doses of insulin (100 pmol/L to 100 nmol/L) for 36 h. B: Time-course effects of forskolin and glucocorticoid. HuH7 cells were transfected transiently with the PFK2/FBP2-Luc plasmid and glucocorticoid receptor expression plasmid (for glucocorticoid experiment) and then treated with forskolin (10 μmol/L) or dexamethasone (100 nmol/L) for up to 48 h. Each value is shown as a percentage of the control (vehicle, open bars; 0) or value at time zero (open bars, 0). In all figures, closed bars represent values of treated groups. *P < 0.05 vs. corresponding control.
Effects of the coexpression of insulin/glucose-related transcription factors on the transcriptional activity of the PFK2/FBP2 gene.
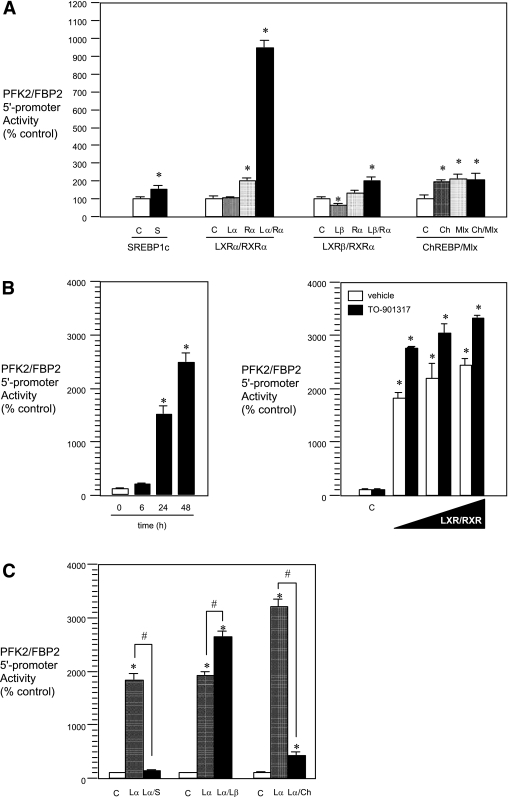
Because the 5′-promoter activity of the PFK2/FBP2 gene is found to be regulated like that of other anabolic/glycolytic enzyme genes, transcription factors under the control of insulin/glucose, such as SREBP1c, LXRα/β, and ChREBP/Mlx, seem to be involved in its transcription. Therefore, we next tried to determine which transcriptional factor(s) is regulating the expression of the PFK2/FBP2 gene by coexpression experiments. The results showed that, among the factors examined, LXRα/RXRα potently (>900% increase) stimulated the 5′-promoter activity of the PFK2/FBP2 gene (Fig. 3A). Other factors also had significant positive effects but much less (~1.5- to 2-fold increase) so than that of LXRα/RXRα. The effect of LXRα/RXRα coexpression was time dependent (Fig. 3B, left panel) and also dose dependent, which was further augmented by a specific LXRα ligand, TO-901317 (Fig. 3B, right panel). These results suggest that LXRα/RXRα is one of the key regulatory factors involved in PFK 2/FBP2 gene transcription. Of interest, however, when LXRα/RXRα and other factors were simultaneously expressed, LXRβ/RXRα augmented, whereas SERBP1c or ChREBP/Mlx rather inhibited, the positive effect of LXRα/RXRα (Fig. 3C). The precise molecular mechanisms and the physiological significance of the interactions among the factors observed here remain unresolved in this study.
FIG. 3.
Effects of insulin/glucose-related transcription factors on the 5′-promoter activity of the PFK2/FBP2 gene. A: Coexpression experiment. HuH7 cells were transfected with the PFK2/FBP2-Luc and each test plasmid [SREBP1c (S), LXRα/β (Lα/Lβ), and/or RXRα (Rα) and ChREBP (Ch) and/or Mlx expression plasmids] [reporter plasmid (μg): expression plasmid(s) (μg) = 1:1], and then the cells were cultured for 24 h. Each value is shown as a percentage of the corresponding control. *P < 0.05 vs. corresponding control [C (open bars); empty plasmid]. B: Time-course and dose-response experiments. HuH7 cells were transfected with PFK2/FBP2-Luc plasmid and LXRα/RXRα expression plasmids [reporter plasmid (μg): expression plasmids (μg) = 1:1] (time-course study) or PFK2/FBP2-Luc plasmid and increasing amounts of LXRα/RXRα expression plasmids [reporter plasmid (μg): expression plasmids (μg) = 1:0, 0.1, 0.5, and 1]. The cells were then cultured for various time intervals (6–48 h) (time-course experiment) or for 48 h without or with TO-901317 (1 μmol/L) (dose-response experiment). *P < 0.05 vs. value at time zero [0 (open bars)] or corresponding control (C; empty plasmid). C: Combination experiment. HuH7 cells were transfected with PFK2/FBP2-Luc, LXRα/RXRα expression plasmids, and SREBP1c (S), LXRβ (Lβ), or ChREBP/Mlx (Ch) expression plasmid(s) [reporter plasmid (μg): expression plasmid(s) (μg) = 1:1], and then the cells were cultured for 48 h. *P < 0.05 vs. corresponding control [C (open bars); empty plasmid]. #P < 0.05 vs. LXRα/RXRα group (Lα). In all figures, hatched, dotted, or closed bars represent values of treated groups.
Effects of TO-901317, glucose, and insulin on the transcriptional activities of PFK2/FBP2 and LXRα genes.
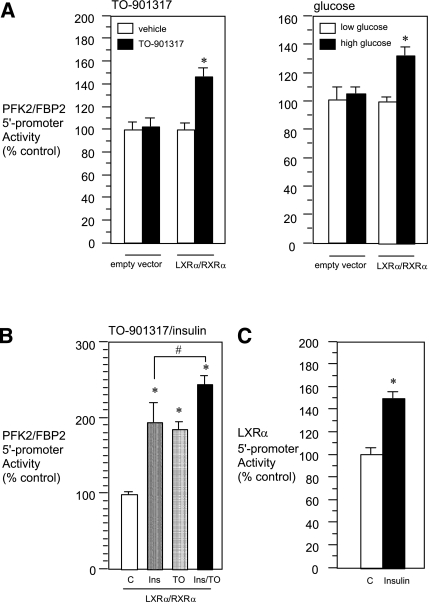
To confirm the involvement of LXRα/RXRα in the regulation of the PFK2/FBP2 gene, we next examined the effects of a synthetic LXRα ligand, TO-901317, and a putative endogenous LXRα activator, glucose (or its metabolite[s]) (20), on PFK2/FBP2 gene transcription. In naïve HuH7, treatment of the cells with high glucose (330 mg/dL) or TO-901317 had no significant effect on PFK2/FBP2 gene 5′-promoter activity. On the other hand, when LXRα/RXRα was coexpressed, mild but significant transcriptional induction was observed by either treatment (Fig. 4A). We then examined the role of insulin in LXRα/RXRα-mediated PFK2/FBP2 gene transcription. Under the condition where LXRα/RXRα was coexpressed, insulin as well as TO-901317 caused significant positive effects, and a further increase was observed when cells were treated with both insulin and TO-901317 (Fig. 4B). These results suggest that a factor(s) other than LXRα/RXRα also is involved in the positive effect of insulin. In addition, the transcriptional activity of the LXRα gene was significantly stimulated by insulin (Fig. 4C), as reported previously (21).
FIG. 4.
Effects of the LXR agonist/activator and insulin on the 5′-promoter activity of the PFK2/FBP2 gene. A: HuH7 cells were transfected with PFK2/FBP2-Luc and LXRα/RXRα expression plasmids or empty plasmid [reporter plasmid (μg): expression plasmid(s) (μg) = 1:1] and then treated with LXR ligand TO-901317 (1 μmol/L) for 24 h (left panel), or cultured with low (100 mg/dL) or high glucose (330 mg/dL) for 24 h (right panel). *P < 0.05 vs. corresponding control. B: HuH7 cells were transfected with PFK2/FBP2-Luc plasmid, insulin receptor expression plasmid, and LXRα/RXRα expression plasmids [reporter plasmid (μg): expression plasmid(s) (μg) = 1:1] and then treated with vehicle, insulin (Ins; 10 nmol/L), TO-901317 (TO; 1 μmol/L), or both for 24 h. Each value is shown as a percentage of the control [C (open bars)]. *P < 0.05 vs. control. #P < 0.05 vs. insulin alone. C: HuH7 cells were transfected with LXRα-Luc plasmid and insulin receptor expression plasmid [reporter plasmid (μg): expression plasmid (μg) = 1:1] and then treated with vehicle or insulin (10 nmol/L) for 24 h. *P < 0.05 vs. control [C (open bars)]. In all figures, hatched, dotted, or closed bars represent values of treated groups.
Deletion/mutation analyses of the LXRα/RXRα-dependent effect on the transcriptional activity of the PFK2/FBP2 gene.
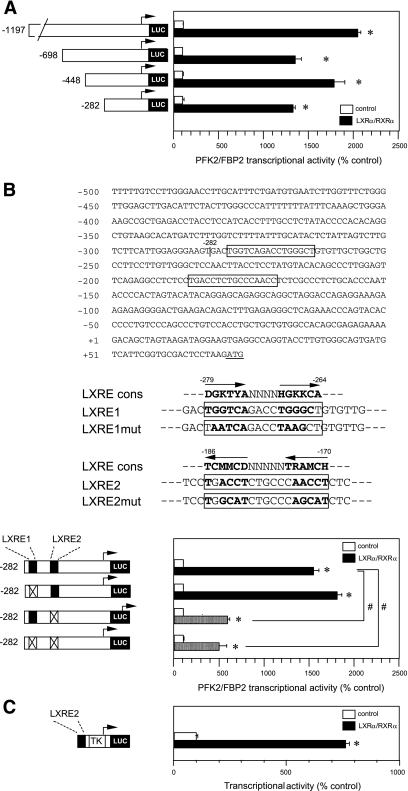
We next tried to identify the LXRE(s) in the PFK2/FBP2 gene 5′-promoter. We first performed deletion analysis. Our initial experiment showed that deletion up to −282 bp did not eliminate LXRα/RXRα responsiveness (Fig. 5A), suggesting that the LXRE seems to be located in the proximal portion of the PFK2/FBP2 gene promoter. We then analyzed this region more precisely using a site-directed mutagenesis technique. Within −282 bp of the promoter, we found two putative LXRE consensus sequences, LXRE1 and LXRE2 (Fig. 5B, upper panel). We introduced mutation in either or both of the elements (Fig. 5B, middle panel) and then examined the effect of LXRα/RXRα coexpression. We found that the mutation of LXRE1 did not affect the responsiveness, whereas the mutation of LXRE2 markedly diminished it, although a significant increase still remained (Fig. 5B, lower panel). In addition, we recognized a significant positive effect of LXRα/RXRα coexpression in a heterologous construct containing only LXRE2 (Fig. 5C). No effect was found in empty plasmid (TK-Luc alone) (data not shown). These results suggest that LXRE2 plays a significant role in the LXRα/RXRα-mediated transcriptional activation of the PFK2/FBP2 gene.
FIG. 5.
Deletion/mutation analyses of the LXRα/RXRα activation of the PFK2/FBP2 gene. A: Deletion analysis. HuH7 cells were transfected with a plasmid containing one of the PFK2/FBP2 gene promoter deletion mutants (−1197, −698, −448, or −282 bp) and LXRα/RXRα expression plasmids [reporter plasmid (μg): expression plasmids (μg) = 1:1], and the cells were cultured for 48 h. Each value is shown as a percentage of the corresponding control (empty plasmid). *P < 0.05 vs. corresponding control. B: Site-directed mutagenesis analysis. The upper panel shows the nucleotide sequences of the proximal region of the human PFK2/FBP2 gene. Two putative LXR response elements are shown in the boxes. The middle panel shows the mutations introduced in either LXRE1 (upstream) or LXRE2 (downstream). The lower panel shows the effects of LXRE1/LXRE2 mutations on the LXRα/RXRα-induced PFK2/FBP2 gene transcription. HuH7 cells were transfected with a plasmid containing wild-type or one of the mutated PFK2/FBP2 gene promoters (LXRE1mut or LXRE2mut) and LXRα/RXRα expression plasmids [reporter plasmid (μg): expression plasmids (μg) = 1:1], and the cells were cultured for 48 h. Each value is shown as a percentage of the corresponding control (empty plasmid). *P < 0.05 vs. corresponding control. #P < 0.05 vs. wild-type PFK2/FBP2 (−282/+33 bp) promoter activity coexpressed with LXRα/RXRα. C: Heterologous construct analysis. HuH7 cells were transfected with the test plasmid containing only LXRE2 of the PFK2/FBP2 gene (−191/−165 bp) fused with TK minimal promoter, and LXRα/RXRα expression plasmids or empty plasmid [reporter plasmid (μg): expression plasmids (μg) = 1:1]. The cells were then cultured for 48 h. *P < 0.05 vs. control (empty plasmid).
Binding of LXRα/RXRα on the LXRE2 of the PFK2/FBP2 gene by EMSA.
We then tried to confirm the LXR binding of the LXRE2 element by EMSA. We found a clear single band, which was completely competed by an excess amount of unlabeled probe containing consensus LXRE element but not by that with the mutated element (Fig. 6A and B, left panel). We also observed a supershift using specific anti-LXR antibody (Fig. 6B, right panel). These data strongly suggest that LXRα/RXRα binds to the LXRE2 located in the proximal promoter of the PFK2/FBP2 gene.
Binding of LXRα/RXRα on the LXRE2 of the PFK2/FBP2 gene by ChIP assay.
Because binding of LXR was recognized by EMSA, we also tried to confirm the binding of LXRα/RXRα on the LXRE2 by ChIP assay using anti-LXRα antibody. We found a PCR product using a primer set spanning LXRE2 but none with negative control primers (Fig. 7). The results confirm the in vivo binding of LXRα on the LXRE2 of the PFK2/FBP2 gene.
Effects of fasting/refeeding on PFK2/FBP2 mRNA expression in the mouse liver.
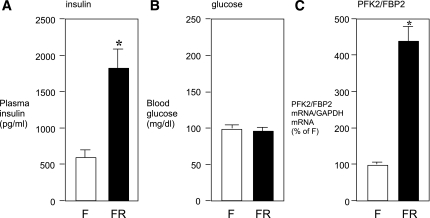
Finally, we performed an in vivo experiment to determine whether nutritional status indeed influences the expression level of PFK2/FBP2 mRNA in the mouse liver by quantitative RT-PCR. We found that fasting/refeeding caused a significant increase in plasma insulin (Fig. 8A), whereas no difference was found in blood glucose levels (Fig. 8B). In this condition, RT-PCR analysis showed that PFK2/FBP2 mRNA was markedly increased in fasting/refeeding compared with fasting alone (Fig. 8C). These in vivo results are in accordance with our in vitro data, suggesting that PFK2/FBP2 gene transcription is regulated in a similar way as other anabolic/glycolytic enzyme genes rather than gluconeogenic enzyme genes.
FIG. 8.
Effects of fasting/refeeding on PFK2/FBP2 mRNA expression in the mouse liver in vivo. C57/BL6 mice were kept fasting for 24 h and then treated without (F) or with (FR) refeeding for 3.5 h. At the end of experiment, mice were killed, trunk blood was obtained, and plasma insulin (A) and blood glucose (B) levels were determined. C: Total mRNA was also extracted from the liver tissue, and the PFK2/FBP2 mRNA levels were determined by quantitative real-time RT-PCR using specific TaqMan probe. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA level determined simultaneously was used as an internal control. *P < 0.05 vs. fasting alone.
DISCUSSION
PFK2/FBP2, a bifunctional enzyme involved in both glycolysis and gluconeogenesis, plays an important role in the regulation of glucose metabolism in the liver. In this study, we comprehensively examined the transcriptional regulation of the PFK2/FBP2 gene, using the HuH7 human hepatocyte–derived cell line in vitro and the mice liver tissue in vivo. We found that the promoter activity of the PFK2/FBP2 gene was stimulated by insulin and inhibited by forskolin/cAMP at the transcriptional level, suggesting that the PFK2/FBP2 gene is regulated in a similar way as other glycolytic enzyme genes rather than gluconeogenic enzyme genes (22–24). Furthermore, among the insulin/glucose-related transcription factors, we found that LXRα/RXRα is one of the key regulators of PFK2/FBP2 gene transcription. In addition, our in vivo experiment also showed that endogenous PFK2/FBP2 mRNA in the liver is induced by fasting/refeeding compared with fasting alone, in agreement with the previous report obtained in the rat liver estimated by Northern blot analysis (8).
Glycolysis/gluconeogenesis is known to be controlled not only by the regulation of enzyme activities but also by their expression levels. Regarding the PFK2/FBP2 gene, previous studies have shown that insulin, a representative anabolic/glycolytic hormone, increased the mRNA of the liver-type PFK2/FBP2 gene in the adult rat hepatocyte in vitro (9), in the rat-derived FAO-1 hepatoma cells in vitro (10), and also in the rat liver in vivo (8). Because insulin and/or glucose and its metabolite(s) is known to play an important role in the regulation of glycolytic enzyme genes, we surveyed the role of the insulin/glucose-related representative transcription factors, such as SREBP1c, LXRα/β, and ChREBP/Mlx, and identified LXRα as one of the key regulatory factors in PFK2/FBP2 gene expression.
LXRα (NR1H3) and LXRβ (NR1H2) belong to the nuclear receptor superfamily and are known to be involved in glucose and fatty acid synthesis as well as intracellular cholesterol metabolism (25–27). Insulin stimulates LXRα expression in hepatocytes (21), and LXRα is known to induce a variety of metabolism-related target genes directly or indirectly (27–30). To confirm that the involvement of LXRα on PFK2/FBP2 gene transcription is functional, we examined the effects of a specific LXR ligand, TO-901317, and found that in the presence of LXRα/RXRα, the ligand significantly increased the transcriptional activity of PFK2/FBP2 gene by 50–100%. A similar but weaker effect was observed when cells were treated with high glucose, a putative activator of LXRα (20). To further confirm that the PFK2/FBP2 gene is a direct target of LXRα, deletion/mutation analyses and an experiment using heterologous promoter (TK) construct were carried out, with the result that LXRE2 was identified as a cis-acting element for LXRα/RXRα. EMSAs and ChIP assays also provided convincing data supporting the direct binding of LXRα/RXRα on the element. Our data do not directly support the idea that LXRα is the predominant factor mediating the insulin stimulation of PFK2/FBP2 gene expression. However, considering the facts that insulin is known to be a potent inducer of LXRα (21), and that the current data show a marked stimulatory effect of LXRα/RXRα on PFK2/FBP2 gene promoter activity, we assume that LXRα is at least partly mediating the positive effect of insulin on PFK2/FBP2 gene transcription.
The present results suggest the possible role of other transcription factors as well. Indeed, coexpression of insulin-related transcription factors, such as SREBP1c and LXRβ, and the glucose metabolite(s)-stimulated factor ChREBP/Mlx also caused a significant increase in the transcriptional activity of the PFK2/FBP2 gene, although the effects were much weaker than that of LXRα/RXRα. Furthermore, simultaneous treatment of insulin and TO-901317 under the coexpression of LXRα/RXRα had a minimal but significant additive effect, suggesting that a factor(s) other than LXRα/RXRα is mediating the additional effect of insulin. The presence of different pathway(s) and/or element(s) also is suggested by the fact that a weaker but significant stimulatory effect of LXRα/RXRα still exists in the mutant promoter construct in which the confirmed LXR binding site (LXRE2) is deleted or mutated. Because no possible LXR binding site was found in the nucleotide sequence of the remaining promoter region, we assume that some unknown factor(s) induced by LXRα, possibly SREBP1c, is responsible for the remaining positive effect observed in the shortest construct (−282/+33 bp), because the expression of SREBP1c is partly dependent on LXRα/RXRα (29,30).
LXRα is expressed dominantly in the liver and regulates genes involved not only in sterol metabolism but also in fatty acid synthesis, such as acetyl CoA carboxylase and fatty acid synthase via LXREs located in the promoter of each gene (24,28,31). It generally is recognized that the glycolytic pathway is a part of the lipogenic pathway, and, indeed, under the control of PFK2/FBP2, PFK1 produces dihydroxyacetone phosphate, which is a precursor of glycrol-3-phospate, a basic component of triacylglycerol. In this sense, it is not surprising that LXRα regulates gene expression related to not only fatty acid synthesis but also to glycolysis. To our knowledge, however, this is the first work showing that a gene (PFK2/FBP2) belonging to the Embden-Meyerhof pathway also is a direct target of LXRα.
In contrast to insulin, the glucagon/cAMP pathway is known to have positive and negative effects on gluconeogenic and glycolytic enzyme gene expression, respectively (32), and, indeed, in this study, we found an inhibitory effect of forskolin on PFK2/FBP2 gene transcription. On the other hand, the effect of glucocorticoid is complex because the hormone facilitates gluconeogenesis during starvation, whereas it exhibits lipogenic effects in the anabolic condition. Regarding the effect of glucocorticoid on the PFK2/FBP2 gene, previous studies show conflicting results: dexamethasone decreased PFK2/FBP2 mRNA in adult rat hepatocytes (9), whereas opposite effects were observed in rat hepatoma-derived FAO and FTO-2B cells (10,11). The human PFK2/FBP2 gene has no clear glucocorticoid response element(s) in the promoter region examined in this study, and we found inhibitory rather than stimulatory effects. Additional studies using human primary hepatocyte cell culture are necessary to clarify the issue.
Finally, from the clinical standpoint, activation of LXRα is assumed to have a beneficial effect on glucose and cholesterol metabolism but at the same time can potentially facilitate fatty acid and triglyceride synthesis by inducing lipogenic genes, such as acetyl CoA carboxylase, fatty acid synthase, and PFK2/FBP2. Indeed, previous animal experiments showed that administration of the LXR ligand, TO-901317, improved diabetes but caused severe fatty liver and obesity (33,34). In contrast, selective inhibition of PFK2 causes decreased PFK1 activity, which decreases adipogenesis, as shown in PFK1-M knockout mice (19), but causes glycogenosis and diabetes, as observed in PFK1-M–deficient humans (18). However, moderate inhibition of both PFK2 and FBP2 activities using LXR inhibitor may possibly be beneficial, because it will suppress both adipogenesis and gluconeogenesis without complete suppression of the mainstream glycolytic pathway. In this context, pharmacological inhibitors of LXRα or PFK2/FBP2 might be a promising therapeutic tool for the prevention of obesity and diabetes in the future.
ACKNOWLEDGMENTS
This work was supported in part by the Kochi University President’s Discretionary Grant, and Grants-in-Aid for Scientific Research (Research on Hypothalamo-Hypophyseal Disorders) from the Ministry of Health, Labor, and Welfare of Japan.
No potential conflicts of interest relevant to this article were reported.
L.-F.Z. performed research, analyzed the data, wrote the manuscript, and contributed to the discussion. Y.I. designed and performed research, analyzed the data, contributed to the discussion, and reviewed and edited the manuscript. M.N., T.T., M.T., M.O., S.N., K.H., and K.M. helped perform research and contributed to the discussion. M.K. performed research. S.F. and Y.T. reviewed the manuscript. Y.I. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors thank Ms. Reiko Matsumoto (Department of Endocrinology, Metabolism, and Nephrology, Kochi Medical School, Kochi University) for her excellent technical support.
REFERENCES
- 1.Randle PJ, Priestman DA, Mistry SC, Halsall A. Glucose fatty acid interactions and the regulation of glucose disposal. J Cell Biochem 1994;55(Suppl.):1–11 [DOI] [PubMed] [Google Scholar]
- 2.Hue L, Rider MH. Role of fructose 2,6-bisphosphate in the control of glycolysis in mammalian tissues. Biochem J 1987;245:313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu C-D, Khan SA, Peng L-J, Lange AJ. Roles for fructose-2,6-bisphosphate in the control of fuel metabolism: beyond its allosteric effects on glycolytic and gluconeogenic enzymes. Adv Enzyme Regul 2006;46:72–88 [DOI] [PubMed] [Google Scholar]
- 4.Okar DA, Manzano A, Navarro-Sabatè A, Riera L, Bartrons R, Lange AJ. PFK-2/FBPase-2: maker and breaker of the essential biofactor fructose-2,6-bisphosphate. Trends Biochem Sci 2001;26:30–35 [DOI] [PubMed] [Google Scholar]
- 5.Rider MH, Bertrand L, Vertommen D, Michels PA, Rousseau GG, Hue L. 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase: head-to-head with a bifunctional enzyme that controls glycolysis. Biochem J 2004;381:561–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pilkis SJ, Granner DK. Molecular physiology of the regulation of hepatic gluconeogenesis and glycolysis. Annu Rev Physiol 1992;54:885–909 [DOI] [PubMed] [Google Scholar]
- 7.Yabaluri N, Bashyam MD. Hormonal regulation of gluconeogenic gene transcription in the liver. J Biosci 2010;35:473–484 [DOI] [PubMed] [Google Scholar]
- 8.Colosia AD, Marker AJ, Lange AJ, et al. Induction of rat liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase mRNA by refeeding and insulin. J Biol Chem 1988;263:18669–18677 [PubMed] [Google Scholar]
- 9.Casado M, Boscá L, Martín-Sanz P. Differential regulation of the expression of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase and pyruvate kinase by cyclic adenosine 3′,5′-monophosphate in fetal and adult hepatocytes. J Cell Physiol 1995;165:630–638 [DOI] [PubMed] [Google Scholar]
- 10.Espinet C, Vargas AM, el-Maghrabi MR, Lange AJ, Pilkis SJ. Expression of the liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase mRNA in FAO-1 cells. Biochem J 1993;293:173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemaigre FP, Lause P, Rousseau GG. Insulin inhibits glucocorticoid-induced stimulation of liver 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase gene transcription. FEBS Lett 1994;340:221–225 [DOI] [PubMed] [Google Scholar]
- 12.Ferré P, Foretz M, Azzout-Marniche D, Bécard D, Foufelle F. Sterol-regulatory-element-binding protein 1c mediates insulin action on hepatic gene expression. Biochem Soc Trans 2001;29:547–552 [DOI] [PubMed] [Google Scholar]
- 13.Iizuka K, Horikawa Y. ChREBP: a glucose-activated transcription factor involved in the development of metabolic syndrome. Endocr J 2008;55:617–624 [DOI] [PubMed] [Google Scholar]
- 14.Aoki Y, Iwasaki Y, Katahira M, Oiso Y, Saito H. Regulation of the rat proopiomelanocortin gene expression in AtT-20 cells: I: effects of the common secretagogues. Endocrinology 1997;138:1923–1929 [DOI] [PubMed] [Google Scholar]
- 15.Ren Y, Satoh T, Yamada M, et al. Stimulation of the preprothyrotropin-releasing hormone gene by epidermal growth factor. Endocrinology 1998;139:195–203 [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto K, Ishida E, Matsumoto S, et al. Carbohydrate response element binding protein gene expression is positively regulated by thyroid hormone. Endocrinology 2009;150:3417–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vora S, Oskam R, Staal GE. Isoenzymes of phosphofructokinase in the rat: demonstration of the three non-identical subunits by biochemical, immunochemical and kinetic studies. Biochem J 1985;229:333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ristow M, Vorgerd M, Möhlig M, Schatz H, Pfeiffer A. Deficiency of phosphofructo-1-kinase/muscle subtype in humans impairs insulin secretion and causes insulin resistance. J Clin Invest 1997;100:2833–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Getty-Kaushik L, Viereck JC, Goodman JM, et al. Mice deficient in phosphofructokinase-M have greatly decreased fat stores. Obesity (Silver Spring) 2010;18:434–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitro N, Mak PA, Vargas L, et al. The nuclear receptor LXR is a glucose sensor. Nature 2007;445:219–223 [DOI] [PubMed] [Google Scholar]
- 21.Tobin KA, Ulven SM, Schuster GU, et al. Liver X receptors as insulin-mediating factors in fatty acid and cholesterol biosynthesis. J Biol Chem 2002;277:10691–10697 [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Iwasaki Y, Zhao L-F, et al. Hormonal regulation of glycolytic enzyme gene and pyruvate dehydrogenase kinase/phosphatase gene transcription. Endocr J 2009;56:1019–1030 [DOI] [PubMed] [Google Scholar]
- 23.Hirota K, Sakamaki J, Ishida J, et al. A combination of HNF-4 and Foxo1 is required for reciprocal transcriptional regulation of glucokinase and glucose-6-phosphatase genes in response to fasting and feeding. J Biol Chem 2008;283:32432–32441 [DOI] [PubMed] [Google Scholar]
- 24.Zhao LF, Iwasaki Y, Zhe W, et al. Hormonal regulation of acetyl-CoA carboxylase isoenzyme gene transcription. Endocr J 2010;57:317–324 [DOI] [PubMed] [Google Scholar]
- 25.Wójcicka G, Jamroz-Wiśniewska A, Horoszewicz K, Bełtowski J. Liver X receptors (LXRs): part I: structure, function, regulation of activity, and role in lipid metabolism. Postepy Hig Med Dosw (Online) 2007;61:736–759 [PubMed] [Google Scholar]
- 26.Jamroz-Wiśniewska A, Wójcicka G, Horoszewicz K, Bełtowski J. Liver X receptors (LXRs): part II: non-lipid effects, role in pathology, and therapeutic implications. Postepy Hig Med Dosw (Online) 2007;61:760–785 [PubMed] [Google Scholar]
- 27.Baranowski M. Biological role of liver X receptors. J Physiol Pharmacol 2008;59(Suppl. 7):31–55 [PubMed] [Google Scholar]
- 28.Joseph SB, Laffitte BA, Patel PH, et al. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J Biol Chem 2002;277:11019–11025 [DOI] [PubMed] [Google Scholar]
- 29.Yoshikawa T, Shimano H, Amemiya-Kudo M, et al. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol Cell Biol 2001;21:2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen G, Liang G, Ou J, Goldstein JL, Brown MS. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci USA 2004;101:11245–11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Talukdar S, Hillgartner FB. The mechanism mediating the activation of acetyl-coenzyme A carboxylase-alpha gene transcription by the liver X receptor agonist T0-901317. J Lipid Res 2006;47:2451–2461 [DOI] [PubMed] [Google Scholar]
- 32.Koo SH, Flechner L, Qi L, et al. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 2005;437:1109–1111 [DOI] [PubMed] [Google Scholar]
- 33.Cao G, Liang Y, Broderick CL, et al. Antidiabetic action of a liver x receptor agonist mediated by inhibition of hepatic gluconeogenesis. J Biol Chem 2003;278:1131–1136 [DOI] [PubMed] [Google Scholar]
- 34.Chisholm JW, Hong J, Mills SA, Lawn RM. The LXR ligand T0901317 induces severe lipogenesis in the db/db diabetic mouse. J Lipid Res 2003;44:2039–2048 [DOI] [PubMed] [Google Scholar]