Abstract
Here we present a highly efficient protocol for on-the-resin coupling of fluorescent dyes or other functional groups to the N-termini of synthetic peptides prior to cleavage and deprotection. The protocol avoids expensive pre-activated dyes and instead employs carboxylated dyes activated by large amounts of coupling reagents. The protocol was used to label peptides with low reactivity such as long hydrophobic peptides and peptides with strong tendencies to form sterically-shielding structures or aggregates in solution. In all cases, the yields far exceeded those from commercially available pre-activated compounds.
Keywords: N-terminal peptide labeling, high yields
The ability to introduce specific chemical moieties into proteins or peptides is critical for basic biophysical studies and for a variety of biotechnological applications. Examples include fluorophore incorporation for bioimaging [1] and structural analysis [2], PEGylation for solubility and bioavailability modulation [3], glycosylation for synthetic vaccine and drug development [4], or immobilization for bioseparations and biosensing [5;6]. One of the most commonly used strategies for such modifications involves labeling amino acid side chains [7]. There are many published protocols for labeling various amino acid side chains using well-documented chemical reactions in solution (see [7] for a review). One such widely used strategy is the chemical modification of cysteine residues with maleimide-containing fluorescent dyes for FRET-based applications [7]. In the past, we have used this approach to conjugate FRET dyes such as fluorescein and rhodamine to long hydrophobic peptides with a single cysteine near the N or the C terminus, with coupling efficiencies higher than 80%, and sometimes as high as 100% [8–10]. Such high labeling yields are critical for quantitative FRET characterization of interactions between long hydrophobic peptides, as the labeled and unlabeled hydrophobic peptides are difficult to separate by HPLC [10–14].
The main limitation of this labeling approach is the availability of a suitable single cysteine residue in the sequence of the peptides, as this approach cannot be used to label peptides with no cysteines or peptides containing multiple cysteines in their sequences. For instance, the transmembrane (TM) domains of two receptor tyrosine kinases that we are studying, FGFR1 (sequence: TSPLYLEIIIYCTGAFLISCMVGSVIVYKMK) and FGRF2 (sequence TASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTK), have multiple cysteine residues in the middle of their sequences.
As an alternative to labeling cysteine side chains in solution, we sought to label the N-terminus on-the-resin prior to cleaving the peptides. Potential advantages of N-terminal on-the-resin labeling include easy purification of the products, and commercial availability of pre-activated and ready-to-use derivatives with the desired chemical functionality. Therefore, we attempted to conjugate the commercially available N-hydroxysuccinimide (NHS) ester derivatives of fluorescein and rhodamine to the N-termini of FGFR1 and FGFR2 TM peptides. As discussed below, however, the yields of the conventional N-terminal modification methods were very low for the long hydrophobic peptides. We therefore present here a highly efficient protocol for N-terminal on-the-resin modification of such peptides that employs large amounts of carboxylated dyes activated by coupling reagents.
FGFR1 and FGFR2 TM peptides were synthesized on a 433A peptide synthesizer (Applied Biosystems) using 9-fluorenylmethoxy-carbonyl (Fmoc) chemistry and a triple coupling protocol. The peptides were produced on a CLEAR-amide resin with a substitution level of 0.4 mmol/g on a 0.1 mmol scale as previously described [15]. After the synthesis, the solid phase coupling to the N-terminus was attempted with two fluorescent dyes: fluorescein-5-EX, succinimidyl ester and 6-[tetramethylrhodamine-5-(and-6)-carboxamido]hexanoic acid, succinimidyl ester [5(6)-TAMRA-X, SE] (Invitrogen, Eugene, OR). The reactions were conducted in a mixture of N,N-dimethylformamide (DMF) (ACS grade, EMD chemicals Inc., Gibbstown, NJ), 100 mM sodium phosphate buffer (pH 7.0) and 6 equivalents of the dye, at 4°C under 4 h of vigorous agitation. The labeling yields determined after cleavage (detailed cleavage protocol given in [15]) were less than 10% when the pre-activated dyes were used. This yield was too low for quantitative biophysical characterization of the peptides.
To improve the yields, we tried several other reaction media, including 2,2,2-trifluoroethanol (Sigma-Aldrich, St. Louis, MO), methanol (Fisher Scientific, Fair Lawn, NJ), and N-methyl-2-pyrrolidone (NMP) (Advanced ChemTech, Louisville, KY). However, these substitutions did not increase the reaction efficiency. Furthermore, changing the buffer and solvent ratios, lengthening the reaction time, and adding more dye did not improve the reaction yield (data not shown).
Previously, Fernández-Carneado and Giralt [16] have proposed an alternative protocol for solid phase coupling of 5(6)-carboxyfluorescein to the N-terminus via activation of the carboxylic group without the use of the commercially available pre-activated derivatives of the fluorescent dyes. Their protocol yielded labeling efficiencies higher than 80% for peptides with up to 12 residues, but only 60% for 18 residue long peptides. With this approach, both FGFR1 and FGFR2 (over 30 residues each) were expected to have labeling efficiencies below 50%. Thus, we had to modify the protocol in order to increase the labeling efficiencies.
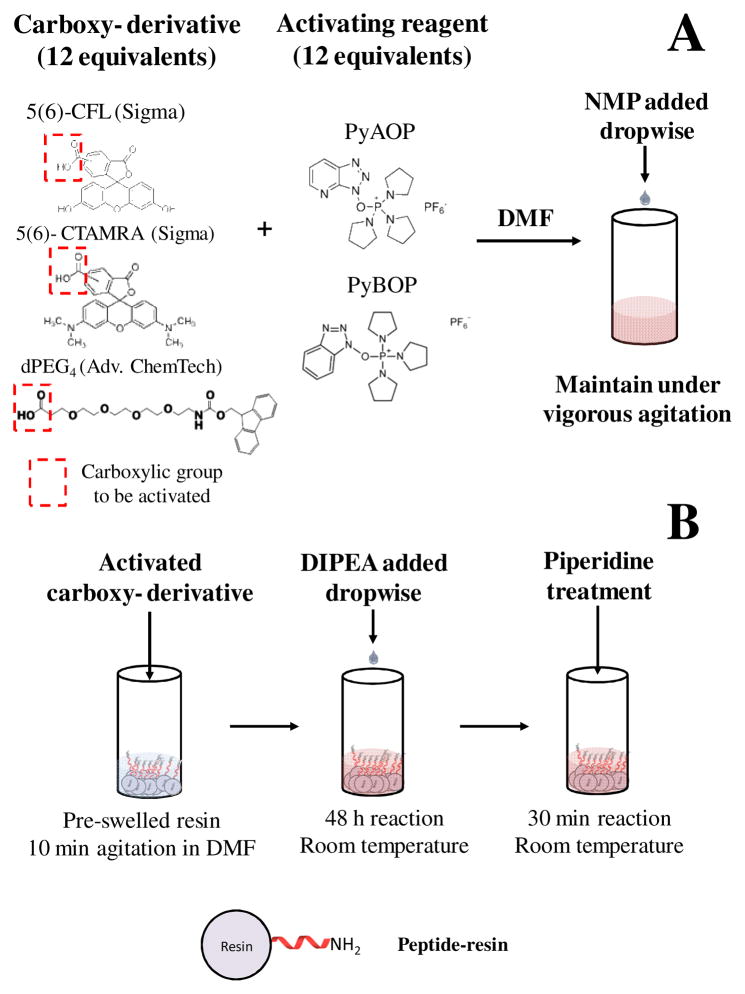
In the first step of the new protocol (Fig. 1A), 12 equivalents of the carboxy-derivative of the fluorescent dye [either carboxyfluorescein (CF) or carboxytetramethylrhodamine (CTAMRA)] were dissolved in DMF under vigorous agitation. This differs from the protocol of Fernández-Carneado and Giralt [16], which uses only 6 equivalents of the dye. The large excess of dye in the new protocol introduced solubility problems (see Fig. S1 in Supplementary Information), which were solved by adding more NMP dropwise until a homogeneous, saturated solution was formed. The carboxylic group of the fluorescent dyes was activated by the addition of 12 equivalents of either 7-Azabenzotriazolyoxytris(pyrrolidino)phosphonium hexafluorophosphate (PyAOP) (Sigma-Aldrich, St. Louis, MO) or benzotriazol-1-yl-oxytripyrrolidinophosphonium hexafluorophosphate (PyBOP) (Sigma-Aldrich, St. Louis, MO).
Fig. 1.
Protocol for the on-the-resin N-terminal modification of long synthetic hydrophobic and self-associating peptides. (A) Mix 12 equivalents (compared to peptide) of a carboxy-derivative of the compound to be conjugated with 12 equivalents of the activating reagent (PyAOP or PyBOP). Resuspend this powder mixture under vigorous agitation in DMF. Add NMP dropwise as needed to ensure that a homogeneous solution is formed. This is typically achieved in about 30 minutes. (B) Transfer the activated compound from (A) to a container with the pre-swelled peptide-resin (swelling is achieved by agitating the peptide-resin in DMF for about 10 minutes). Under vigorous agitation, add DIPEA dropwise, allowing complete homogenization between additions. This process usually lasts for 30–60 min depending on the amount to be conjugated. The reaction is then carried out for 48 h. Wash the peptide-resin several times with DMF and centrifuge between washes to avoid the loss of peptide-resin. Finally, add piperidine in DMF [1:4 (v/v)] to the peptide-resin and agitate the mixture for 30 minutes. Wash the peptide-resin before proceeding to cleavage.
Slow addition of DIPEA to the reaction mixture turned out to be critical for high reaction yield. In the original protocol, DIPEA is mixed together with PyAOP (or PyBOP); however, this resulted in phase separation with the heavy phase exhibiting gel-type consistency (Fig. S2 in Supplementary Information). Such phase separation limits mass transfer during the coupling reaction. To overcome this problem, we modified the protocol by adding DIPEA dropwise over a period of 1 h to an agitating mixture that contained the peptide-resin and the pre-mixed carboxy-derivative (Fig. 1B). The reaction was carried out for 48 h at room temperature (in the presence of PyAOP) or longer (with PyBOP), since PyBOP is known to generate a less active intermediate than PyAOP [17;18].
Due to the large excess of dyes and activators, a concern arose that single peptides may be labeled with multiple dyes through a phenolic ester formation mechanism that has been reported in the literature [19]. Therefore, the peptide-resin was treated with piperidine/DMF (1:4, v/v) for 30 minutes to remove ester-bound dyes. After the piperidine treatment, the resin was filtered out and washed with DMF. The peptide was subsequently cleaved and purified via reverse-phase HPLC. The molecular weights of the peptides were confirmed by MALDI-TOF mass spectrometry (Voyager DE-STR, Applied Biosystems, Foster City, CA). The labeling efficiencies were determined by assessing the peptide and dye concentrations using circular dichroism and absorbance measurements, respectively, as described previously [11]. For both CF and CTAMRA labeling of FGFR1 and FGFR2, the coupling efficiencies were higher than 85% (Table 1).
Table 1.
On-the-resin peptide labeling protocol and yields
| Peptidyl-Resin* | Fluorescent Label | Activators | Product | Yield |
|---|---|---|---|---|
| FGRF1-resin | Fluorescein-5-EX, SE or 5(6)-TAMRA-X, SE | - | Fluorescein-FGRF1 | <10%a |
| TAMRA-FGRF1 | <10%a | |||
| CF or CTAMRA | PyAOP, DIPEA | CF-FGRF1 | 86%b | |
| CTAMRA-FGRF1 | 87%b | |||
| FGRF2-resin | Fluorescein-5-EX, SE or 5(6)-TAMRA-X, SE | - | Fluorescein-FGRF2 | <10%a |
| TAMRA-FGRF2 | <10%a | |||
| CF or CTAMRA | PyAOP, DIPEA | CF-FGRF2 | ~100%b | |
| CTAMRA-FGRF2 | ~100%b | |||
| CMP9-resin | CF | PyAOP, DIPEA | CF-CMP9 | 94%a |
| CMP8-resin | dPEG4 | PyBOP, DIPEA | dPEG-CMP8 | ~100%a |
Sequences:
FGRF1: TSPLYLEIIIYCTGAFLISCMVGSVIVYKMK
FGRF2: TASPDYLEIAIYCIGVFLIACMVVTVILCRMKNTTK
CMP9: G3-(GPP)9
CMP8: G3-(POG)8, O: hydroxyproline
Yield obtained by MALDI-TOF and HPLC
Yield obtained by CD and UV-Vis absorbance
After developing the new protocol for long hydrophobic peptide modification, we assessed its utility to modify soluble peptides with strong tendencies to associate and form secondary structures. Collagen mimetic peptides (CMPs) contain X-Y-glycine amino acid repeat sequences (X and Y are typically proline or hydroxyproline) that induce three CMP strands to intertwine into a collagen-like triple helix [20]. The strong propensity to form this CMP triple helix in a wide range of solvent conditions presents a steric hindrance for N-terminal modification in solution. Yet, high yield of these functionalized peptides are important for work employing fluorescently labeled CMPs for facile modification of collagen tissue scaffolds and for fabrication of CMP-hybridized PEG hydrogels [21–24].
G3-(GPP)9, a 30-residue CMP was labeled with CF on-the-resin using PyAOP and the newly developed protocol. The efficiency of the labeling reached 94% after 24 h, confirming the success of the protocol for labeling the mimetic peptides with fluorescent tags. The on-the-resin labeling was also highly effective in coupling CMP to other functional chemical species, such as a PEG oligomer (dPEG4) (Advanced ChemTech). Near 100% efficiency was achieved for coupling CMPs to dPEG4 using the described on-the-resin coupling strategy employing PyBOP activation. This on-the-resin coupling strategy far exceeded the reaction efficiency (about 10%) of attempts to couple NHS-activated PEG to the peptide’s N-terminus. Thus, the presented labeling protocol is highly efficient and versatile, and we hope that it will enable the biophysical characterization and technological applications of long, hydrophobic, or strongly associating peptides.
Supplementary Material
Acknowledgments
This work was supported by NSF MCB-0718841 and DMR-1003441 awarded to K.H. and by NSF DMR-0645411 and NIH R01AR060484 awarded to S.M.Y.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Marks JR, Placone J, Hristova K, Wimley WC. Spontaneous membrane-translocating peptides by orthogonal high-throughput screening. J Am Chem Soc. 2011;133:8995–9004. doi: 10.1021/ja2017416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Togashi DM, Ryder AG. Assessing protein-surface interactions with a series of multi-labeled BSA using fluorescence lifetime microscopy and Forster Energy Resonance Transfer. Biophys Chem. 2010;152:55–64. doi: 10.1016/j.bpc.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Carter PJ. Introduction to current and future protein therapeutics: A protein engineering perspective. Experim Cell Res. 2011;317:1261–1269. doi: 10.1016/j.yexcr.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Payne RJ, Wong CH. Advances in chemical ligation strategies for the synthesis of glycopeptides and glycoproteins. Chemical Communications. 2010;46:21–43. doi: 10.1039/b913845e. [DOI] [PubMed] [Google Scholar]
- 5.Diaz-Rodriguez A, Davis BG. Chemical modification in the creation of novel biocatalysts. Current Opinion in Chemical Biology. 2011;15:211–219. doi: 10.1016/j.cbpa.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Berrade L, Garcia AE, Camarero JA. Protein Microarrays: Novel Developments and Applications. Pharmaceu Res. 2011;28:1480–1499. doi: 10.1007/s11095-010-0325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basle E, Joubert N, Pucheault M. Protein Chemical Modification on Endogenous Amino Acids. Chem & Biol. 2010;17:213–227. doi: 10.1016/j.chembiol.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 8.Li E, Hristova K. Imaging FRET Measurements of Transmembrane Helix Interactions in Lipid Bilayers on a Solid Support. Langmuir. 2004;20:9053–9060. doi: 10.1021/la048676l. [DOI] [PubMed] [Google Scholar]
- 9.Li E, You M, Hristova K. SDS-PAGE and FRET suggest weak interactions between FGFR3 TM domains in the absence of extracellular domains and ligands. Biochemistry. 2005;44:352–360. doi: 10.1021/bi048480k. [DOI] [PubMed] [Google Scholar]
- 10.Schick S, Chen LR, Li E, Lin J, Koper I, Hristova K. Assembly of the M2 Tetramer Is Strongly Modulated by Lipid Chain Length. Biophys J. 2010;99:1810–1817. doi: 10.1016/j.bpj.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.You M, Li E, Wimley WC, Hristova K. FRET in liposomes: measurements of TM helix dimerization in the native bilayer environment. Analytical Biochemistry. 2005;340:154–164. doi: 10.1016/j.ab.2005.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Merzlyakov M, Cohen T, Shai Y, Hristova K. Energetics of ErbB1 transmembrane domain dimerization in lipid bilayers. Biophys J. 2009;96:4622–4630. doi: 10.1016/j.bpj.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merzlyakov M, Hristova K. Forster Resonance Energy Transfer Measurements of Transmembrane Helix Dimerization Energetics. Methods in Enzymology: Fluorescence Spectroscopy. 2008;450:107–127. doi: 10.1016/S0076-6879(08)03406-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You M, Spangler J, Li E, Han X, Ghosh P, Hristova K. Effect of pathogenic cysteine mutations on FGFR3 transmembrane domain dimerization in detergents and lipid bilayers. Biochemistry. 2007;46:11039–11046. doi: 10.1021/bi700986n. [DOI] [PubMed] [Google Scholar]
- 15.Iwamoto T, You M, Li E, Spangler J, Tomich JM, Hristova K. Synthesis and initial characterization of FGFR3 transmembrane domain: Consequences of sequence modifications. Biochim Biophys Acta. 2005;1668:240–247. doi: 10.1016/j.bbamem.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez-Carneado J, Giralt E. An efficient method for the solid-phase synthesis of fluorescently labelled peptides. Tetrahedron Letters. 2004;45:6079–6081. [Google Scholar]
- 17.Albericio F, Cases M, Alsina J, Triolo SA, Carpino LA, Kates SA. On the use of PyAOP, a phosphonium salt derived from HOAt, in solid-phase peptide synthesis. Tetrahedron Letters. 1997;38:4853–4856. [Google Scholar]
- 18.Albericio F, Carpino LA. Coupling reagents and activation. Solid-Phase Peptide Synthesis. 1997;289:104–126. doi: 10.1016/s0076-6879(97)89046-5. [DOI] [PubMed] [Google Scholar]
- 19.Fischer R, Mader O, Jung G, Brock R. Extending the applicability of carboxyfluorescein in solid-phase synthesis. Bioconjugate Chemistry. 2003;14:653–660. doi: 10.1021/bc025658b. [DOI] [PubMed] [Google Scholar]
- 20.Baum J, Brodsky B. Folding of peptide models of collagen and misfolding in disease. Cur Opinion Struc Biol. 1999;9:122–128. doi: 10.1016/s0959-440x(99)80016-5. [DOI] [PubMed] [Google Scholar]
- 21.Wang AY, Mo X, Chen CS, Yu SM. Facile modification of collagen directed by collagen mimetic peptides. J Am Chem Soc. 2005;127:4130–4131. doi: 10.1021/ja0431915. [DOI] [PubMed] [Google Scholar]
- 22.Wang AY, Leong S, Liang YC, Huang RCC, Chen CS, Yu SM. Immobilization of Growth Factors on Collagen Scaffolds Mediated by Polyanionic Collagen Mimetic Peptides and Its Effect on Endothelial Cell Morphogenesis. Biomacromolecules. 2008;9:2929–2936. doi: 10.1021/bm800727z. [DOI] [PubMed] [Google Scholar]
- 23.Wang AY, Foss CA, Leong S, Mo X, Pomper MG, Yu SM. Spatio-temporal modification of collagen scaffolds mediated by triple helical propensity. Biomacromolecules. 2008;9:1755–1763. doi: 10.1021/bm701378k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stahl PJ, Romano NH, Wirtz D, Yu SM. PEG-Based Hydrogels with Collagen Mimetic Peptide-Mediated and Tunable Physical Cross-Links. Biomacromolecules. 2010;11:2336–2344. doi: 10.1021/bm100465q. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.