Abstract
Glycosylation is highly sensitive to the biochemical environment and has been implicated in many diseases including cancer. Glycan compositional profiling of human serum with mass spectrometry has already identified potential biomarkers for several types of cancer and diseases; however, composition alone does not fully describe glycan stereo- and regioisomeric diversity. The vast structural heterogeneity of glycans presents a formidable analytical challenge. We have developed a method to identify and quantify isomeric native glycans using nanoflow liquid chromatography (nano-LC)/mass spectrometry. A microfluidic chip packed with graphitized carbon was used to chromatographically separate the glycans. To determine the utility of this method for structure-specific biomarker discovery, we analyzed serum samples from two groups of prostate cancer patients with different prognoses. More than 300 N-glycan species (including isomeric structures) were identified, corresponding to over 100 N-glycan compositions. Statistical tests established significant differences in glycan abundances between patient groups. This method provides comprehensive, selective, and quantitative glycan profiling.
Keywords: glycan, isomer, quantitation, cancer biomarker, nano-LC, mass spectrometry
INTRODUCTION
Aberrant glycosylation has been observed for decades in nearly all types of human cancer and is now quite well established as an indicator of cell oncogenic transformation.1, 2 Glycans are important determinants of protein function, and many cancer-related processes such as apoptosis,3, 4 angiogenesis,5, 6 growth factor receptor binding,7-9 or integrin-cadherin function10, 11 are profoundly affected by changes in glycosylation. The glycosylation machinery involves competition between a set of glycosyltransferase enzymes such that up- or down-regulation of an individual transferase is amplified across the entire glycan biosynthetic pathway.2 Examples of aberrant glycosylation associated with disease states are rampant in the literature and include variations in the amount of specific saccharide residues, expression of incomplete or truncated glycan structures, and increases or decreases in the relative abundance of certain glycan species.1, 12, 13
Glycans, consisting of short saccharide chains of two to twenty units, are the only post-translational modification with significant structural complexity. Whereas sequence is sufficient to determine the primary structures of linear biomolecules such as nucleic acids and proteins, glycans are branched, with linkage and anomeric characters. For example, a standard hexose has five different sites at which it may be connected to other saccharide units. When anomeric character is considered, the possibilities increase to ten unique potential linkages. In addition, branching is theoretically possible at every residue. The plethora of stereo- and regio-isomeric possibilities greatly increases the complexity of oligosaccharide analysis.
Until now, the search for glycan biomarkers has focused mainly on glycan compositional profiling.14-20 However, structure-specific glycan profiling may uncover more robust glycan markers with higher specificity than compositional profiling alone. Changes in specific saccharide linkages have been associated with diseases such as pancreatic cancer and Alzheimer’s disease.21, 22 Since the number of glycan structures greatly surpasses the number of compositions, structure-specific profiling provides more biological information and thus serves as a significantly larger source of potential glycan biomarkers.23, 24 In order to gain structure-specific access to individual glycan species, isomer separation is required.
Glycan chromatographic profiling enables not only the discovery of structure-specific markers but also unprecedented structural analysis of the glycome. This method complements our previously-developed strategies of glycan mass profiling by high resolution mass spectrometry, which provides rapid compositional analysis based on accurate mass.14-17 Each mass or composition identified by non-chromatographic MS analysis is made up of several distinct structures. Chromatographic profiling, coupled with mass spectrometry, allows these structures to be separated and specific sugar-sugar linkage types examined.
Limited attempts at glycan isomeric differentiation and characterization have been reported. Isailovic et al used ion mobility mass spectrometry to interrogate derivatized serum glycans from cancer patients.25 Peak perturbations in the ion mobility spectrum were attributed to partial separation of glycan isomers or conformers. Nakagawa et al employed a more traditional reversed phase LC/MS-based approach to analyze N-glycan isomers from rheumatoid arthritis patients; however, glycans had to be desialylated and then derivatized prior to analysis.26 Bones et al attempted a non-MS approach involving ultra performance LC and fluorescence detection of labeled N-glycans.27 In contrast, Prien et al employed a wholly non-chromatographic approach using MSn to differentiate structural isomers for a small number of derivatized glycans isolated from tumor cells.28 Studies on isomeric separation of native glycans are essentially non-existent in the literature. However, for biomarker-related applications, minimal sample manipulation and processing are preferred, so as to preserve the original glycan profile and maximize method reproducibility. Thus, for this study, native, underivatized glycans were analyzed.
Our group was first to use microfluidic chip-based nano-LC for the global separation of serum glycans.29 Nano-LC/MS provides significantly greater sensitivity compared to conventional LC/MS or MALDI-MS.18, 19 In addition, nano-LC-ESI/MS generally produces low energy ions and therefore yields less in-source fragmentation than MALDI. The microfluidic chip is the latest evolution of nano-LC, providing unmatched retention time reproducibility.30 Coupling chip-based nano-LC technology with nano-ESI and a high performance time-of-flight (TOF) MS detector provides the added benefits of high mass accuracy and a wide instrumental dynamic range.31-34
In this study we developed and optimized a method to chromatographically separate and profile native serum N-glycans by porous graphitized carbon (PGC) nano-LC/TOF-MS for the purposes of identifying structure-specific glycan biomarkers. Using this method, serum N-glycan samples from prostate cancer patients were profiled both by overall compositional abundance and in relation to specific isomers. Patients with poor prognoses (P group) and good prognoses (G group) were compared according to individual glycan abundances. Statistical tests were performed to establish significant differences. Isomer-specific glycan chromatographic profiling will supplement and augment glycan compositional profiling achieved by MS analysis by identifying the specific glycan structures that vary according to disease.
EXPERIMENTAL
Collection of human sera
Serum samples were collected from patients who had been diagnosed with prostate cancer and were seen at the UC Davis Cancer Center. Their samples were procured by the Cancer Center Biorepository via an IRB-approved protocol. Patients were divided into two groups based on PSA (prostate-specific antigen) levels. The P group (n = 4) consisted of patients with poor prognoses based on elevated PSA levels post-RRP (radical retropubic prostatectomy), while the G group (n = 4) consisted of patients with good prognoses based on undetectable PSA levels post-RRP. Serum samples were collected by standard venous phlebotomy into a clot-activator vacuum blood collection tube (BD Diagnostics, Franklin Lakes, NJ).
Enzymatic release of N-glycans
Denaturation buffer consisting of 200 mM ammonium bicarbonate and 10 mM dithiothreitol was prepared. A 100 μL aliquot of serum was added to 100 μL of denaturation buffer and heated to 100 °C in order to denature the serum proteins. After cooling, 2.0 μL of peptide N-glycosidase F (New England Biolabs, Ipswich, MA) were added and the mixture was incubated in a water bath at 37 °C for 12 hours in order to enzymatically release the N-glycans. Finally, 800 μL of cold ethanol were added and the mixture was chilled at −2 0 °C for 1 hour in order to precipitate out the deglycosylated proteins. Following centrifugation, released N-glycans were isolated in the supernatant fraction and dried in vacuo.
N-glycan enrichment with graphitized carbon SPE
Released N-glycans were purified by graphitized carbon (GC) solid-phase extraction (SPE). GC cartridges were washed with 0.05% (v/v) trifluoroacetic acid in 80% acetonitrile/water (v/v) followed by conditioning with water. The N-glycan fractions were reconstituted in water and then washed with water at a flow rate of approximately 1 mL/min to remove salts and buffer. Serum N-glycans were eluted stepwise with 10% acetonitrile/water (v/v), 20% acetonitrile/water (v/v), and 0.05% (v/v) trifluoroacetic acid in 40% acetonitrile/water (v/v). Samples were dried in vacuo prior to MS analysis.
Chromatographic separation of the serum N-glycome
N-glycan fractions were reconstituted in water and analyzed using an Agilent HPLC-Chip/Time-of-Flight (Chip/TOF) MS system equipped with a microwell-plate autosampler (maintained at 6 °C), capillary sample loading pump, nano pump, HPLC-Chip/MS interface, and the Agilent 6210 TOF MS detector. The chip used consisted of a 9 × 0.075 mm i.d. enrichment column and a 43 × 0.075 mm i.d. analytical column, both packed with 5 μm porous graphitized carbon (PGC) as the stationary phase, with an integrated nano-ESI spray tip. For sample loading, the capillary pump delivered 0.1% formic acid in 3.0% acetonitrile/water (v/v) isocratically at 4.0 μL/min. Injection volume was 2.0 μL for each sample. A nano pump gradient was delivered at 0.3 μL/min using (A) 0.1% formic acid in 3.0% acetonitrile/water (v/v) and (B) 0.1% formic acid in 90.0% acetonitrile/water (v/v). Samples were eluted with 0% B, 0.00-2.50 min; 0 to 16% B, 2.50-20.00 min; 16 to 44% B, 20.00-30.00 min; 44 to 100% B, 30.00-35.00 min; and 100% B, 35.00-45.00 min. This was followed by a quick gradient from 0 to 100% B in order to wash out any remaining compounds, and finally re-equilibration at 0% B. The drying gas temperature was set at 325 °C with a flow rate of 4 L/min (2 L of filtered nitrogen gas and 2 L of filtered dry compressed air). MS spectra were acquired in the positive ionization mode over a mass range of m/z 400-2500 with an acquisition time of 1.5 seconds per spectrum. Mass correction was enabled using reference masses of m/z 622.029, 922.010, 1221.991, 1521.971, 1821.952, and 2121.933 (ESI-TOF Calibrant Mix G1969-85000, Agilent Technologies, Santa Clara, CA).
To minimize possible bias due to injection order and/or instrumental drift, samples were injected in randomized order, using the same solvents, over the course of a single instrument session. The random sample sequence was repeated three times such that all samples were injected in triplicate.
RESULTS AND DISCUSSION
Method optimization
Serum N-glycans are a complex mixture with large structural diversity and dynamic range. Incorporation of chromatographic separation into established mass spectral methods of glycomic analysis allows us to distinguish between isomeric compounds of the same glycan composition. The chip-based nano-LC/TOF-MS (Chip/TOF) system provides high sensitivity, large instrumental dynamic range, minimal ion suppression, and low sample consumption.29, 30 These attributes are uniquely suited to the analysis of serum N-glycans.
In order to ensure accurate, quantitative, and reproducible data that would span the serum N-glycan dynamic range, method optimization was necessary. Optimal instrumental parameters for high ionization efficiency and low in-source fragmentation had already been established by our previous work with the Chip/TOF system.29, 30 To complement this information, we examined chromatographic loading capacity and separation capabilities of the chip-based nano-LC. Starting from an initial concentration (henceforth a “1x dilution”) corresponding to 4 μL serum per 2 μL injection, samples were diluted to final concentrations corresponding to 400 nL serum/injection (10x dilution); 40 nL serum/injection (100x dilution); 15 nL serum/injection (300x dilution); and 9 nL serum/injection (500x dilution). Sample dilutions were compared in order to optimize chromatographic separation and detection of both low- and high-abundance serum N-glycans.
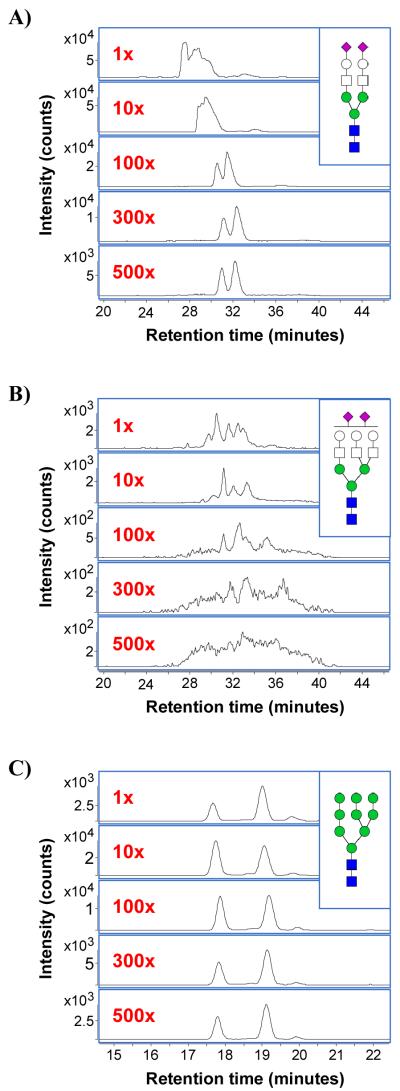
In order to evaluate glycan isomer separation capabilities, representative N-glycans were selected for analysis based on characteristics such as structure, abundance, and interaction strength with PGC. The high mass accuracy of the TOF MS detector enabled us to confidently predict the expected m/z values of our selected N-glycans. The m/z values associated with charge states 1 < z < 4 of each selected N-glycan composition were computed, and extracted ion chromatograms (XICs) showing the abundance vs. time of the selected N-glycans were plotted. Visual inspection of the XICs was sufficient to identify saturated as well as low-abundance peaks (Figure 1).
Figure 1.
a) XICs of m/z 1112.40, i.e. the doubly protonated ion of the high-abundance complex biantennary disialylated glycan (NB2) at dilutions of 1x, 10x, 100x, 300x, and 500x (top to bottom). b) XICs of the low-abundance complex triantennary disialylated glycan (NT2) at dilutions of 1x, 10x, 100x, 300x, and 500x. c) XICs of m/z 942.33, i.e. the doubly protonated ion of the high-mannose glycan composition Hex9-HexNAc2, at dilutions of 1x, 10x, 100x, 300x, and 500x.
N-glycan abundances exhibited a large dynamic range spanning over five orders of magnitude. As a result, high- and low-concentration analyses often returned complementary information. At the extreme edges of the dynamic range, lower sample concentrations were more suitable for effectively separating high-abundance glycans, whereas higher sample concentrations were more suitable for detecting low-abundance glycans. Sample concentration had to be optimized in order to maintain high chromatographic resolution and baseline peak separation of isomers. Figure 1a shows the XICs of m/z 1112.40, i.e. the doubly protonated [M+2H]+2 ion of the biantennary disialylated N-glycan (NB2) consisting of Hex5-HexNAc4-NeuAc2 at dilutions of 1x, 10x, 100x, 300x, and 500x. In human serum, NB2 is reported as the most abundant N-glycan.35 Accordingly, the chromatograms of the 1x and 10x dilutions show heavy saturation, with severe peak tailing, poor resolution, and retention time shifts. In contrast, XICs of the low-abundance complex triantennary disialylated N-glycan NT2 (Hex6-HexNAc5-NeuAc2) (Figure 1b) show that at the 1x dilution, nano-LC/MS is able to resolve the glycans into several distinct peaks. However, as the dilution factor increases, competive ionization leads to dramatic decreases in MS sensitivity.
For the majority of compounds, however, a wide range of injection concentrations are suitable for quantitative analysis. Figure 1c shows the XICs of m/z 942.33, i.e. the doubly protonated ion of the high-mannose glycan Hex9-HexNAc2, at the same series of dilutions (1x to 500x). Each dilution shows the same major and minor peaks with good resolution and Gaussian peak distributions. In addition, retention times are highly reproducible, with extremely slight deviation even at vastly different sample concentrations. One issue of note is that, whereas the 10x to 500x dilutions follow the expected trend of decreasing XIC abundances with increasing dilution factors, the abundances from the 1x dilution are in fact lower than their 10x dilution equivalents. This phenomenon is also evident for the other high-mannose glycan compositions, and is likely related to the previously noted column saturation at the 1x dilution. As evidenced by their early retention times, high-mannose glycans have a relatively weak interaction with porous graphitized carbon in comparison to complex/hybrid sialylated compounds. Therefore, in cases of high column saturation, porous graphitized carbon preferentially binds to complex/hybrid rather than high-mannose glycans. Since each distinct glycan structure has a different interaction with porous graphitized carbon, this explanation would also account for the dynamic isomer ratios observed throughout the dilution series.
High- and low-concentration analyses returned complementary information; thus, the samples were eventually analyzed at both the 1x and 300x dilutions in order to maximize glycan coverage. This strategy effectively extended the experimental dynamic range of the nano-LC/MS method and allowed accurate, quantitative analysis of both high- and low-abundance aspects of the serum N-glycome.
Method reproducibility and sample variance
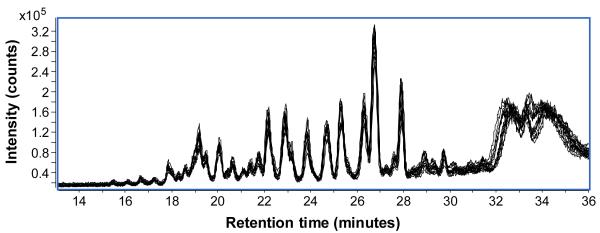
Serum N-glycan samples (n = 8) were analyzed in triplicate by nano-LC/MS. Peak intensities and retention times were examined in order to establish the quantitative and qualitative reproducibility of the method as well as the variance within the sample set. Figure 2 shows 24 overlaid total ion chromatograms (TICs) of all serum samples from both patient groups, injected in triplicate at a 300x dilution. Simple visual inspection highlights the low sample-to-sample variance even across different patient groups and the need for exceedingly precise measurements in order to differentiate between them.
Figure 2.
Overlaid total ion chromatograms (TICs) (n=24) of all serum samples from both patient groups, injected in triplicate at a 300x dilution. Average retention time difference between chromatograms was only 5.0 seconds ± 2.7 seconds.
Statistical tests were used to establish the quantitative precision, sensitivity, and reproducibility of the method. Ion abundances corresponding to selected representative N-glycans were subjected to two-factor analysis of variance (ANOVA), with patient group (P or G) as one factor and batch effect (first, second, or third replicate) as the other factor. Using a 95% confidence level (p < 0.05), patient group was found to be a significant factor for several N-glycans. Since patient groups corresponded to disease prognosis, these glycans could be considered disease biomarkers within the scope of this sample set.
In contrast to patient group, ANOVA results showed batch effect to be an insignificant factor (p > 0.05) in all tested glycans; i.e. each replicate was not significantly different from the others. These results affirmed that instrumental drift was not a significant issue and the collected data were indeed statistically independent of sample injection order. As a result, batch effect was disregarded for subsequent statistical analyses, allowing the sole use of T-tests.
The quantitative precision of the method was further tested by dividing the patient groups into individual patients (n = 4) and subjecting the glycan abundance data to one-factor ANOVA. In both patient groups, for all glycans tested, the individual patient was found to be a significant factor (p < 0.05); i.e. the method was precise enough to differentiate even among individual patients from the same patient group.
Qualitative aspects of the method reproducibility, such as retention time, were analyzed by computer algorithm. Every single X-Y coordinate (n = 600) of the TIC function was compared to its corresponding X-Y coordinate in every other TIC function (n = 24), and the distance between each was recorded. From this data, average differences in the X axis (retention time) were calculated. In general, chromatograms of 300x diluted samples ion exhibited less run-to-run variance than chromatograms of undiluted (1x) samples. For the 300x dilution chromatograms, average retention time difference was only 5.0 seconds ± 2.7 seconds, while for the undiluted 1x chromatograms, average retention time difference increased to 7.9 seconds ± 3.7 seconds. The loss of retention time reproducibility from the 300x to 1x runs emphasize the importance of column loading to precise glycan analysis.
Aside from column loading, much of the observed variation stemmed from the extreme sensitivity of highly sialylated, acidic glycans such as NB2 to gradient changes. Indeed, when NB2 was removed from the comparison, retention time reproducibility for the 300x dilution increased dramatically to 3.7 seconds ± 1.5 seconds. Work is ongoing to optimize the elution gradient and stabilize retention times for this particular glycan class.
Detection and identification of N-glycans
In order to obtain N-glycan profiles of each serum sample, computerized algorithms first extracted a generalized list of compound peaks in the sample and then identified the N-glycan compositions by accurate mass.
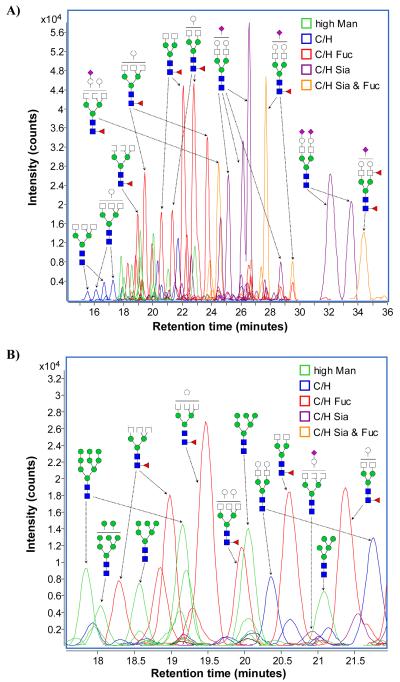
Raw LC/MS data was filtered with a signal-to-noise ratio of 5.0 and analyzed using the Molecular Feature Extractor algorithm included in the MassHunter Qualitative Analysis software (Version B.03.01, Agilent Technologies). Taking into account expected charge carriers, potential neutral mass losses, and a predicted isotopic distribution, the total ion chromatogram (TIC) was divided into a number of extracted compound chromatograms (ECCs) (Figure 3). Each ECC represents the summed chromatograms of all ion species associated with a single compound (e.g. the singly protonated, doubly protonated, singly dehydrated, etc. ions). Thus, each individual ECC peak could be taken to represent the total ion count associated with a single distinct compound.
Figure 3.
a) Extracted compound chromatograms (ECCs) of glycans found in a representative serum sample. b) Magnified view of a short segment of the glycan elution profile, showing the high sensitivity and resolution achieved by nano-LC separation. Colors denote different glycan classes.
The deconvoluted mass, retention time, abundance (in summed ion counts), and observed charge states associated with each extracted compound were exported for further analysis. N-glycans were identified by accurate mass using a set of in-house software tools. A mass error tolerance of 20 ppm was used. As the sample set originated from human serum, only N-glycan compositions containing hexose (Hex), N-acetylhexosamine (HexNAc), fucose (Fuc), and N-acetylneuraminic acid (NeuAc) were considered. To aid these types of analyses, our laboratory has developed theoretical N-glycan mass libraries that cover all possible complex, hybrid, and high-mannose glycan compositions based on known biological synthesis pathways and glycosylation patterns36. Using these search criteria, N-glycan compositions were identified and correlated with abundances and retention times.
On average, our nano-LC method was able to resolve and identify over 300 N-linked glycan compound peaks with over 100 distinct N-linked glycan compositions spanning five orders of magnitude in abundance (Figure 3). Each of the identified compositions included two or more peaks corresponding to structural and/or linkage isomers. The high reproducibility of the nano-LC means that a specific glycan structure will have the same retention time in different samples across multiple chromatographic runs. While each peak/retention time has not yet undergone full structural elucidation, libraries that match chip-based PGC nano-LC retention times to specific glycan structures are currently under development in our laboratory.
Glycan separation by porous graphitized carbon (PGC)
PGC has been used as a stationary phase to separate glycans based on size, polarity, and three-dimensional structure.37, 38 Traditionally, LC separation of native glycans has been done using PGC as well as amine/amide and anion-exchange stationary phases.39, 40 PGC columns offer distinct advantages over these other stationary phases such as low column-to-column variability, stability over a large pH range, and the ability to separate neutral and acidic glycans within a single run.41, 42 In particular, the PGC nano-LC system presented herein has been successfully demonstrated in the past to separate glycan isomers.29, 30
Nano-LC separation of serum N-glycans by PGC exhibited a number of consistent trends (Figure 3). In most cases, smaller, simpler glycans eluted earlier, while larger, more complex glycans eluted later. In particular, sialylation increased retention time additively, with higher degrees of sialylation associated with correspondingly later retention times. Core fucosylation, i.e. addition of a fucose monosaccharide to the N-glycan core, was also observed to increase retention times relative to equivalent non-fucosylated compounds. Addition of a Hex or HexNAc increased retention times in biantennary N-glycans; however, the effect on the retention times of triantennary N-glycans was less predictable. Notably, the triantennary high-mannose glycans eluted in order from largest (Hex9-HexNAc2) to smallest (Hex5-HexNAc2).
The demonstrated ability of PGC to separate isomeric glycans is related to differences in sugar-sugar linkages which lead to changes in the three-dimensional structure of compositionally identical glycans.37, 38 A triantennary N-glycan has a significantly different three-dimensional structure from a biantennary glycan of similar size and composition.43 As a result, it exhibits modified interactions with both the porous graphitized carbon stationary phase as well as the acetonitrile/water mobile phase. These account for the modified elution orders seen with triantennary glycans.
Glycan compositional profiling for biomarker discovery
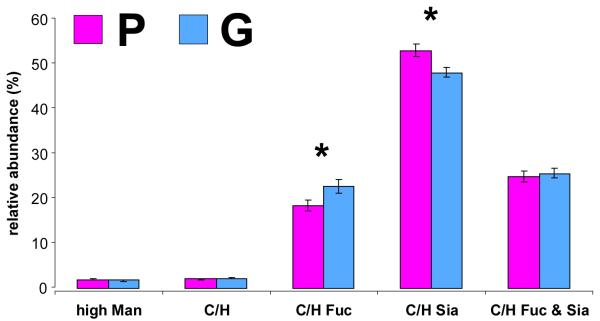
In order to simplify the analysis and obtain a generalized view of the cancer serum glycome, compositional N-glycan profiling was applied as a data-reduction method.
An overall perspective of the N-glycan content of serum from prostate cancer patients was gained by grouping together N-glycans of similar composition or structure and analyzing them as a correlated set. Individual glycan compounds were sorted into several N-glycan classes— 1) high-mannose glycans; 2) undecorated complex/hybrid glycans; 3) fucosylated (but nonsialylated) complex/hybrid glycans; 4) sialylated (but non-fucosylated) complex/hybrid glycans; and 5) fucosylated and sialylated complex/hybrid glycans. Abundances were calculated relative to the total ion abundance of all N-glycans in a particular nano-LC run; i.e. relative abundance. Sialylated, non-fucosylated complex/hybrid glycans were found to be the most abundant N-glycan class in both patient groups, at 52.8% relative abundance in patients with poor prognoses (P group) and 47.9% relative abundance in patients with good prognoses (G group). High abundances were also found of fucosylated and sialylated complex/hybrid glycans (24.8% in the P group; 25.5% in the G group) as well as fucosylated, non-sialylated complex/hybrid glycans (18.5% in the P group; 22.6% in the G group). In contrast, low abundances were found of undecorated complex/hybrid glycans (2.05% in the P group; 2.17% in the G group) and high-mannose glycans (1.87% in the P group; 1.78% in the G group). Figure 4 summarizes the different relative abundances of each N-glycan class found in the P and G patient groups. In order to determine whether differences between the P and G groups were significant, standard T-tests were performed.44 Fucosylated, non-sialylated complex/hybrid glycans were significantly more abundant in the G group than in the P group (p = 0.0495). In addition, sialylated, nonfucosylated complex/hybrid glycans were significantly more abundant in the P group than in the G group (p = 0.00758). These results provide a macro perspective of the glycosylation changes associated with prostate cancer and may help highlight glycan synthesis pathways that are affected by disease. However, when searching for clinically significant biomarkers, a more detailed and compound-specific approach would be desired, e.g. compositional or isomer-specific profiling.
Figure 4.
Relative abundances of high-mannose glycans, undecorated complex/hybrid (C/H) glycans, fucosylated (but non-sialylated) C/H glycans, sialylated (but non-fucosylated) C/H glycans, and finally, fucosylated and sialylated C/H glycans found in the P and G patient groups. Asterisks denote statistically significant differences (p < 0.05) between patient groups.
A more detailed analysis of the serum N-glycan profile was obtained by separately considering each individual N-glycan composition. Table 1 summarizes the relative abundances of the top twenty most abundant N-glycans found in serum from prostate cancer patients. N-glycans from patients with poor prognoses (P group) were observed at different relative abundances compared to sera from patients with good prognoses (G group). T-tests were again used to determine the significance of these differences. Relative as well as absolute abundances were compared using data from both the 1x and 300x dilutions. Various levels of significance were tested, with 48 N-glycan compositions found at p < 0.100, 36 N-glycan compositions found at p < 0.050, 18 N-glycan compositions found at p < 0.010, and 6 N-glycan compositions found at p < 0.001. N-glycan compositions with statistically significant differences between the P and G groups included fucosylated, sialylated, and high-mannose type glycans. These results are summarized in Table 2.
Table 1.
Relative abundances of the top twenty most abundant glycans found in serum from prostate cancer patients
| Glycan Mass (Da) | Composition | Relative Abundance (%) | ||||
|---|---|---|---|---|---|---|
| Hex | HexNAc | Fuc | NeuAc | G | P | |
| 2222.78 | 5 | 4 | 0 | 2 | 22.0 | 24.1 |
| 1931.69 | 5 | 4 | 0 | 1 | 18.1 | 18.4 |
| 2077.75 | 5 | 4 | 1 | 1 | 10.3 | 9.17 |
| 1786.65 | 5 | 4 | 1 | 0 | 6.82 | 5.15 |
| 1624.60 | 4 | 4 | 1 | 0 | 6.33 | 5.47 |
| 2368.84 | 5 | 4 | 1 | 2 | 4.14 | 4.55 |
| 2426.88 | 5 | 5 | 2 | 1 | 3.11 | 2.25 |
| 1462.54 | 3 | 4 | 1 | 0 | 2.51 | 1.96 |
| 2587.92 | 6 | 5 | 0 | 2 | 1.85 | 3.12 |
| 2879.01 | 6 | 5 | 0 | 3 | 1.62 | 1.84 |
| 2296.82 | 6 | 5 | 0 | 1 | 1.49 | 1.94 |
| 1948.70 | 6 | 4 | 1 | 0 | 1.13 | 1.20 |
| 1640.59 | 5 | 4 | 0 | 0 | 0.943 | 0.953 |
| 2572.94 | 5 | 5 | 3 | 1 | 0.770 | 0.890 |
| 1234.43 | 5 | 2 | 0 | 0 | 0.715 | 0.714 |
| 1827.68 | 4 | 5 | 1 | 0 | 0.586 | 0.565 |
| 2792.01 | 6 | 6 | 2 | 1 | 0.236 | 0.360 |
| 2953.05 | 7 | 6 | 0 | 2 | 0.218 | 0.413 |
| 2880.03 | 6 | 5 | 2 | 2 | 0.110 | 0.330 |
| 2369.86 | 5 | 4 | 3 | 1 | 0.0808 | 0.146 |
Table 2.
Complex/hybrid and high-mannose type N-glycans which showed statistically significant differences in relative abundance
| Glycan Mass (Da) | Composition | T-Test | |||
|---|---|---|---|---|---|
| Hex | HexNAc | Fuc | NeuAc | ||
| High mannose | |||||
| 1396.486 | 6 | 2 | 0 | 0 | 0.014648 |
| 1720.592 | 8 | 2 | 0 | 0 | 1.7987 × 10−4 |
| 1882.645 | 9 | 2 | 0 | 0 | 0.024594 |
| 2044.698 | 10 | 2 | 0 | 0 | 0.016121 |
| Complex/hybrid (non-fucosylated, non-sialylated) | |||||
| 1478.539 | 4 | 4 | 0 | 0 | 0.056212 |
| 1519.566 | 3 | 5 | 0 | 0 | 2.9086 × 10−6 |
| 1681.619 | 4 | 5 | 0 | 0 | 7.3339 × 10−4 |
| 1843.672 | 5 | 5 | 0 | 0 | 0.047356 |
| 2005.724 | 6 | 5 | 0 | 0 | 0.029473 |
| Complex/hybrid (fucosylated) | |||||
| 1462.544 | 3 | 4 | 1 | 0 | 0.0081408 |
| 1624.597 | 4 | 4 | 1 | 0 | 0.0076610 |
| 1786.650 | 5 | 4 | 1 | 0 | 1.8114 × 10−4 |
| 2110.756 | 7 | 4 | 1 | 0 | 0.012047 |
| 1665.624 | 3 | 5 | 1 | 0 | 0.021612 |
| 1989.729 | 5 | 5 | 1 | 0 | 0.039760 |
| 2313.835 | 7 | 5 | 1 | 0 | 0.011292 |
| 2192.809 | 5 | 6 | 1 | 0 | 0.012267 |
| 2094.761 | 6 | 4 | 2 | 0 | 0.041820 |
| 2281.845 | 5 | 5 | 3 | 0 | 0.026727 |
| 2809.030 | 7 | 6 | 3 | 0 | 0.0086584 |
| 2630.982 | 5 | 6 | 4 | 0 | 0.0061516 |
| 2793.035 | 6 | 6 | 4 | 0 | 8.8975 × 10−4 |
| Complex/hybrid (sialylated) | |||||
| 2093.740 | 6 | 4 | 0 | 1 | 0.0044658 |
| 2296.820 | 6 | 5 | 0 | 1 | 0.0024734 |
| 2661.952 | 7 | 6 | 0 | 1 | 0.0013869 |
| 2222.783 | 5 | 4 | 0 | 2 | 0.069364 |
| 2587.915 | 6 | 5 | 0 | 2 | 0.011771 |
| 2790.995 | 6 | 6 | 0 | 2 | 0.042393 |
| 2953.047 | 7 | 6 | 0 | 2 | 0.0051923 |
| 3082.090 | 6 | 6 | 0 | 3 | 0.076477 |
| Complex/hybrid (fucosylated and sialylated) | |||||
| 1874.666 | 5 | 3 | 1 | 1 | 0.0052378 |
| 1915.693 | 4 | 4 | 1 | 1 | 0.0098378 |
| 2077.745 | 5 | 4 | 1 | 1 | 0.042098 |
| 2239.798 | 6 | 4 | 1 | 1 | 0.055126 |
| 2118.772 | 4 | 5 | 1 | 1 | 3.4185 × 10−4 |
| 2604.930 | 7 | 5 | 1 | 1 | 0.035766 |
| 2321.851 | 4 | 6 | 1 | 1 | 0.0076360 |
| 2020.724 | 5 | 3 | 2 | 1 | 0.070091 |
| 2061.751 | 4 | 4 | 2 | 1 | 0.048719 |
| 2467.909 | 4 | 6 | 2 | 1 | 0.059209 |
| 3157.147 | 7 | 7 | 2 | 1 | 0.062040 |
| 3042.084 | 7 | 5 | 2 | 2 | 0.012021 |
| 3286.190 | 6 | 7 | 2 | 2 | 0.083454 |
| 2660.957 | 5 | 4 | 3 | 2 | 0.061295 |
| 3885.396 | 7 | 7 | 3 | 3 | 0.0056551 |
| 2906.083 | 4 | 6 | 5 | 1 | 0.068077 |
| 4177.512 | 7 | 7 | 5 | 3 | 0.095601 |
Glycan chromatographic profiling for biomarker discovery
The main advantage of PGC nano-LC/MS analysis is the ability to distinguish between isomeric glycans. Identifying specific isomers which exhibit significant differences in abundance provides a window into the up- and down-regulation of glycosyltransferase activity under different biological conditions. While the nano-LC methodology described herein is easily capable of separating out and resolving glycan isomers, the associated data analysis is more time- and labor-intensive due to the computational complexities associated with peak recognition algorithms and retention time alignment. The present analysis focused on representative fucosylated, sialylated, and high-mannose type glycans as well as statistically interesting glycans (p < 0.01) identified during compositional profiling. Nano-LC was able to separate up to six different isomers for each composition.
Computerized algorithms identified the beginning and end of each chromatographic peak and calculated absolute abundances of each isomer in terms of ion counts. From this data, relative abundances of each isomer were calculated by dividing the ion abundance of a particular isomer over the total ion abundance of all isomers with that composition. High retention time reproducibility allowed peaks from different samples and runs to be aligned and compared. Both absolute and relative abundances were tested for significant differences.
Overall, 38 isomers from 19 N-glycan compositions showed significant differences between the P and G groups. Differences in isomer absolute abundances were found in 33 isomers from 17 N-glycan compositions, while differences in isomer relative abundances were found in 15 isomers from 9 N-glycan compositions. Several glycan isomers showed differences in both absolute and relative abundances. Results are summarized in Supplemental Table 1.
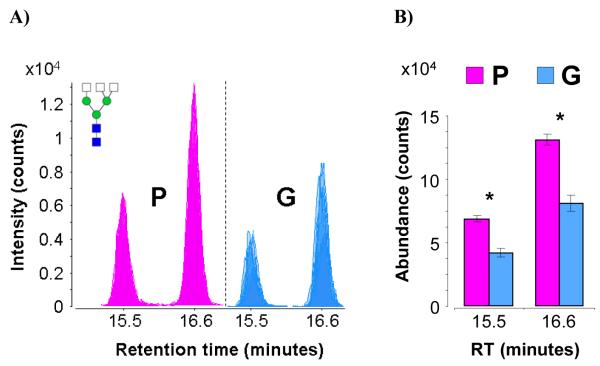
Figure 5a shows the overlaid raw extracted compound chromatograms (ECCs) for the complex triantennary N-glycan composition Hex3-HexNAc5. Two isomers are observed in the sera of both patient groups. However, the absolute abundances of both isomers are higher in the P group than the G group. Figure 5b represents the same data in bar graph form. T-tests of absolute abundances show that the isomer eluting at 15.5 minutes is indeed significantly more abundant in the P group, with a p-value of 6.50×10−5, while the isomer eluting at 16.6 minutes is also significantly more abundant in the P group, with a p-value of 5.25×10−5. However, comparisons of relative abundances show that both isomers appeared at more or less the same ratios in both groups, exhibiting no significant difference between the P and G groups.
Figure 5.
a) Overlaid chromatograms of the isomers of complex triantennary glycan composition Hex3-HexNAc5. Overlaps are represented by varying degrees of translucency. b) Bar graph representation of average abundances and standard error for the isomers of Hex3- HexNAc5. Asterisks denote statistically significant differences between patient groups.
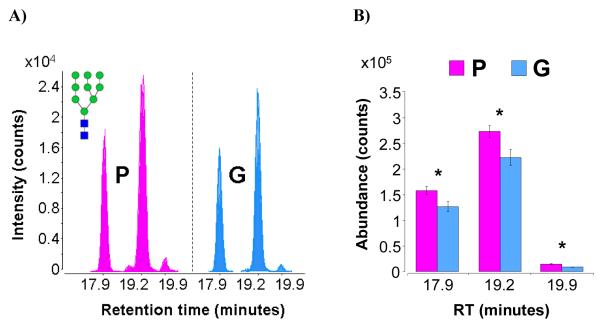
Figure 6 shows the raw ECCs and representative bar graphs for the high-mannose glycan composition Hex9-HexNAc2. Three isomers are observed in the sera of both patient groups. T-tests of absolute abundances show that the isomers at 17.9 minutes (p = 0.0351), 19.2 minutes (p = 0.0392), and 19.9 minutes (p = 0.00180) are significantly more abundant in the P group than in the G group. T-tests of relative abundances show that the isomers at 17.9 minutes and 19.9 minutes appeared at more or less the same ratios in both groups, exhibiting no significant difference between the P and G groups, whereas the isomer at 19.2 minutes appeared at a significantly lower ratio in the P group than in the G group, with a p-value of 0.0281.
Figure 6.
a) Overlaid chromatograms of the isomers of high-mannose glycan composition Hex9-HexNAc2. Overlaps are represented by varying degrees of translucency. b) Bar graph representation of average abundances and standard error for the isomers of Hex9-HexNAc2. Asterisks denote statistically significant differences between patient groups.
It should be noted that, in some cases, the glycan isomers separated by nano-LC may include alpha and beta anomers. However, in our experience, anomeric separation does not always occur, or may be insignificant when it does occur. This is supported by the existence of multiple glycan masses for which an odd number of isomers have been detected. Given the exceptionally high retention time reproducibility demonstrated by this nano-LC method, detection and identification of any potential anomer peaks may easily be addressed in the future with the use of retention time libraries currently under development by our lab.
CONCLUSION
We have demonstrated for the first time comprehensive, isomer-specific chromatographic profiling of serum N-glycans for the purposes of biomarker discovery. Chip-based porous graphitized carbon nano-LC/MS was applied to interrogate native serum N-glycans. Nano-LC run conditions were optimized, resulting in highly reproducible glycan retention times and abundances across all samples and sample replicates. Over 300 N-linked glycan compound peaks with over 100 distinct N-linked glycan compositions were identified. General glycan elution trends revealed complex interactions with porous graphitized carbon based not only on saccharide composition and associated functional groups but also the three-dimensional geometry of the glycan branches and linkages.
The developed method was applied to serum samples from prostate cancer patients with good (n = 4) and bad (n = 4) prognoses. Chromatographic profiling was performed with respect to N-glycan composition as well as individual isomer-specific structure. Overall compositional profiling was applied first as a data-reduction method. Results compared favorably with previously reported compositional profiling methods,14-20 with high sensitivity and a wide dynamic range. Relative abundances of fucosrylated, sialylated, and high-mannose type glycans were determined. Isomer-specific analysis revealed that PGC nano-LC was able to separate up to six different isomers for each N-glycan composition. Highly reproducible abundances allowed compositional as well as isomer-specific statistical analysis. A number of significant differences were found between N-glycan abundances in patients with poor versus good prognoses.
Glycan chromatographic profiling was shown to be a powerful new technique for characterization of the serum N-glycome. In addition to highly sensitive compositional profiling, isomer-specific glycan quantitation will add a new dimension to existing glycomic analyses. Work is ongoing to further increase the speed, sensitivity, and dynamic range of this method. The combination of chromatographic profiling with structure-specific retention time and tandem MS libraries currently being developed by our lab will allow rapid and accurate identification of N-glycan structure. From a biomarker discovery perspective, application of high-throughput glycan chromatographic profiling to larger sample sets will greatly improve the statistical power of these comparisons and enable the identification of many more potential biomarkers.
Supplementary Material
ACKNOWLEDGMENT
We would like to thank Rudolph Grimm (Agilent Technologies Inc) for instrumentation and technical support. We also thank Lauren Dimapasoc and Cynthia Williams for their assistance. Financial support was provided by the National Institutes of Health (RO1GM049077 for C.B.L.).
ABBREVIATIONS
- MALDI
matrix-assisted laser desorption/ionization
- ESI
electrospray ionization
- MS
mass spectrometry
- HPLC
High performance liquid chromatography
- TOF
time-of-flight
- PGC
porous graphitized carbon
- SPE
solid phase extraction
- TIC
total ion chromatogram
- XIC
extracted ion chromatogram
- ECC
extracted compound chromatogram
- Hex
hexose (e.g. mannose, glucose, or galactose)
- HexNAc
N-acetylhexosamine
- Fuc
fucose
- NeuAc
N-acetylneuraminic acid
Footnotes
Hyun Joo An and Carlito B. Lebrilla contributed equally to this paper as co-corresponding authors.
REFERENCES
- 1.Dube DH, Bertozzi CR. Nat Rev Drug Discov. 2005;4:477–488. doi: 10.1038/nrd1751. [DOI] [PubMed] [Google Scholar]
- 2.Fuster MM, Esko JD. Nat Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 3.Hauptmann P, Riel C, Kunz-Schughart LA, Fröhlich K-U, Madeo F, Lehle L. Molecular Microbiology. 2006;59:765–778. doi: 10.1111/j.1365-2958.2005.04981.x. [DOI] [PubMed] [Google Scholar]
- 4.Rapoport E, Pendu J. Glycobiology. 1999;9:1337–1345. doi: 10.1093/glycob/9.12.1337. [DOI] [PubMed] [Google Scholar]
- 5.Saito T, Miyoshi E, Sasai K, Nakano N, Eguchi H, Honke K, Taniguchi N. Journal of Biological Chemistry. 2002;277:17002–17008. doi: 10.1074/jbc.M200521200. [DOI] [PubMed] [Google Scholar]
- 6.Pili R, Chang J, Partis RA, Mueller RA, Chrest FJ, Passaniti A. Cancer Res. 1995;55:2920–2926. [PubMed] [Google Scholar]
- 7.Duchesne L, Tissot B, Rudd TR, Dell A, Fernig DG. Journal of Biological Chemistry. 2006;281:27178–27189. doi: 10.1074/jbc.M601248200. [DOI] [PubMed] [Google Scholar]
- 8.Wang X-Q, Sun P, O’Gorman M, Tai T, Paller AS. Glycobiology. 2001;11:515–522. doi: 10.1093/glycob/11.7.515. [DOI] [PubMed] [Google Scholar]
- 9.Triantis V, Saeland E, Bijl N, Oude-Elferink RP, Jansen PLM. Hepatology. 2010;9999 doi: 10.1002/hep.23708. NA. [DOI] [PubMed] [Google Scholar]
- 10.Guo H-B, Lee I, Kamar M, Akiyama SK, Pierce M. Cancer Res. 2002;62:6837–6845. [PubMed] [Google Scholar]
- 11.Nita-Lazar M, Noonan V, Rebustini I, Walker J, Menko AS, Kukuruzinska MA. Cancer Res. 2009;69:5673–5680. doi: 10.1158/0008-5472.CAN-08-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brockhausen I. Biochimica et Biophysica Acta (BBA) - General Subjects. 1999;1473:67–95. doi: 10.1016/s0304-4165(99)00170-1. [DOI] [PubMed] [Google Scholar]
- 13.Arnold JN, Wormald MR, Sim RB, Rudd PM, Dwek RA. Annual Review of Immunology. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 14.An HJ, Miyamoto S, Lancaster KS, Kirmiz C, Li B, Lam KS, Leiserowitz GS, Lebrilla CB. Journal of Proteome Research. 2006;5:1626–1635. doi: 10.1021/pr060010k. [DOI] [PubMed] [Google Scholar]
- 15.de Leoz MLA, An HJ, Kronewitter S, Kim J, Beecroft S, Vinall R, Miyamoto S, de Vere White R, Lam KS, Lebrilla C. Disease Markers. 2008;25:243–258. doi: 10.1155/2008/515318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leiserowitz GS, Lebrilla C, Miyamoto S, An HJ, Duong H, Kirmiz C, Li B, Liu H, Lam KS. International Journal of Gynecological Cancer. 2008;18:470–475. doi: 10.1111/j.1525-1438.2007.01028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirmiz C, Li B, An HJ, Clowers BH, Chew HK, Lam KS, Ferrige A, Alecio R, Borowsky AD, Sulaimon S, Lebrilla CB, Miyamoto S. Molecular & Cellular Proteomics. 2007;6:43–55. doi: 10.1074/mcp.M600171-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Bereman MS, Williams TI, Muddiman DC. Analytical Chemistry. 2009;81:1130–1136. doi: 10.1021/ac802262w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bereman MS, Young DD, Deiters A, Muddiman DC. Journal of Proteome Research. 2009;8:3764–3770. doi: 10.1021/pr9002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang ZQ, Varghese RS, Bekesova S, Loffredo CA, Hamid MA, Kyselova Z, Mechref Y, Novotny MV, Goldman R, Ressom HW. Journal of Proteome Research. 2010;9:104–112. doi: 10.1021/pr900397n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, Simeone DM, Heidt D, Anderson MA, Lubman DM. Journal of Proteome Research. 2006;5:1792–1802. doi: 10.1021/pr060034r. [DOI] [PubMed] [Google Scholar]
- 22.Reggi M, Capon C, Gharib B, Wieruszeski J-M, Michel R, Fournet B. European Journal of Biochemistry. 1995;230:503–510. doi: 10.1111/j.1432-1033.1995.tb20589.x. [DOI] [PubMed] [Google Scholar]
- 23.Fernandez-Rodriguez J, Dwir O, Alon R, Hansson GC. Glycoconjugate Journal. 2001;18:925–930. doi: 10.1023/a:1022208727512. [DOI] [PubMed] [Google Scholar]
- 24.Schulz BL, Sloane AJ, Robinson LJ, Sebastian LT, Glanville AR, Song Y, Verkman AS, Harry JL, Packer NH, Karlsson NG. Biochem. J. 2005;387:911–919. doi: 10.1042/BJ20041641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isailovic D, Kurulugama RT, Plasencia MD, Stokes ST, Kyselova Z, Goldman R, Mechref Y, Novotny MV, Clemmer DE. Journal of Proteome Research. 2008;7:1109–1117. doi: 10.1021/pr700702r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakagawa H, Hato M, Takegawa Y, Deguchi K, Ito H, Takahata M, Iwasaki N, Minami A, Nishimura S-I. Journal of Chromatography B. 2007;853:133–137. doi: 10.1016/j.jchromb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Bones J, Mittermayr S, O’Donoghue N, Guttman A. s., Rudd PM. Analytical Chemistry. 2010;82:10208–10215. doi: 10.1021/ac102860w. [DOI] [PubMed] [Google Scholar]
- 28.Prien JM, Huysentruyt LC, Ashline DJ, Lapadula AJ, Seyfried TN, Reinhold VN. Glycobiology. 2008;18:353–366. doi: 10.1093/glycob/cwn010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chu CS, Niñonuevo MR, Clowers BH, Perkins PD, An HJ, Yin H, Killeen K, Miyamoto S, Grimm R, Lebrilla CB. Proteomics. 2009;9:1939–1951. doi: 10.1002/pmic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ninonuevo M, An H, Yin H, Killeen K, Grimm R, Ward R, German B, Lebrilla C. Electrophoresis. 2005;26:3641–3649. doi: 10.1002/elps.200500246. [DOI] [PubMed] [Google Scholar]
- 31.Fortier M-H, Bonneil E, Goodley P, Thibault P. Analytical Chemistry. 2005;77:1631–1640. doi: 10.1021/ac048506d. [DOI] [PubMed] [Google Scholar]
- 32.Yin H, Killeen K, Brennen R, Sobek D, Werlich M, van de Goor T. Analytical Chemistry. 2004;77:527–533. doi: 10.1021/ac049068d. [DOI] [PubMed] [Google Scholar]
- 33.Niñonuevo M, An H, Yin H, Killeen K, Grimm R, Ward R, German B, Lebrilla C. Electrophoresis. 2005;26:3641–3649. doi: 10.1002/elps.200500246. [DOI] [PubMed] [Google Scholar]
- 34.Yin H, Killeen K. Journal of Separation Science. 2007;30:1427–1434. doi: 10.1002/jssc.200600454. [DOI] [PubMed] [Google Scholar]
- 35.Morelle W, Flahaut C, Michalski J-C, Louvet A, Mathurin P, Klein A. Glycobiology. 2006;16:281–293. doi: 10.1093/glycob/cwj067. [DOI] [PubMed] [Google Scholar]
- 36.Kronewitter SR, An HJ, Leoz M. L. d., Lebrilla CB, Miyamoto S, Leiserowitz GS. Proteomics. 2009;9:2986–2994. doi: 10.1002/pmic.200800760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koizumi K, Okada Y, Fukuda M. Carbohydrate Research. 1991;215:67–80. doi: 10.1016/0008-6215(91)84013-5. [DOI] [PubMed] [Google Scholar]
- 38.Davies M, Smith KD, Harbin A-M, Hounsell EF. Journal of Chromatography A. 1992;609:125–131. doi: 10.1016/0021-9673(92)80155-n. [DOI] [PubMed] [Google Scholar]
- 39.Mechref Y, Novotny MV. Journal of Chromatography B. 2006;841:65–78. doi: 10.1016/j.jchromb.2006.04.049. [DOI] [PubMed] [Google Scholar]
- 40.Packer NH, Lawson MA, Jardine DR, Redmond JW. Glycoconjugate Journal. 1998;15:737–747. doi: 10.1023/a:1006983125913. [DOI] [PubMed] [Google Scholar]
- 41.Davies MJ, Smith KD, Carruthers RA, Chai W, Lawson AM, Hounsell EF. Journal of Chromatography A. 1993;646:317–326. doi: 10.1016/0021-9673(93)83344-r. [DOI] [PubMed] [Google Scholar]
- 42.Ruhaak L, Deelder A, Wuhrer M. Analytical and Bioanalytical Chemistry. 2009;394:163–174. doi: 10.1007/s00216-009-2664-5. [DOI] [PubMed] [Google Scholar]
- 43.Cooper CA, Harrison MJ, Wilkins MR, Packer NH. Nucl. Acids Res. 2001;29:332–335. doi: 10.1093/nar/29.1.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Markowski CA, Markowski EP. The American Statistician. 1990;44:322–326. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.