Abstract
Mice have become a preferred model system for bone research because of their genetic and pathophysiological similarities to humans: a relatively short reproductive period, leading to relatively low cost of maintenance and the availability of the entire mouse genome sequence information. The success in producing the first transgenic mouse line that expressed rabbit β-globin protein in mouse erythrocytes three decades ago marked the beginning of the use of genetically engineered mice as model system to study human diseases. Soon afterward the development of cultured pluripotent embryonic stem cells provided the possibility of gene replacement or gene deletion in mice. These technologies have been critical to identify new genes involved in bone development, growth, remodeling, repair, and diseases, but like many other approaches, they have limitations. This review will introduce the approaches that allow the generation of transgenic mice and global or conditional (tissue-specific and inducible) mutant mice. A list of the various promoters used to achieve bone-specific gene deletion or overexpression is included. The limitations of these approaches are discussed, and general guidelines related to the analysis of genetic mouse models are provided.
Keywords: Transgenic, Knock-out, Knock-in, Conditional knock-out, Gene deletion, Mouse model, Cre-lox system, Cre recombinase, Bone promoters
Transgenic approaches
Transgenic construct
Transgenesis consists in the introduction in the genome of a transgene to study the biological function of a gene of interest by overexpressing it, globally or in a tissue/cell-restricted manner. Since the insertion site of the transgene is random, two additional components, a promoter that drives transgene expression and a poly A tail that allows protein translation, must be included to ensure efficient expression of the transgene. The transgene is usually a wildtype (WT) coding DNA sequence (cDNA) or genomic DNA sequence containing exons and introns. It can also be a mutated coding DNA sequence (cDNA) that will act as a dominant negative product or that will mimic genetic mutations observed in human diseases, or an anti-sense RNA. Suicide genes, such as the diphtheria receptor (DTR), can also be expressed to eliminate the cells expressing it [1,2]. Over-expression can be targeted to specific cell type, for instance, osteoblasts, osteoclasts, or osteocytes and is achieved by placing a transgene downstream of a cell-specific promoter. Alternatively, global transgene overexpression can be achieved by using a house keeping gene promoter or a viral promoter. If desired, the minimal transgenic construct can be modified 1) by using inducible promoters instead of constitutively active ones to provide temporal regulation of gene expression, 2) by adding enhancers and introns to increase the level of transgene expression, or 3) by linking an epitope tag sequence to the transgene to distinguish the transgene from endogenous expression.
Production of transgenic mice
The final transgenic construct is usually microinjected into the pronuclei of fertilized mouse eggs, which are then implanted in the uterus of a surrogate mother. The transgene will be expressed by some of the offspring, named transgenic founders, which are bred to WT mice to select transgenic founders in which the transgene is transmitted through the germ line. The transgenic founders are then mated with WT mice to establish mouse lines with stable DNA integration and expression of the transgene.
Advantages and limitations
The main advantage of this technique lies in its short development time. Its main drawbacks are random integration sites and variable copy number of the transgene that lead to variable levels of expression, depending on the overall transcriptional activity of the genomic region where the transgene is integrated. The analysis of two or more independent transgenic lines, each deriving from a different founder, allows one to select the desired level of expression and to verify that the observed phenotypes are caused by the effect of the transgene rather than disruption of another gene due to random integration of the transgene. Caution must indeed be taken when breeding transgenic lines to a homozygous state since the random site of integration of the transgene may alter the gene it falls into. Homozygous transgenic lines may cause a phenotype independent of the targeted gene or lethality if the site of insertion is in a gene critical for survival. In addition, the sometime high copy number of the transgene, when brought to homozygosity, increases the chances of DNA recombination and chromosomal rearrangements, which can also generate a phenotype independent of the targeted or overexpressed gene. Importantly, promoters used to direct expression of a transgene are rarely strictly specific to any cell type, and thus careful analysis of transgene expression in multiple tissues, and at the cellular level, is essential for the validation of a founder line. Lastly, the supraphysiological levels of gene expression achievable in this approach, as well as its non-regulated nature, may not be biologically relevant, and thus interpretation of results must be done carefully.
“Targeted” transgenic mice
To overcome such drawbacks, a “knock-in” strategy based on targeted insertion of a transgene to an exact genomic locus using homologous recombination in embryonic stem (ES) cells can be used. This approach allows one to target a transgene to a transcriptionally active DNA region and to control copy number to obtain stable expression with no disruption of unknown genes whose alteration may contribute to the phenotype. The experimental design of a gene knock-in project uses the same basic principles as for a knock-out (see below), and is thus more tedious and time consuming compared to the generation of a “basic” transgenic line [3]. A new approach based on the use of engineered zinc finger nucleases, commercialized by Sigma, facilitates this approach. This technology uses an artificial zinc finger nuclease (ZFN) that consists of a designed zinc finger protein (ZFP) fused to the cleavage domain of the Fok I restriction enzyme. This ZFN can be redesigned to cleave new targets by developing ZFPs with new sequence specificities, thus allowing genome editing [4].
Global gene deletion
Global gene targeting consists of the introduction of specific mutations into a gene of interest by homologous recombination using ES cell technology. This approach is feasible thanks to the availability of the entire mouse genome sequence and to the ability of ES cells to be cultured and manipulated in vitro without losing their totipotency. It results in the generation of a null allele i.e., knockout (KO), allowing gene inactivation after breeding of the animals as homozygotes. The gene modification generated is constitutive and present as soon as the endogenous promoter regulating the targeted gene is turned on.
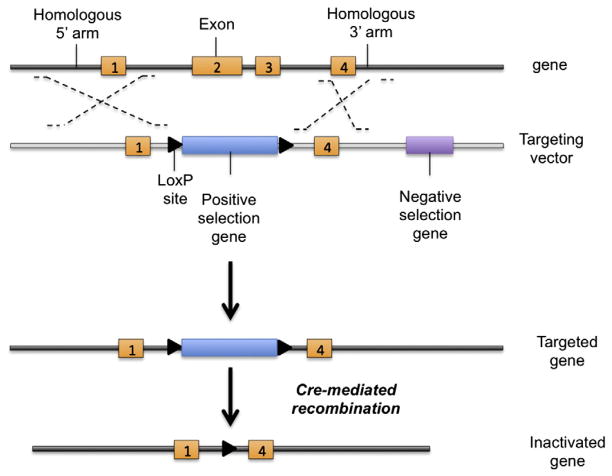
Target DNA construct
Targeting by homologous recombination requires the generation of a targeting DNA construct that requires a more complicated and time-consuming multistep strategy than generating a transgenic construct. Commercially available Bacterial Artificial Chromosome (BAC) clones are usually used to subclone the flanking upstream and downstream genomic DNA sequences (arms) of the gene of interest, which will be used to induce homologous recombination. Identical mouse backgrounds, such as 129/SV, are usually used between the BAC clone utilized for generating the construct and ES cells to avoid a decrease in recombination efficiency caused by DNA polymorphisms. Successive cloning steps are used to progressively clone the different functional entities of the construct into a single construct. The neo gene, which confers resistance to neomycin (G-418), is commonly used to disrupt the endogenous targeted gene and to select for ES cell clones that have integrated the targeting construct (Fig. 1). In addition, because random integration via nonhomologous recombination occurs more frequently than targeted integration via homologous recombination, the TkHSV gene is also usually placed outside of the homology region of the targeting construct. This gene, if inserted via nonhomologous recombination into the genome of ES cells, encodes a viral thymidine kinase that converts the nucleotide analogue ganciclovir into a cytotoxic product, thus allowing for negative selection of clones characterized by random insertions (cells with random insertion only are sensitive to ganciclovir).
Fig. 1.
Schematic presentation of a conventional targeting construct. Several kilobases of genomic DNA on either side of the target gene are cloned around a drug-selection marker. LoxP sequences flank the positive drug-selection gene in the targeting construct. After the cloned DNA (targeting vector) is introduced into ES cells, positive and negative drug selection is performed in culture. Cre recombinase can delete the DNA sequence between the IoxP sites, thereby deleting part of a targeted gene in ES cells.
Restriction digestions and ligations can be used for subcloning of the targeting vector, but this strategy is complicated by the length of the genomic DNA to be manipulated, and is limited by the presence and position of adapted restriction sites within the sequence to be handled. An alternative to this strategy is the recombineering technique, which exploits homologous recombination in Escherichia coli. In this technique, DNA is electroporated in a strain of bacteria containing an integrated defective and temperature inducible prophage carrying recombination genes that will integrate this exogenous DNA. Regardless of the cloning strategy used, the selection of a DNA probe that allows for the targeted alleles to be distinguished from the WT allele is of critical importance and should be considered very early during the cloning strategy.
Production of knockout mice
Subsequently, the engineered targeting construct is introduced into ES cells by electroporation and the normal gene on the chromosome is replaced with the targeting construct by homologous recombination. The ES cells that have incorporated the construct into their DNA can be selected in vitro by their resistance to antibiotics (G-418 for instance). ES cells containing nonhomologous recombined DNA (and thus the TK cassette) can be selected by ganciclovir. Selected totipotent selected ES cells are then injected in the blastocysts of foster mothers from a different mouse strain, such as C57BL6. The pups born from these mothers are referred to as chimeras as they are composed of cells derived from two different strains. Chimerism is visible by the presence of coat patches of different colors since cells derived from 129/SV and C57BL/6 mice give rise to an agouti and black coat colors, respectively. If a chimeric embryo possesses mutant ES-derived germ cells, the genetic alteration can be propagated to its offspring to generate heterozygote and eventually homozygote mutant mice.
Advantages and drawbacks
Global gene targeting in the bone field greatly extended the knowledge derived from the analysis of naturally occurring mutations and has allowed for the reproduction of several human bone diseases such as osteogenesis imperfecta, various types of achondroplasiae, and others. However, this strategy also has limitations, the most important being the possible lethality of KO mice that reflects the non-redundant role of the targeted gene but precludes further analyses of its function in later stages of development. Global gene targeting also does not allow one to track gene function at specific developmental stages or cell differentiation stages. Lastly, some phenotypes caused by such genetic alterations may be difficult to interpret because they may be due to cell-autonomous, cell non-autonomous, or systemic effects.
Conditional gene targeting
The molecular “switch” enabled by conditional gene targeting allows the creation of mutations in a tissue-specific and time-specific manner and thus addresses some of the limitations of the global gene deletion strategy.
DNA construct
This strategy consists of introducing in the gene to be deleted two or more short DNA tag sequences that can be recognized by recombinases. The Cre/Lox recombinase system derived from bacteriophage P1 is the most widely used and will be discussed in detail below, but the Flp/FRT system derived from Saccharomyces cerevisiae can also be used. Both recombinases recognize a distinct 34-bp consensus sequence (Lox and FRT sites, respectively) and can excise the sequence between two of these sites and ligate the two generated extremities, thus “popping-out” the intervening sequence (Fig. 1) [5]. The orientation of Lox sites in a single gene and their respective positions on different chromosomes also allows one to invert sequences of DNA and to generate chromosomal rearrangements that can be useful to recapitulate, in mice, human genetic diseases. In a targeting construct, the two Lox sites are usually inserted in introns flanking exonic DNA sequence(s) crucial for gene function. The structure of this construct allows for the “floxed” gene’s WT activity to be retained, whereas its cre-recombination leads to gene inactivation. It is particularly important not to interrupt endogenous gene function by the insertion of the Lox sites, and splice sites within introns as well should be avoided if possible when cloning the Lox sequences.
Production of the conditional knockout mice
This approach requires two mouse lines to be crossed, a transgenic mouse line that overexpresses the Cre recombinase in a specific tissue or cell type, and a mouse strain that contains the target gene of interest flanked by two Lox sites. Recombination and consequently inactivation of the target gene occurs only in those cells expressing the Cre recombinase. Hence, the target gene remains active in all cells and tissues that do not express the Cre recombinase. Alternatively to the use of a Cre transgenic mouse line, the Cre recombinase can be delivered to the mouse strain containing the floxed gene by viral strategies in vitro or in vivo to transmit the recombinase to “floxed” cells.
Alternative uses of conditional knockout mice
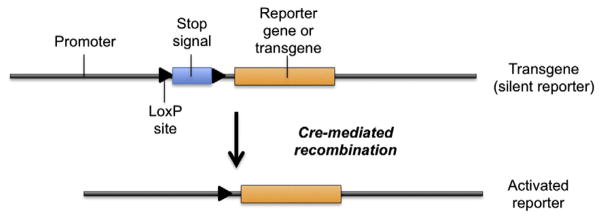
In addition to conditional gene inactivation, this approach is commonly used to determine the site of Cre recombinase activity in vivo [6]. This is achieved by crossing a Cre transgenic mouse line with the Cre recombinase targeted to a specific tissue or cell type, and a second mouse strain that has a reporter gene such as LacZ, placental alkaline phosphatase (pALP) or a variety of fluorescent proteins (GFP, YFP…), whose expression is blocked by a floxed stop cassette sequence (which can be polyadenylation sequences, false translation initiation codons and/or stop codons) incorporated between the 5′ regulatory elements and reporter gene. In this setting the reporter transgene is not expressed until the Cre recombinase is expressed, since the excision of the stop signal occurs only in those cells expressing the Cre recombinase, depending on the tissue specificity of the promoter used (Fig. 2). These “reporter” mouse lines are also very useful for lineage tracing studies because activation of such reporter indicates that Cre recombinase activity is present in that particular cell or was present in its progenitor [7].
Fig. 2.
The Cre/LoxP system can be used for cell tracking purpose or to conditionally active expression of a transgene. The transgenic construct contains a stop cassette/signal flanked by two LoxP sites between a promoter and a reporter gene such as LacZ or GFP, or a transgene such as a suicide gene. The cross between Reporter and Cre-expressing mice leads to excision of the STOP cassette and transcription of the reporter gene or transgene.
Limitations and precautions
Although chromogenic or fluorescent “reporter” mice are widely used and convenient (see below), one must keep in mind that this method has limitations. Endogenous LacZ or AP activity along with non-specific background may prevent the detection of weak Cre activity that may still have significant consequences in the progeny of these Cre-positive cells if they divide, since recombination of the target floxed gene is non-reversible and permanent. Transient Cre-recombinase activity is also likely to be missed in studies focusing on a limited number of developmental stages. Therefore, complementary approaches must be utilized. These include genomic PCR from tissues or extracted cells to assess with more sensitivity recombination of the floxed alleles per se, or ex vivo measurement of the proliferation, differentiation, function or gene expression pattern of targeted cells versus non-targeted cells.
Since conditional gene deletion relies on transgenic expression of the Cre recombinase, the potency of each transgenic promoter to drive expression of the Cre recombinase is critical to achieve efficient gene recombination, and depends on several parameters mentioned in section 1 above. Transgene copy number is usually, but not always, proportional to the strength of expression. This variability can be caused by a chromosomal position effect and can relate to the site of insertion of the transgene(s), which can be in a transcriptionally more or less active chromosomal region. Comparing several founders is always useful to select for the most appropriate line with optimal level of expression.
Another important parameter to measure in conditional knockout animals is the efficiency of recombination. As mentioned above, Cre transgenic lines have differing levels of expression of the Cre recombinase, and each floxed gene has differing chromosomal accessibility to be clipped by the Cre recombinase, leading to variable recombination efficiency. Recombination efficiency can be assessed and semi-quantified by genomic PCR or Southern blot using tissues or cells extracted, and possibly purified, from the mutant mice. Enrichment of the Cre-positive cells to quantify recombination efficiency limits contamination by cre-negative cells and the artefactual decrease in recombination efficiency it generates.
Inducible gene targeting
An inducible system allows one to turn gene expression off or on by using an exogenous inducer administered to the mice. The Cre-ERT system [8,9] and the tet-ON/tet-OFF systems [10,11] have been most widely used to provide a temporal control of gene recombination.
The Cre-ERT system
In this system the Cre recombinase sequence is fused with a mutated ligand-binding domain from either the progesterone or the estrogen receptor (ER). These modified ligand-binding domains become localized to the nucleus only in the presence of synthetic steroids (RU486 and tamoxifen or 4-hydroxytamoxifen) but not in the presence of endogenous steroids.
The CreERT recombinase system has been demonstrated to be functional in vivo, allowing widespread time-dependent recombination when CreERT expression is placed under the control of a strong promoter. In this system, tamoxifen can be administered systemically but also topically in studies involving skin or fracture for instance, and can be given to pregnant mothers as well to study gene inactivation in developing mouse embryos. Toxic effects of tamoxifen may be a limitation for some studies, especially studies in embryos. To alleviate this problem, a second generation of CreERT mice (Cre-ERT2) that is about 10 times more sensitive to 4OH-Tamoxifen in vivo than the original CreERT [12] has been generated and used in several fields, including the bone field [13,14]. A drawback of this system is that it may be somewhat leaky, resulting in weak constitutive activation. In addition, recombination mediated by inducible promoters is generally mosaic and its efficiency has been dependent on tissue types, despite nearly toxic doses of inducer.
The progesterone-binding domain of the human progesterone receptor has been mutated to one that binds the synthetic steroid RU486 (a glucocorticoid receptor antagonist) but not endogenous progesterone or glucocorticoids. This inducible Cre system is very similar to the CreERT system but has not, to our knowledge, been very much used in the bone field.
tet-ON/tet-OFF systems
The tet systems rely on two components, i.e., a tetracycline-controlled transactivator (tTA or rtTA) and a tTA/rtTA-dependent promoter that controls expression of a downstream cDNA in a tetracycline-dependent manner. In the tet-OFF system, the presence of tetracycline or its derivatives, such as doxycycline (Dox), prevents tTA to bind its target and the system is inactive. The tet-ON system utilizes a mutated tTA, i.e., reverse tTA (rtTA). In this case the presence of doxycycline allows the system to be turned on. In vivo, the system requires the breeding of two independent types of transgenic mice, i.e., transactivator mice, in which tTA or rtTA is expressed under the control of a tissue-specific promoter, and responder mice, in which expression of the gene of interest is under the control of the tetO minimal promoter. Addition or no addition of doxycycline to the food or drinking water of the double-transgenic mice allows in vivo spatiotemporal control of cDNA expression.
The tet-ON system requires higher doxycycline concentrations to be active compared with the concentration tet-OFF requires to be inactive [15]. The advantages of tet-ON compared with tet-OFF are that the transgene is not expressed until doxycycline is given to the animals and that upregulation in vivo is faster than downregulation. Using the tet systems, one can achieve a tight control of gene expression in a tissue-specific manner. It is also possible to return to a control situation by simply discontinuing doxycycline administration. The major disadvantage of the tet systems is that control of the expression of the acceptor construct is often leaky because of strong positional effects on the tetO minimal promoter.
In all of these systems, the inducible transgene can be a WT protein to be overexpressed (including the Cre-recombinase), a mutated, truncated or reporter protein, a suicide protein, or silencing RNAs.
Tissue-selective promoters used in bone cells for conditional recombination/overexpression
Skeletal morphogenesis involves sequential steps that include patterning, condensation, and organogenesis of the membranous and endochondral embryonic skeletons [16]. Three lineages give rise to the skeleton structures during early development. Neural crest cells will form the branchial arch derivatives of the craniofacial skeleton; the paraxial mesoderm will contribute to the craniofacial skeleton and form most of the axial skeleton following division of somites, and cells of the lateral plate mesoderm will give rise to limb skeleton. During mouse embryogenesis, between E10 and E12 days post-coitum (dpc), a group of homogenous cells condenses, acquires positional properties, and starts differentiate to form the mold of the future skeleton. Condensed mesenchymal cells can differentiate directly into osteoblasts and form flat bones such as skull bones as well as the subperiosteal region of long bones (intramembranous ossification). Condensed mesenchymal cells can also differentiate into chondrocytes to form a cartilage anlagen that is eventually replaced by bone during the process of endochondral ossification [17]. The characterization of the molecular machinery controlling these intricate processes relied heavily on the use of mutant mice, and its future refinement will depend on the optimal use of new genetic tools to target genes in specific cells and developmental time points.
Promoters driving Cre expression in specific skeletal cell types used in the bone field are described below. They are classified by the targeted cell lineage, including mesenchymal stem cell/osteochondroprogenitors, chondrocytes, osteoblasts, osteocytes and osteoclasts. Table 1 summarizes the promoter-cre lines, the reported site(s) of leakage of each promoter (if addressed), the original study describing each transgenic line, and some references describing the use of each line, when available. The Mouse Genome Informatics website (http://www.informatics.jax.org/) is a valuable resource to obtain an updated list of cre-lines for each tissues, including the skeletal system, and references of studies that have used inventoried cre lines.
Table 1.
Main Cre transgenic lines used for the analysis of bone-related genes. This list is not exhaustive.
| Cell type | Promoter | Reported leakage | Examples of gene targeted | Original description |
|---|---|---|---|---|
| Osteochondro progenitors | Prx-Cre | Tendons, interlimb flank [18] | Nf1 [19] | [18] |
| Dermo1-Cre (mouse) | Fgfr2 [20] | [20] | ||
| Z/EG, Ai9 [21] | ||||
| Catnby [22] | ||||
| Sox9-Cre (mouse) | Notochord, node, endodermal linings at E8, skin at E10.5, gonads, intestin, spinal cord, pancreas, tendons and synoviums at E17.0 | Tace, Osx [7] | [23] | |
| Chondrocytes | Col2A1-Cre (human) | Developing eye, epidermis, heart myocardium, endoderm of the developing yolk sac and cranial mesenchymal cells [24] | V-Egf [24] | [24] |
| Fosl2 [25] | ||||
| Col2a1-Cre (mouse) | Liver [26] | Trsp [26] | [30] | |
| Pten [27, 28] | ||||
| Nf1 [29] | ||||
| Col2a1CreERT (mouse) | [31] | |||
| Col2a1CreERT2 (mouse) | Osteochondroprogenitors | [32] | ||
| Col2a1CreERT (rat) | Osteochondroprogenitors | Smo [33] | [33] | |
| Col2a1 DOX-Cre (mouse) | Ext1 [34] | [35] | ||
| Col10a1-Cre (BAC mouse) | Subchondral and endochondral bone marrow cells | [36] | ||
| Col10a1-Cre (1kb mouse) | Skin, perichondrium, some trabecular osteoblasts | [37] | ||
| Acg-CreERT2 (mouse) | [38] | |||
| Osteoblasts | Runx2-CreERT (mouse) | Cartilage | Vegf164 [39] | [40] |
| Tet-OSX-Cre (mouse) | P53/Rb [41] | |||
| A-Raf/B-Raf [42] | [44] | |||
| Cxcr4 [43] | ||||
| OSX-CreERT2 (mouse) | Vegf164 [39] | [45] | ||
| 2.3kb Col1A1 -Cre (mouse) | Weak activity at E14.5 in skin and digits [46] | Adrβ2 [47] | [46] | |
| Hdac8 [48] | ||||
| Lrp5 [49] | ||||
| Nf1 [50] | ||||
| 2.3kb Col1A1-Cre ERT2 (mouse) | Osx [51] | [13] | ||
| 2.3kb Col1A1 (rat) | Sporadic activity in chondrocytes before E18 [52] | [52] | ||
| 3.2kb Col1-CreERT2 (mouse) | [45] | |||
| 3.6kb Col1A1-Cre (rat) | Tendons | Rb1[53] | [52] | |
| Sporadic activity in chondrocytes before E18 | ||||
| Strong activity in articular chondrocytes [52] | ||||
| OCN-Cre (human) | Igf [54] | [54] | ||
| Notch1 [55] | ||||
| Cnb1 [56] | ||||
| Smad4 [57] | ||||
| Pten [58] | ||||
| Osteocytes | 15kb Dmp1-Cre (mouse) | Pkd1 [59] | [60] | |
| 10kb Dmp1-creERT (mouse) | Small % of osteoblasts | Ppr [61] | [61] | |
| Osteoclasts | Trap-Cre (mouse) | Chondrocytes, liver, spleen and stomach, heart, liver and lung [62] | Ilk [63] | [62] |
| Lysozyme-Cre (mouse) | Gr [64] | [67] | ||
| ER-a [65] | ||||
| JunB [66] | ||||
| Cat K-Cre (mouse) | Stomach, salivary gland (line 1) and liver (line 8) [62] | Bcl-x [68] | [62, 70] | |
| Dicer [69] | ||||
| CD11b-Cre (human) | Peritoneal mcrophages, bone marrow granulocytes | S1P1 [72] | [71] | |
| Low level in spleen, testis, liver, kidney, brain, heart, and lung [71] | Ikkβ [73] |
Mesenchymal stem cell promoters
Floxed genes in mesenchymal or osteochondroprogenitor cells can be targeted by using the Prx1, Dermo1 or Sox-9 cre transgenic mice.
Prx1 promoter
Prx1 is a paired-related homeobox gene that plays an essential role in regulating skeletal development in the limb [74]. Using transgenic mice in which a 2.4 kb Prx1 promoter directs Cre expression (Prx1-Cre transgenic mice), Cre recombinase activity was observed in mesenchymal cells during limb bud development, in the lateral plate mesoderm, and the cranial mesenchyme, but not in the axial skeleton [18,75]. Cre recombinase activity in this line was first observed in limb buds at 9.5 dpc. By E16.5, Cre recombinase activity was detected in the entire limb bud and interlimb flank, in the craniofacial mesenchyme, and around the eyes.
As a note of caution, partially penetrant germline recombination was evident in the offspring of female (but not male) Prx1-Cre heterozygous mice, however, the penetrance of this apparent germline recombination event level varied depending on the particular gene flanked by loxP sites [18]. Therefore, males from this transgenic line should be used for targeting mesenchymal cells.
It is possible to use this promoter in an inducible fashion in vivo and also to purify and track cells where this promoter is active by using the Prx1-Cre-ERT-GFP transgenic mice [76]. While the expression of CreERT allows timed recombination of the floxed allele, the expression of GFP allows isolation of transgene-expressing cells from the periosteum by cell sorting as well as cell tracking. In this line, Tamoxifen injection into pregnant dams at E9 induced Cre recombinase activity and recombination at E17 in chondrocytes, osteoblasts, periosteal, and perichondrial cells, indicating that the transgene was efficiently expressed in limb bud osteochondroprogenitors. When tamoxifen was injected at E15.5 and E16.5, Cre recombinase activity persisted in perichondrial and periosteal cells, but was not detected in chondrocytes anymore. This strategy thus allows one to achieve a different level of specificity. In adult mice, daily tamoxifen injection between P19 and P23 induced Cre recombinase activity in the periosteum and perichondrium, but not in epiphyseal chondrocytes at P26 and up to P35. Recombined cells were predominantly found in the inner cambium layer of the periosteum in regions where the periosteum showed morphologically distinct inner and outer layers. Some of the articular chondrocytes and tendon cells also showed recombination.
Dermo1 promoter
Dermo1 (also named Twist2) is a transcription factor highly expressed in condensed mesenchyme during skeletal development and later in perichondrial cells. Because the regulatory elements of the Dermo1 promoter were not initially characterized, Dermo1-cre mice were generated by inserting the Cre-recombinase gene within the first exon of Dermo1[20]. Cre recombinase activity in this line was detected as early as E9.5 in mesodermal tissues such as branchial arches and somites, but also in the skin. During embryonic endochondral ossification, Cre recombinase activity was first detected at E11.5 in the condensed mesenchyme from which both chondrocytes and osteoblasts are derived. Later in development (E16.5), Cre recombinase activity was detected in chondrocytes in growth plate cartilage and in osteoblasts and osteocytes in the perichondrium, periosteum, and endosteum;, but not bone marrow cells and osteoclasts. During intramembranous ossification in developing sutures, Cre recombinase activity was present at the osteogenic fronts and in surrounding mesenchymal tissues. Growth plate chondrocytes along with bone trabecular and cortical osteoblasts were recombined in adult mice.
Sox9 promoter
Sox9 is a transcription factor with a high-mobility group DNA-binding domain, expressed in all chondroprogenitors and having an essential role in chondrogenesis. The mouse Sox9-Cre transgenic mouse line was initially generated via a knock-in strategy to follow the fate of Sox9 expressing cells, [7] and has not been used yet as a tool to target genes in osteochondroprogenitors [24]. Sox9-cre recombinase activity was detected in limb bud mesenchyme in E10.5 embryos and in all cells in cartilage primordia and perichondrium at E13.5. In E17.0 limb buds all chondrocytes as well as perichondrial, periosteal, and osteoblast cells, displayed Cre recombinase activity, indicating that Sox9-expressing limb bud mesenchymal cells give rise to both chondrogenic and osteogenic cell lineages, like Prx1-expressing cells. Comparison with the Prx1-cre line indicated that Prx1-expressing limb bud mesenchymal cells give rise to Sox9-expressing osteochondroprogenitors and that the Prx-cre line can recombine floxed genes in a broader limb bud-derived mesenchymal cell population than the Sox9-cre line. Cre recombinase activity was also detected in notochord, node, and endodermal linings at E8 and skin at E10.5. In E17 embryos, gonads (Sertoli cells and Leydig cells but not germ-cells), intestinal epithelial cells, neurons and glia in the spinal cord, endocrine and exocrine pancreatic cells, tendons, and synoviums also showed Cre recombinase activity. Similarly to the Prx-1 and Dermo1 lines, the broad spectrum of Cre recombinase expression in this non-inducible transgenic line might limit the use of this model for developmental studies.
Precautions
In all three lines, the multipotent nature of the cells where these promoters are active makes the pattern of Cre recombinase activity broad and therefore the interpretation of results relatively difficult. More specifically, because chondrocytes, osteoblasts, and osteocytes derive from common mesenchymal osteochondroprogenitor cells, Cre recombinase expression, even transient, under the control of these three promoters in osteochondroprogenitor cells will be followed by gene recombination in osteoblasts, osteocytes and chondrocytes, and also possibly in adipocytes, muscles or endothelial cells. Gene recombination in any of these descendants might indirectly and distinctly contribute to the observed bone phenotypes.
Chondrocyte promoters
Gene targeting in chondrocytes has mostly been done by using the Col2a1 promoter, which encodes Type II collagen, a major constituent of the growth plate made by chondrocytes. Mouse, rat, and human Col2a1 promoter Cre lines have been generated and inducible lines are also available.
Col2α1 promoter
The mouse Col2α1 promoter contains a small enhancer region located within the first intron of the Col2α1 gene that in conjunction with 1 kb of the Col2α1 promoter region efficiently drives transgenic reporter expression and recapitulates the tissue-specific patterns of Col2α1 gene expression in vivo [77]. Based on this feature, the mouse Col2α1–Cre transgenic line has been generated and exhibited gene excision activity starting at E9 in the notochord, cranial mesenchyme, and in all cartilage primordia [26,27,30]. It is less well known in the bone field, however, that the Cre recombinase activity in the Col2α1–Cre line is also observed in osteoblastic cells within the primary spongiosa, as well as in trabecular osteoblasts and osteocytes in adult mice [27,29,33].
The human Col2α1 promoter/enhancer Cre transgenic line has a very similar pattern of Cre recombinase activity and recombines floxed genes in chondrocytes but also in the developing eye, epidermis, heart myocardium and atria, cranial mesenchymal cells, the endoderm of the developing yolk sac and the spinal cord [24].
Two mouse Tamoxifen (TM)-inducible Col2α1–Cre lines have been created, and show similar properties to a rat TM-inducible Col2α1–Cre line [31,32]. In the mouse Col2α1–CreERT line [31], weak activity could be detected in facial condensations, rostral sclerotomes, limb buds, and base of the skull as early as E10, 1 day after TM injection. Recombination time course experiments indicated that Cre recombinase activity could be first detected 8 h post-TM injection; mosaic recombination was evident 12 h post-injection and extensive recombination was detected 24 h post-injection. In this line, Cre recombinase activity was found in the perichondrium (E13.5), a major source of periosteum and osteoblast progenitors giving rise to the bone collar including both cortical and trabecular bone in the spongiosa [78]. TM injection after E13.5 avoided recombination in periosteum and bone. Cre recombinase activity could be induced post-natally until day 21, but declined thereafter to become essentially undetectable in 12-week-old mice.
In an attempt to increase sensitivity to TAM, another CreERT recombinase (CreERT2) was used in association with the 1 kb mouse Col2α1 promoter to generate the mouse Col2α1–CreERT2 transgenic line [32]. The CreERT2 recombinase contains a G400V/M543A/L544A triple mutation in the ER ligand binding domain (LDB) and is more sensitive to 4-OHT than is the mutant ER LBD with a single G521R substitution. In this line, upon TM injection at E15.5, Cre recombinase activity was detected 3 days later in cartilaginous areas, but not in bones formed by intramembranous formation. Upon TM injection at post-natal stages (2 and 8 week-old mice), recombination was demonstrated in cells from the resting, proliferating, and hypertrophic zones of the growth plate cartilage and in articular cartilage (1 and 6 months post-TM injection, [14]).
Cre recombinase activity in the inducible rat Col2α1–CreERT line is localized in columnar and articular chondrocytes, as well as perichondrial cells. The inducible nature of this CreERT here again enables one to control Cre recombinase activity in specific growth plate compartments by choosing a proper developmental stage at which TM switches on Cre recombinase activity. For instance, this strategy was used to induce Cre recombinase activity in chondrocytes, but not perichondrial cells by starting TM injections at or after E12.5 [33]. TM injections in this model can also be used post-natally to recombine genes efficiently in columnar and articular chondrocytes. The tetON system is also available to induce Cre recombinase activity in a temporal fashion selectively in chondrocytes via the mouse Col2α1 promoter [35]. In these Col2α1–DOX–Cre transgenic mice, Cre recombinase activity was observed in the cartilaginous elements of E14.5 embryos whose mother was given doxycycline since conception. P7 pups also displayed Cre recombinase activity within the entire femoral growth plate (from articular surfaces to proliferating chondrocytes) when fed milk from mothers given doxycycline. One-month-old mice also showed Cre recombinase activity throughout the growth plate when given doxycycline, however, 3 month-old mice showed Cre recombinase activity solely in articular chondrocytes upon doxycycline treatment.
Col10α1 promoter
Although not widely used yet, the mouse Col10α1-Cre transgenic line is another option to manipulate genes in the growth plate, based on the selective expression of Type X collagen in growth plate hypertrophic chondrocytes. Based on the observation that a 4.6 kb promoter sequence of the mouse Col10α1 gene including a 500 bp enhancer element was sufficient to drive specific expression of a LacZ reporter gene in all hypertrophic cartilage zones of transgenic mice [79], a transgenic line was created by inserting the Cre recombinase into the Col10α1 gene, using a BAC recombineering technique [36]. In this line, Cre recombinase activity was detected as early as E13.5 in long bones where hypertrophic cartilage develop and in long bone and vertebra growth plate hypertrophic chondrocytes of E16.5 and P1 mice. Cre recombinase activity was also detected in the subchondral bone marrow zone adjacent to the cartilage–bone border and in endochondral bone trabeculae. Whether this activity comes from residual LacZ activity, chondrocytes surviving in the cartilaginous core of endochondral bone trabeculae, or from leakage in osteoblasts is unclear.
Another less characterized Col10α1-Cre transgenic line was generated using the 1 kb proximal promoter of the mouse Col10α1 gene. This line displayed Cre recombinase activity in E14.5 ribs cartilage primordiae and in some, but not all, hypertrohic chondrocytes [37].
Aggrecan promoter
Aggrecan is a major extracellular matrix (ECM) protein of both growth plate and articular cartilage. Inducible Acg-CreERT2 transgenic mice were generated by introducing a tamoxifen-inducible CreERT2 cassette in the 3′-untranslated region of the endogenous aggrecan gene [38]. The use of an IRES sequence and insertion downstream of the aggrecan gene stop codon left the endogenous gene intact in this line. The offspring of crosses between Acg-CreERT2 and Rosa26 reporter mice injected with tamoxifen displayed efficient Cre reporter expression in the hyaline cartilage of the growth plate and articular cartilage as well as the fibrocartilage of the meniscus, trachea, and intervertebral disks of growing and adult mice. Positive X-gal cells were first detected in the forelimb of E13.0 Acg-CreERT2; R26R embryos harvested from pregnant mothers injected with 4OH-tamoxifen at E12.0. Positive X-gal staining was observed in the proliferating and hypertrophic chondrocytes. An important feature of this line is the prolonged expression of Cre recombinase in adult mice, compared to Col2α1–Cre based mice.
Precautions
It is important to emphasize that in most of these “chondrocyte-specific” transgenic lines, Cre recombination is evident in a significant proportion of osteoblasts, especially in adults [29]. This irreversible gene alteration is likely caused by a transient Cre activity in a subset of perichondrial osteochondroprogenitor cells that give rise to osteoblasts. Regardless of the site and time of Cre expression, this observation indicates that any osteoblastic-like or bone-like phenotype observed in mutants using these Cre lines may be mediated by a direct effect of gene deletion in the osteoblast lineage. The induction of Cre recombinase activity after E12.5 in inducible Col2α1–Cre lines can limit this “leakage” to osteoblasts.
Osteoblast promoters
There are a number of Cre lines to manipulate gene expression in either immature or mature osteoblasts that are quite widely used in the bone field.
Runx2 promoter
Runx2 is a critical transcription factor of the Runx family that is required for determination of the osteoblast lineage. Runx2-Cre transgenic mice were created by inserting the Cre recombinase in the Runx2 locus at the translational start site of the bone-specific distal promoter (P1) [40]. A cross with ROSA26 reporter mice confirmed efficient recombination at all sites of endochondral and intramembranous bone formation in neonatal mice, particularly in periosteal cells, osteoblasts, and osteocytes, but not in osteoclasts or any other mesenchymal tissues such as fat or muscles. Although this model has not been fully characterized or used extensively yet compared to other osteoblast “specific” Cre mice such as the mouse 2.3 kb Col1α1-Cre mice (see below), its specificity and early pattern of expression during differentiation of the osteoblast lineage may prove very useful to study osteoprogenitor fate. Expression of Cre in the hypertrophic chondroctyes in cartilage of this model needs, however, to be taken into account.
Osterix promoter
Immature osteoblasts can be targeted by using the Osx–Cre transgenic mice as well. Osterix (Osx) is a zinc finger-containing transcription factor expressed in osteoblasts of all endochondral and membranous bones. The mouse Osx–Cre transgenic line expresses the Cre recombinase in committed osteoblast progenitors in both endochondral and membranous-derived bones [44] in a manner that follows that of endogenous Osx [80]. Cre activity was observed during embryonic development from E14.5 and post-natally (P10) in the inner bone-forming perichondrium adjacent to hypertrophic chondrocytes, sporadically in hypertrophic chondrocytes, in the periosteum, and primary spongiosa. This Osx–Cre line is inducible. Doxycycline administration prevents Cre recombination and stopping the treatment relieves the brake on Cre recombinase expression, leading to Cre-mediated recombination. Another characteristic of this model is that Cre is fused to eGFP, which allows direct visualization of the site of expression of the transgene in vivo as well as lineage tracing. Osx–Cre mice display a slight growth delay [41] and a high prevalence of teeth malocclusion in adults of the C57BL6 strain irrespective of the targeted floxed gene (Dr. Louise Purton, personal communication).
The Osx–CreERT2 line, generated by inserting the Cre-ERT2 fusion cDNA and downstream poly-A element (pA) into a BAC containing the Osterix (Sp7) (Osx) gene promoter, was recently reported [45]. 4OH-tamoxifen treatment in pregnant mothers at E12.5–13.5 triggers recombination in embryos’ mandible, the lateral calvarial bones, the proximal part of the ribs and the proximal limb bones one day later, as assessed following a cross with ROSA26 reporter mice. Cells in the perichondrium were also recombined, and tracing experiments indicated that these Osx–LacZ+ perichondrial cells predominantly moved inside the bone to the trabecular regions to become trabecular and endosteal osteoblasts as well as osteocytes, whereas Col-LacZ+ cells predominantly remain on the cortex (see below). Mosaic labeling in the pre- and early hypertrophic chondrocyte zones was also observed in Osx–CreET2 mice, in line with the endogenous expression of Osx in prehypertrophic chondrocytes.
Col1α1 promoter
Mature osteoblasts on the other hand can be targeted based on their rich expression of type I collagen, the main constituent of bones. Type I collagen is also expressed in fibroblasts and other mesenchymal cells, but an osteoblast-specific enhancer has been identified in the mouse and rat 2.3 kb proximal Col1a1 promoters which allows efficient transgene expression in osteoblasts and odontoblasts specifically [81,82].
The mouse 2.3 kb Col1α1-Cre transgenic line is quite widely used in the bone field because of its robust promoter activity and transgene expression specificity to mature osteoblasts [46]. This line displays Cre recombinase activity from E14.5 in long bones and skull, and in all bones (intramembranous and endochondral-derived) at E16.5 and P5. Weak staining in digit and face skin was observed.
The rat 2.3 kb Col1α1-Cre transgenic line has very similar characteristics. In calvaria and long bones, Cre recombinase activity was observed in mature osteoblasts and osteocytes (cortical and trabecular cells in long bones), but not in suture mesenchyme nor cells of the inner and outer periosteum at E18 and P5.
The rat 3.6 Col1α1-Cre transgenic line is also available to target osteoblasts, but the specificity of this Cre line is different from the 2.3 kb Col1α1-Cre transgenic line [52]. Ex vivo analysis of bone marrow stromal cell and primary calvarial osteoblast cultures have shown that the 3.6 kb Col1α1-Cre is expressed early during osteogenic differentiation, whereas the 2.3 kb Col1α1-Cre is activated later and its activity is restricted to maturing osteoblasts [83,84]. Accordingly, in contrast to the 2.3 kb Col1α1-Cre line, the 3.6 kb Col1α1-Cre line displays in vivo Cre recombinase activity broadly in cells of the osteoblast lineage and in suture mesenchyme. During development (E18), 2.3 kb Col1α1-Cre activity was observed in long bones in cells of the inner layer of the perichondrium, which are destined to become osteoblasts, while 3.6 kb Col1α1-Cre recombinase activity was detected in the entire perichondrium. Sporadic activity was observed in growth plate chondrocytes in both 2.3 kb Col1α1-Cre and 3.6 kb Col1α1-Cre mice. Surprisingly, strong Cre activity was observed in the articular cartilage of long bones in 3.6 kb Col1α1-Cre mice. 3.6 kb Col1α1-Cre activity was also observed in tendons. These studies thus indicate that the 2.3 kb and the 3.6 kb Col1α1-Cre transgenic mice can be used to target different populations of osteoblasts. Importantly, Dr. Kream and collaborators have observed that Col1α1-Cre mediated gene rearrangement can occur in the germ line by transmission of Cre mRNA or protein in oocytes and sperm irrespective of inheritance of the Cre transgene [52]. This occurred more frequently with females than with males. They thus suggested that Col1α1-Cre transgenes should be transmitted through the male breeder and that genotyping should be done to identify animals that display gene rearrangement in the absence of inheritance of the Cre transgene. One study also showed that the 3.6 kb Col1α1 promoter unexpectedly directs the expression of transgenes in the osteoclast lineage, and this effect must be considered when utilizing this promoter [85].
Temporal regulation of Cre recombinase activity can be achieved selectively in mature osteoblasts thanks to an inducible mouse 2.3 kb Col1α1-Cre transgenic line that was generated by using the CreERT2 recombinase [13]. In this line, Cre recombinase activity was detected following 4-OHT in E18.5 long bone, calvaria, rib, and vertebra osteoblasts. No Cre activity was observed in other organs such as heart, lung, liver, and kidney of embryos whose dams were treated with 4-OHT. Cre recombination was also observed post-natally in osteoblasts in 18 day-old mice upon 4-OHT treatment. The advantage of this transgenic mouse line over the non-inducible 2.3 kb Col1α1-Cre mouse line is to offer the possibility to study the function of a gene in osteoblasts after birth, without affecting bone development since the targeted floxed gene can be inactivated post-natally upon 4-OHT injection.
The Col1-CreERT2 transgenic line was generated by cloning the Cre-ERT2 fusion protein cDNA and downstream poly-A element (pA) behind a 3.2 kb fragment of the mouse type I collagen regulatory sequences [45]. The pups of Col1-CreERT2; ROSA26 reporter mothers injected with 4OH-Tamoxifen at E13.5 displayed, at E14.5, a staining pattern similar to OSX–CreERT2; ROSA26 mice, although a reduced distribution throughout the skeleton was observed, such as absence of staining in the hind limbs. Further studies confirmed that Osx–CreERT2 was expressed earlier in development than Col1-CreERT2. Mosaic perichondrial recombination was observed in this model, and these cells were shown to predominantly remain in the cortex, as opposed to Osx–CreERT2 perichondrial cells that migrate to trabecular regions. No recombination was observed in cells of the growth plate.
Osteocalcin promoter
The most mature/differentiated osteoblasts can be targeted thanks to the selective expression of Osteocalcin (Ocn, also called bone γ-carboxyglutamic acid protein, or Bgp) in differentiated osteoblasts [86]. The human OCN promoter is first activated in fetal life just prior to birth [87,88], so it is not suitable for inactivating a gene during early development. The 1.7 kb OCN promoter does not drive Cre recombinase expression at a level sufficient to induce gene rearrangement in mature osteoblasts [46]. However, the 3.5 kb human OCN promoter has been used successfully to inactivate gene expression in mature osteoblasts using the OCN-cre transgenic mice that display Cre recombinase activity in calvaria osteoblasts and osteocytes starting at E17 [54] and in trabecular osteoblasts and osteocytes in 6 week-old mice [57].
Osteocyte promoters
Osteocytes represent a population of fully differentiated osteoblasts embedded within the bone matrix that is relatively difficult to study due to the absence of good cell lines, their tissue localization, and limited proliferative capacity. It is important to realize that gene deletion at any time point during differentiation of the osteoblast lineage will eventually give rise to KO osteocytes since these cells are the ultimate differentiation stage of this lineage. Osteocytes can, however, be distinguished from osteoblasts by their expression of Dentin Matrix Protein 1 (DMP1), an extracellular matrix protein that is expressed in all mineralized tissues [89,90]. Consequently, several DMP1 transgenic lines have been generated. GFP expression driven by a 7.9-kb mouse Dmp1 promoter was found mainly in osteocytes [91]. When Cre-expression was driven by a 9.6-kb mouse Dmp1 promoter, Cre recombinase activity was found in both dentin odontoblasts and bone osteocytes post-natally [60]. A mouse Dmp1-Cre-ERT2 is available, which allows floxed gene recombination in osteocyte post-natally only (and not during development) following tamoxifen administration [61]. The osteocyte specificity of this transgenic line seems very good, since only 1% and 9% β-gal-positive osteoblasts were detected in calvaria and femoral bones, respectively, following crossing to the ROSA26R reporter mice.
The 9.6 Dmp1 promoter has been used to drive the expression of the diphtheria toxin receptor, a suicide gene that allowed specific ablation of osteocytes [2], as well as GPF for tracing studies [91,92]. Although the GFP moiety of the Dmp1-GFP mice can theoretically be used to purify osteocytes using FACS, the extraction of these cells from bone and their subsequent culture in vitro are technically challenging. An Immortomouse/Dmp1-GFP derived bone cell line (IDG-SW3) is available as an alternative to study some aspects of osteocyte biology in culture [93]. These cells express a temperature-sensitive mutant of the SV40 large tumor antigen under the control of the interferon-γ-inducible H-2Kb promoter (H-2Kb-tsA58) at 33 °C in the presence of γ-IFN, inducing continuous proliferation and immortalization. Switch to 37 °C in absence of γ-IFN allows these cells to behave as differentiated osteoblasts capable of differentiating to osteocyte-like cells under proper differentiation conditions.
Osteoclast promoters
Trap promoter
TRAP is indeed highly expressed in differentiated osteoclasts [94] and this characteristic was used to generate the mouse Trap-Cre transgenic line. These mice display Cre recombinase activity in long bones, vertebrae, ribs, and calvaria multinucleated osteoclasts, but also display some activity in chondrocytes. The latter is consistent with TRAP mRNA localization in murine chondrocytes [62,95]. Non-osseous activity was also observed in liver, spleen, stomach, heart, liver, and lung. Although the Trap-Cre transgenic line has not been used to create gene deletion yet, the TRAP promoter has been used successfully to drive the expression of multiple genes to osteoclasts in vivo, including c-fos [96], src [97], the SV40 T antigen [98], and to ablate macrophages by overexpressing the Diphteria toxin receptor [1].
M lysozyme promoter
M lysozyme, whose function is to cleave peptidoglycan off intruding bacteriae, is expressed in myeloid cells and monocytes/macrophages giving rise to osteoclasts. Mouse LysM-Cre transgenic mice were generated by a knock-in strategy inserting the Cre cDNA into the endogenous mouse M lysozyme gene [67]. Strong Cre recombinase activity in this line was detected by genomic Southern blot analyses in purified F4/80+ peritoneal macrophages and Gr1+ peritoneal neurophils. Weak Cre activity was detected in sorted CD11c+ dendritic cells.
Cathepsin K (Ctsk) promoter
Ctsk is more selectively expressed in osteoclasts [99–101]. The mouse Ctsk-Cre transgenic lines showed Cre recombinase activity in long bones, calvaria, and ribs [62,70]. Non-osseous activity was detected in stomach, salivary gland and liver [62]. Another mouse Ctsk-cre line was created by a knock-in strategy [70]. In this line, Cre activity was detected in skull and long bones at E16.5 and P7, consistent with the appearance and skeletal localization of functionally mature osteoclasts, and in vertebra multinuclear osteoclasts in adult mice.
CD11b promoter
CD11b encodes the α-subunit of the Mac-1 leukocyte integrin heterodimer, a major adhesion molecule and a commonly used hematopoietic surface marker. This gene is upregulated during myeloid differentiation, with the highest levels in mature monocytes, macrophages, and neutrophils [102]. CD11b is also expressed along the osteoclast differentiation pathway from mononucleated early progenitor cells until mature polynucleated osteoclasts [71,103,104]. Two human CD11b-Cre transgenic mouse lines have been generated. Both lines display Cre recombinase activity in hematopoietic organs including bone marrow, spleen and thymus. Cre recombinase activity was detected in peritoneal macrophages and bone marrow granulocytes [71]. B220-positive lymphocytes subpopulations and macrophages showed low level of Cre recombinase activity in the spleen. Strong Cre recombinase activity was detected in ex vivo generated TRAP-positive multinucleated osteoclasts. Very low level of recombination was detectable in testis, liver, kidney, brain, heart, and lung, possibly consistent with peripheral blood contamination. Cre recombinase activity was observed in the brain in several microglia-like cells, which derive from common precursor with macrophages.
Phenotypic analyses and interpretations
The availability of mouse models that allow gain or loss-of-function of specific genes in vivo has profoundly transformed the field of bone biology for the last 10 years. This “genetic” approach is now widely used to demonstrate the contribution of a given gene in a variety of biological processes such as bone development, bone remodeling, bone cancer metastasis, or bone repair. Although it is usually considered as the “gold standard” to ascribe a function to a specific gene in vivo, it is not devoid of limitations, especially if one loses sight of what actually happens in these mutant models, and thus can lead to serious misinterpretations and erroneous conclusions. Things become even more complex when a gene is more widely expressed or is involved in the regulation of multiple physiological processes. The literature is nowadays very much loaded with sometime erroneous conclusions and interpretations from global mutant mice. Science is an evolving business and every scientist will likely be proven wrong about something in the future, but the more general use of conditional KO models and their comparison to global and other Cre models should bring substantial information in the near future, if one keeps in mind the limitations of genetic models.
The cases of mutations or loss of function of a specific gene whose expression is restricted to bone cells is a priori the least prone to ambiguity, although the gene of interest might have distinct role in immature versus fully differentiated cells. Therefore, although a global KO would seems sufficient to address the role of this gene in bone homeostasis, the conditional approach brings a critical advantage to dissect putative distinct roles at various time of differentiation at the cell level or at various times of development. Examples of this would be the cases of NF1, a gene that is mutated in patients with neurofibromatosis type I, or β-catenin. Knock out of Nf1 in mature osteoblasts, using the 2.3 kb Col1α1-Cre mice revealed the contribution of this gene to collagen synthesis, Rankl expression and bone mineralization, but did not affect osteoblast differentiation, leading to a high bone mass associated with high bone turn over [50]. In contrast, deletion of Nf1 in immature osteoprogenitors revealed its role in promoting early differentiation, and led to a low bone mass phenotype [19,29].
β-catenin is a key component of the canonical Wnt signaling pathway and plays a crucial role in multiple steps during chondrogenesis and chondrocyte maturation. Global β-catenin deficiency resulted in embryonic lethality prior to formation of the skeletal elements [105]. Conditional deletion of Catnby, the gene encoding β-catenin, in mesenchymal precursors promoted chondrogenesis [22,106], suggesting that β-catenin inhibits early mesenchymal cell differentiation into cartilage. However, conditional deletion of Catnby in chondrocytes using the Col2α1–Cre mice decreased chondrocyte proliferation and delayed chondrocyte maturation [107]. Thus, β-catenin has opposite roles depending on the stage of development. It inhibits chondrogenesis, but once cartilage has formed, it promotes the maturation of growth plate chondrocytes.
Another consideration to keep in mind is that although a gene may seem cell specific, various physiological or pathological conditions may switch on or off the expression of a gene in adults. A typical example would be stress-responsive genes like Atf4 for instance. The use of promoter specific for mature or differentiated bone cells is thus advantageous for its cell selectivity, but data from these models should be interpreted with caution since the function of the targeted gene in earlier differentiation stages is not taken into account. On the other hand, promoters active in progenitor cells that give rise to multiple sublineages will likely give a more biologically relevant picture of the phenotype, but the low specificity of such promoters makes the analysis of the results more complex. Interpretation of results in studies limited to a single Cre transgenic line to delete a gene of interest must thus be done with caution and comparison of the results obtained following deletion the same floxed gene with different Cre lines is the best approach to understand the gene function in a given lineage.
Importantly, even a restricted and transient expression of the Cre recombinase during development or later will irreversibly recombine a floxed gene in cells that may proliferate and later constitute a significant part of the cell population in the targeted tissue of mutant mice. The Col2α1–Cre lines represent a good example where Cre recombinase activity is expressed, likely transiently, in a pool of progenitor cells giving rise to chondrocytes but also osteoblasts, and possibly other mesenchymal cells. Crosses with reporter mice may not detect this event due to its transient nature or because of the sensitivity of the reporter system (most often time β-gal staining). In addition, authors interested in growth plate development may seldom report leaky expression in the marrow space or in adults. Interpretation of the results obtained from such conditional KO mice must thus be done with caution, keeping in mind that the system is never perfect and that other cell types than the one thought to be targeted may contribute to the observed phenotypes or mechanisms.
Another important issue is the genetic background of the mutant mice under study. Because bone parameters, including bone mass, differ significantly between mouse strains, it is crucial to use WT controls that originate from the same mutant colony, and if possible to have a colony backcrossed to a pure background. When using conditional knockouts, “WT” control animals can be Cre-negative; flox/flox or Cre-positive; +/+. The first control takes into account any possible interference from the introduction of the Lox sequences, while the second takes into account the possible effects of Cre recombinase activity on the genome.
Lastly, gender and age are important factors that should be considered when interpreting results. Studies too often report absence of phenotype using a single age time point and gender, which can lead to erroneous conclusions if the gene under study regulates processes that are slow, rapid, compensated by redundant mechanism(s), or gender-specific.
On a more general note, obviously and despite similar bone biology between mice and human, there are a number of differences between the two species that make extrapolations of mouse results to human conditions, diseases and treatment hazardous. Those include different tissue organization, continuous growth in mice versus growth plate closure in humans, differences of posture and patterns of mechanical loading, different remodeling activities, distinct genetic evolution and others. This being said, the findings obtained in genetically modified mutant mice shed light onto important biological mechanisms that will always eventually increment our knowledge of bone physiology, regardless of whether the results in mutant mice can be directly extrapolated to humans physiopathology or not.
References
- 1.Goren I, Allmann N, Yogev N, Schurmann C, Linke A, Holdener M, et al. A transgenic mouse model of inducible macrophage depletion: effects of diphtheria toxin-driven lysozyme M-specific cell lineage ablation on wound inflammatory, angiogenic, and contractive processes. Am J Pathol. 2009;175:132–47. doi: 10.2353/ajpath.2009.081002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tatsumi S, Ishii K, Amizuka N, Li M, Kobayashi T, Kohno K, et al. Targeted ablation of osteocytes induces osteoporosis with defective mechanotransduction. Cell Metab. 2007;5:464–75. doi: 10.1016/j.cmet.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Roebroek AJ, Wu X, Bram RJ. Knockin Approaches. 2002:187–200. doi: 10.1385/1-59259-340-2:187. [DOI] [PubMed] [Google Scholar]
- 4.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–46. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 5.Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- 6.Soriano P. Generalized lacZ expression ith the ROSA26 Cre reporter strain. Nat Genet. 1999;14:670–89. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 7.Akiyama H, Kim JE, Nakashima K, Balmes G, Iwai N, Deng JM, et al. Osteochondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci USA. 2005;102:14665–70. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brocard J, Warot X, Wendling O, Messaddeq N, Vonesch JL, Chambon P, et al. Spatio-temporally controlled site-specific somatic mutagenesis in the mouse. Proc Natl Acad Sci USA. 1997;94:14559–63. doi: 10.1073/pnas.94.26.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Metzger D, Chambon P. Site- and time-specific gene targeting in the mouse. Methods. 2001;24:71–80. doi: 10.1006/meth.2001.1159. [DOI] [PubMed] [Google Scholar]
- 10.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–51. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–9. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 12.Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, et al. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999;27:4324–7. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JE, Nakashima K, de Crombrugghe B. Transgenic mice expressing a ligand-inducible cre recombinase in osteoblasts and odontoblasts: a new tool to examine physiology and disease of postnatal bone and tooth. Am J Pathol. 2004;165:1875–82. doi: 10.1016/S0002-9440(10)63240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu M, Chen M, Lichtler AC, O’Keefe RJ, Chen D. Tamoxifen-inducible Cre-recombination in articular chondrocytes of adult Col2a1-CreER(T2) transgenic mice. Osteoarthritis Cartilage. 2008;16:129–30. doi: 10.1016/j.joca.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kistner A, Gossen M, Zimmermann F, Jerecic J, Ullmer C, Lubbert H, et al. Doxycycline-mediated quantitative and tissue-specific control of gene expression in transgenic mice. Proc Natl Acad Sci USA. 1996;93:10933–8. doi: 10.1073/pnas.93.20.10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horton WA. Skeletal development: insights from targeting the mouse genome. Lancet. 2003;362:560–9. doi: 10.1016/S0140-6736(03)14119-0. [DOI] [PubMed] [Google Scholar]
- 17.Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- 18.Logan M, Martin JF, Nagy A, Lobe C, Olson EN, Tabin CJ. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 19.Kolanczyk M, Kossler N, Kuhnisch J, Lavitas L, Stricker S, Wilkening U, et al. Multiple roles for neurofibromin in skeletal development and growth. Hum Mol Genet. 2007;16:874–86. doi: 10.1093/hmg/ddm032. [DOI] [PubMed] [Google Scholar]
- 20.Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–74. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Wang L, Fatahi R, Kronenberg M, Kalajzic I, Rowe D, et al. Isolation of murine bone marrow derived mesenchymal stem cells using Twist2 Cre transgenic mice. Bone. 2010;47:916–25. doi: 10.1016/j.bone.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–50. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 23.Horiuchi K, Kimura T, Miyamoto T, Miyamoto K, Akiyama H, Takaishi H, et al. Conditional inactivation of TACE by a Sox9 promoter leads to osteoporosis and increased granulopoiesis via dysregulation of IL-17 and G-CSF. J Immunol. 2009;182:2093–101. doi: 10.4049/jimmunol.0802491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haigh JJ, Gerber HP, Ferrara N, Wagner EF. Conditional inactivation of VEGF-A in areas of collagen2a1 expression results in embryonic lethality in the heterozygous state. Development. 2000;127:1445–53. doi: 10.1242/dev.127.7.1445. [DOI] [PubMed] [Google Scholar]
- 25.Karreth F, Hoebertz A, Scheuch H, Eferl R, Wagner EF. The AP1 transcription factor Fra2 is required for efficient cartilage development. Development. 2004;131:5717–25. doi: 10.1242/dev.01414. [DOI] [PubMed] [Google Scholar]
- 26.Downey CM, Horton CR, Carlson BA, Parsons TE, Hatfield DL, Hallgrimsson B, et al. Osteo-chondroprogenitor-specific deletion of the selenocysteine tRNA gene, Trsp, leads to chondronecrosis and abnormal skeletal development: a putative model for Kashin-Beck disease. PLoS Genet. 2009;5:e1000616. doi: 10.1371/journal.pgen.1000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ford-Hutchinson AF, Ali Z, Lines SE, Hallgrimsson B, Boyd SK, Jirik FR. Inactivation of Pten in osteo-chondroprogenitor cells leads to epiphyseal growth plate abnormalities and skeletal overgrowth. J Bone Miner Res. 2007;22:1245–59. doi: 10.1359/jbmr.070420. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh SC, Chen NT, Lo SH. Conditional loss of PTEN leads to skeletal abnormalities and lipoma formation. Mol Carcinog. 2009;48:545–52. doi: 10.1002/mc.20491. [DOI] [PubMed] [Google Scholar]
- 29.Wang W, Nyman JS, Ono K, Stevenson DA, Yang X, Elefteriou F. Mice lacking Nf1 in osteochondroprogenitor cells display skeletal dysplasia similar to patients with neurofibromatosis type I. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr310. [Electronic publication ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26:145–6. [PubMed] [Google Scholar]
- 31.Nakamura E, Nguyen MT, Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn. 2006;235:2603–12. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- 32.Chen M, Lichtler AC, Sheu TJ, Xie C, Zhang X, O’Keefe RJ, et al. Generation of a transgenic mouse model with chondrocyte-specific and tamoxifen-inducible expression of Cre recombinase. Genesis. 2007;45:44–50. doi: 10.1002/dvg.20261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hilton MJ, Tu X, Long F. Tamoxifen-inducible gene deletion reveals a distinct cell type associated with trabecular bone, and direct regulation of PTHrP expression and chondrocyte morphology by Ihh in growth region cartilage. Dev Biol. 2007;308:93–105. doi: 10.1016/j.ydbio.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones KB, Piombo V, Searby C, Kurriger G, Yang B, Grabellus F, et al. A mouse model of osteochondromagenesis from clonal inactivation of Ext1 in chondrocytes. Proc Natl Acad Sci USA. 2010;107:2054–9. doi: 10.1073/pnas.0910875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grover J, Roughley PJ. Generation of a transgenic mouse in which Cre recombinase is expressed under control of the type II collagen promoter and doxycycline administration. Matrix Biol. 2006;25:158–65. doi: 10.1016/j.matbio.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Gebhard S, Hattori T, Bauer E, Schlund B, Bosl MR, de Crombrugghe B, et al. Specific expression of Cre recombinase in hypertrophic cartilage under the control of a BAC-Col10a1 promoter. Matrix Biol. 2008;27:693–9. [Google Scholar]
- 37.Yang G, Cui F, Hou N, Cheng X, Zhang J, Wang Y, et al. Transgenic mice that express Cre recombinase in hypertrophic chondrocytes. Genesis. 2005;42:33–6. doi: 10.1002/gene.20120. [DOI] [PubMed] [Google Scholar]
- 38.Henry SP, Jang CW, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Generation of aggrecan-CreERT2 knockin mice for inducible Cre activity in adult cartilage. Genesis. 2009;47:805–14. doi: 10.1002/dvg.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maes C, Goossens S, Bartunkova S, Drogat B, Coenegrachts L, Stockmans I, et al. Increased skeletal VEGF enhances beta-catenin activity and results in excessively ossified bones. EMBO J. 2010;29:424–41. doi: 10.1038/emboj.2009.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rauch A, Seitz S, Baschant U, Schilling AF, Illing A, Stride B, et al. Glucocorticoids suppress bone formation by attenuating osteoblast differentiation via the monomeric glucocorticoid receptor. Cell Metab. 2010;11:517–31. doi: 10.1016/j.cmet.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev. 2008;22:1662–76. doi: 10.1101/gad.1656808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Provot S, Nachtrab G, Paruch J, Chen AP, Silva A, Kronenberg HM. A-raf and B-raf are dispensable for normal endochondral bone development, and parathyroid hormone-related peptide suppresses extracellular signal-regulated kinase activation in hypertrophic chondrocytes. Mol Cell Biol. 2008;28:344–57. doi: 10.1128/MCB.00617-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu W, Liang G, Huang Z, Doty SB, Boskey AL. Conditional inactivation of the CXCR4 receptor in osteoprecursors reduces postnatal bone formation due to impaired osteoblast development. J Biol Chem. 2011;286:26794–805. doi: 10.1074/jbc.M111.250985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–44. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- 45.Maes C, Kobayashi T, Selig MK, Torrekens S, Roth SI, Mackem S, et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev Cell. 2010;19:329–44. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dacquin R, Starbuck M, Schinke T, Karsenty G. Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 2002;224:245–51. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- 47.Hinoi E, Gao N, Jung DY, Yadav V, Yoshizawa T, Myers MG, Jr, et al. The sympathetic tone mediates leptin’s inhibition of insulin secretion by modulating osteocalcin bioactivity. J Cell Biol. 2008;183:1235–42. doi: 10.1083/jcb.200809113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haberland M, Mokalled MH, Montgomery RL, Olson EN. Epigenetic control of skull morphogenesis by histone deacetylase 8. Genes Dev. 2009;23:1625–30. doi: 10.1101/gad.1809209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato M, Patel MS, Levasseur R, Lobov I, Chang BH, Glass DA, II, et al. Cbfa1-independent decrease in osteoblast proliferation, osteopenia, and persistent embryonic eye vascularization in mice deficient in Lrp5, a Wnt coreceptor. J Cell Biol. 2002;157:303–14. doi: 10.1083/jcb.200201089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Elefteriou F, Benson MD, Sowa H, Starbuck M, Liu X, Ron D, et al. ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metab. 2006;4:441–51. doi: 10.1016/j.cmet.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baek WY, de Crombrugghe B, Kim JE. Postnatally induced inactivation of Osterix in osteoblasts results in the reduction of bone formation and maintenance. Bone. 2010;46(4):920–8. doi: 10.1016/j.bone.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu F, Woitge HW, Braut A, Kronenberg MS, Lichtler AC, Mina M, et al. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol. 2004;48:645–53. doi: 10.1387/ijdb.041816fl. [DOI] [PubMed] [Google Scholar]
- 53.Gutierrez GM, Kong E, Sabbagh Y, Brown NE, Lee JS, Demay MB, et al. Impaired bone development and increased mesenchymal progenitor cells in calvaria of RB1−/− mice. Proc Natl Acad Sci USA. 2008;105:18402–7. doi: 10.1073/pnas.0805925105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang M, Xuan S, Bouxsein ML, von Stechow D, Akeno N, Faugere MC, et al. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem. 2002;277:44005–12. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]
- 55.Zanotti S, Smerdel-Ramoya A, Stadmeyer L, Durant D, Radtke F, Canalis E. Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology. 2008;149:3890–9. doi: 10.1210/en.2008-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeo H, Beck LH, Thompson SR, Farach-Carson MC, McDonald JM, Clemens TL, et al. Conditional disruption of calcineurin B1 in osteoblasts increases bone formation and reduces bone resorption. J Biol Chem. 2007;282:35318–27. doi: 10.1074/jbc.M702435200. [DOI] [PubMed] [Google Scholar]
- 57.Tan X, Weng T, Zhang J, Wang J, Li W, Wan H, et al. Smad4 is required for maintaining normal murine postnatal bone homeostasis. J Cell Sci. 2007;120:2162–70. doi: 10.1242/jcs.03466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X, Bruxvoort KJ, Zylstra CR, Liu J, Cichowski R, Faugere MC, et al. Lifelong accumulation of bone in mice lacking Pten in osteoblasts. Proc Natl Acad Sci USA. 2007;104:2259–64. doi: 10.1073/pnas.0604153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xiao Z, Dallas M, Qiu N, Nicolella D, Cao L, Johnson M, et al. Conditional deletion of Pkd1 in osteocytes disrupts skeletal mechanosensing in mice. FASEB J. 2011;25:2418–32. doi: 10.1096/fj.10-180299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu Y, Xie Y, Zhang S, Dusevich V, Bonewald LF, Feng JQ. DMP1-targeted Cre expression in odontoblasts and osteocytes. J Dent Res. 2007;86:320–5. doi: 10.1177/154405910708600404. [DOI] [PubMed] [Google Scholar]
- 61.Powell WF, Jr, Barry KJ, Tulum I, Kobayashi T, Harris SE, Bringhurst FR, et al. Targeted ablation of the PTH/PTHrP receptor in osteocytes impairs bone structure and homeostatic calcemic responses. J Endocrinol. 2011;209:21–32. doi: 10.1530/JOE-10-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chiu WS, McManus JF, Notini AJ, Cassady AI, Zajac JD, Davey RA. Transgenic mice that express Cre recombinase in osteoclasts. Genesis. 2004;39:178–85. doi: 10.1002/gene.20041. [DOI] [PubMed] [Google Scholar]
- 63.Dossa T, Arabian A, Windle JJ, Dedhar S, Teitelbaum SL, Ross FP, et al. Osteoclast-specific inactivation of the integrin-linked kinase (ILK) inhibits bone resorption. J Cell Biochem. 2010;110:960–7. doi: 10.1002/jcb.22609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim HJ, Zhao H, Kitaura H, Bhattacharyya S, Brewer JA, Muglia LJ, et al. Glucocorticoids suppress bone formation via the osteoclast. J Clin Invest. 2006;116:2152–60. doi: 10.1172/JCI28084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martin-Millan M, Almeida M, Ambrogini E, Han L, Zhao H, Weinstein RS, Jilka RL, O’Brien CA, Manolagas SC. The Estrogen Receptor-{alpha} in Osteoclasts Mediates the Protective Effects of Estrogens on Cancellous But Not Cortical Bone. Mol Endocrinol. 2010;24:323–34. doi: 10.1210/me.2009-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kenner L, Hoebertz A, Beil T, Keon N, Karreth F, Eferl R, et al. Mice lacking JunB are osteopenic due to cell-autonomous osteoblast and osteoclast defects. J Cell Biol. 2004;164:613–23. doi: 10.1083/jcb.200308155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Forster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–77. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 68.Iwasawa M, Miyazaki T, Nagase Y, Akiyama T, Kadono Y, Nakamura M, et al. The antiapoptotic protein Bcl-xL negatively regulates the bone-resorbing activity of osteoclasts in mice. J Clin Invest. 2009;119:3149–59. doi: 10.1172/JCI39819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mizoguchi F, Izu Y, Hayata T, Hemmi H, Nakashima K, Nakamura T, et al. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J Cell Biochem. 2009;109(5):866–75. doi: 10.1002/jcb.22228. [DOI] [PubMed] [Google Scholar]
- 70.Nakamura T, Imai Y, Matsumoto T, Sato S, Takeuchi K, Igarashi K, et al. Estrogen prevents bone loss via estrogen receptor alpha and induction of Fas ligand in osteoclasts. Cell. 2007;130:811–23. doi: 10.1016/j.cell.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 71.Ferron M, Vacher J. Targeted expression of Cre recombinase in macrophages and osteoclasts in transgenic mice. Genesis. 2005;41:138–45. doi: 10.1002/gene.20108. [DOI] [PubMed] [Google Scholar]
- 72.Ishii M, Egen JG, Klauschen F, Meier-Schellersheim M, Saeki Y, Vacher J, et al. Sphingosine-1-phosphate mobilizes osteoclast precursors and regulates bone homeostasis. Nature. 2009;458:524–8. doi: 10.1038/nature07713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Otero JE, Dai S, Foglia D, Alhawagri M, Vacher J, Pasparakis M, et al. Defective osteoclastogenesis by IKKbeta-null precursors is a result of receptor activator of NF-kappaB ligand (RANKL)-induced JNK-dependent apoptosis and impaired differentiation. J Biol Chem. 2008;283:24546–53. doi: 10.1074/jbc.M800434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu MF, Cheng HT, Lacy AR, Kern MJ, Argao EA, Potter SS, et al. Paired-related homeobox genes cooperate in handplate and hindlimb zeugopod morphogenesis. Dev Biol. 1999;205:145–57. doi: 10.1006/dbio.1998.9116. [DOI] [PubMed] [Google Scholar]
- 75.Martin JF, Olson EN. Identification of a prx1 limb enhancer. Genesis. 2000;26:225–9. [PubMed] [Google Scholar]
- 76.Kawanami A, Matsushita T, Chan YY, Murakami S. Mice expressing GFP and CreER in osteochondro progenitor cells in the periosteum. Biochem Biophys Res Commun. 2009;386:477–82. doi: 10.1016/j.bbrc.2009.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cheah KS, Levy A, Trainor PA, Wai AW, Kuffner T, So CL, et al. Human COL2A1-directed SV40 T antigen expression in transgenic and chimeric mice results in abnormal skeletal development. J Cell Biol. 1995;128:223–37. doi: 10.1083/jcb.128.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Colnot C, Lu C, Hu D, Helms JA. Distinguishing the contributions of the perichondrium, cartilage, and vascular endothelium to skeletal development. Dev Biol. 2004;269:55–69. doi: 10.1016/j.ydbio.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 79.Gebhard S, Poschl E, Riemer S, Bauer E, Hattori T, Eberspaecher H, et al. A highly conserved enhancer in mammalian type X collagen genes drives high levels of tissue-specific expression in hypertrophic cartilage in vitro and in vivo. Matrix Biol. 2004;23:309–22. doi: 10.1016/j.matbio.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 80.Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng J-M, Behringer R, et al. The novel zinc-finger-containing transcription factor Osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 81.Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–69. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rossert J, Eberspaecher H, de Crombrugghe B. Separate cis-acting DNA elements of the mouse Pro-alpha 1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J Cell Biol. 1995;129:1421–32. doi: 10.1083/jcb.129.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kalajzic Z, Liu P, Kalajzic I, Du Z, Braut A, Mina M, et al. Directing the expression of a green fluorescent protein transgene in differentiated osteoblasts: comparison between rat type I collagen and rat osteocalcin promoters. Bone. 2002;31:654–60. doi: 10.1016/s8756-3282(02)00912-2. [DOI] [PubMed] [Google Scholar]
- 84.Kalajzic I, Kalajzic Z, Kaliterna M, Gronowicz G, Clark SH, Lichtler AC, et al. Use of type I collagen green fluorescent protein transgenes to identify subpopulations of cells at different stages of the osteoblast lineage. J Bone Miner Res. 2002;17:15–25. doi: 10.1359/jbmr.2002.17.1.15. [DOI] [PubMed] [Google Scholar]
- 85.Boban I, Jacquin C, Prior K, Barisic-Dujmovic T, Maye P, Clark SH, et al. The 3. 6 kb DNA fragment from the rat Col1a1 gene promoter drives the expression of genes in both osteoblast and osteoclast lineage cells. Bone. 2006;39:1302–12. doi: 10.1016/j.bone.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 86.Hauschka PV, Lian JB, Cole DE, Gundberg CM. Osteocalcin and matrix Gla protein: vitamin K-dependent proteins in bone. Physiol Rev. 1989;69:990–1047. doi: 10.1152/physrev.1989.69.3.990. [DOI] [PubMed] [Google Scholar]
- 87.Frenkel B, Capparelli C, Van Auken M, Baran D, Bryan J, Stein JL, et al. Activity of the osteocalcin promoter in skeletal sites of transgenic mice and during osteoblast differentiation in bone marrow-derived stromal cell cultures: effects of age and sex. Endocrinology. 1997;138:2109–16. doi: 10.1210/endo.138.5.5105. [DOI] [PubMed] [Google Scholar]
- 88.Quarles LD, Siddhanti SR, Medda S. Developmental regulation of osteocalcin expression in MC3T3-E1 osteoblasts: minimal role of the proximal E-box cis-acting promoter elements. J Cell Biochem. 1997;65:11–24. [PubMed] [Google Scholar]
- 89.Feng JQ, Huang H, Lu Y, Ye L, Xie Y, Tsutsui TW, et al. The Dentin matrix protein 1 (Dmp1) is specifically expressed in mineralized, but not soft, tissues during development. J Dent Res. 2003;82:776–80. doi: 10.1177/154405910308201003. [DOI] [PubMed] [Google Scholar]
- 90.George A, Sabsay B, Simonian PA, Veis A. Characterization of a novel dentin matrix acidic phosphoprotein. Implications for induction of biomineralization. J Biol Chem. 1993;268:12624–30. [PubMed] [Google Scholar]
- 91.Kalajzic I, Braut A, Guo D, Jiang X, Kronenberg MS, Mina M, et al. Dentin matrix protein 1 expression during osteoblastic differentiation, generation of an osteocyte GFP-transgene. Bone. 2004;35:74–82. doi: 10.1016/j.bone.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 92.Yang W, Lu Y, Kalajzic I, Guo D, Harris MA, Gluhak-Heinrich J, et al. Dentin matrix protein 1 gene cis-regulation: use in osteocytes to characterize local responses to mechanical loading in vitro and in vivo. J Biol Chem. 2005;280:20680–90. doi: 10.1074/jbc.M500104200. [DOI] [PubMed] [Google Scholar]
- 93.Woo SM, Rosser J, Dusevich V, Kalajzic I, Bonewald LF. Cell line IDG-SW3 replicates osteoblast-to-late-osteocyte differentiation in vitro and accelerates bone formation in vivo. J Bone Miner Res. 2011 doi: 10.1002/jbmr.465. [Electronic publication ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Takahashi S, Goldring S, Katz M, Hilsenbeck S, Williams R, Roodman GD. Down-regulation of calcitonin receptor mRNA expression by calcitonin during human osteoclast-like cell differentiation. J Clin Invest. 1995;95:167–71. doi: 10.1172/JCI117634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hayman AR, Bune AJ, Cox TM. Widespread expression of tartrate-resistant acid phosphatase (Acp 5) in the mouse embryo. J Anat. 2000;196(Pt 3):433–41. doi: 10.1046/j.1469-7580.2000.19630433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beedles KE, Sharpe PT, Wagner EF, Grigoriadis AE. A putative role for c-Fos in the pathophysiology of Paget’s disease. J Bone Miner Res. 1999;14:21–8. doi: 10.1002/jbmr.5650140206. [DOI] [PubMed] [Google Scholar]
- 97.Schwartzberg PL, Xing L, Hoffmann O, Lowell CA, Garrett L, Boyce BF, et al. Rescue of osteoclast function by transgenic expression of kinase-deficient Src in src−/− mutant mice. Genes Dev. 1997;11:2835–44. doi: 10.1101/gad.11.21.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Boyce BF, Wright K, Reddy SV, Koop BA, Story B, Devlin R, et al. Targeting simian virus 40 T antigen to the osteoclast in transgenic mice causes osteoclast tumors and transformation and apoptosis of osteoclasts. Endocrinology. 1995;136:5751–9. doi: 10.1210/endo.136.12.7588333. [DOI] [PubMed] [Google Scholar]
- 99.Drake FH, Dodds RA, James IE, Connor JR, Debouck C, Richardson S, et al. Cathepsin K, but not cathepsins B, L, or S, is abundantly expressed in human osteoclasts. J Biol Chem. 1996;271:12511–6. doi: 10.1074/jbc.271.21.12511. [DOI] [PubMed] [Google Scholar]
- 100.Rantakokko J, Aro HT, Savontaus M, Vuorio E. Mouse cathepsin K: cDNA cloning and predominant expression of the gene in osteoclasts, and in some hypertrophying chondrocytes during mouse development. FEBS Lett. 1996;393:307–13. doi: 10.1016/0014-5793(96)00907-6. [DOI] [PubMed] [Google Scholar]
- 101.Saftig P, Hunziker E, Wehmeyer O, Jones S, Boyde A, Rommerskirch W, et al. Impaired osteoclastic bone resorption leads to osteopetrosis in cathepsin-K-deficient mice. Proc Natl Acad Sci USA. 1998;95:13453–8. doi: 10.1073/pnas.95.23.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rosmarin AG, Weil SC, Rosner GL, Griffin JD, Arnaout MA, Tenen DG. Differential expression of CD11b/CD18 (Mo1) and myeloperoxidase genes during myeloid differentiation. Blood. 1989;73:131–6. [PubMed] [Google Scholar]
- 103.Fujikawa Y, Sabokbar A, Neale S, Athanasou NA. Human osteoclast formation and bone resorption by monocytes and synovial macrophages in rheumatoid arthritis. Ann Rheum Dis. 1996;55:816–22. doi: 10.1136/ard.55.11.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shalhoub V, Elliott G, Chiu L, Manoukian R, Kelley M, Hawkins N, et al. Characterization of osteoclast precursors in human blood. Br J Haematol. 2000;111:501–12. doi: 10.1046/j.1365-2141.2000.02379.x. [DOI] [PubMed] [Google Scholar]
- 105.Haegel H, Larue L, Ohsugi M, Fedorov L, Herrenknecht K, Kemler R. Lack of beta-catenin affects mouse development at gastrulation. Development. 1995;121:3529–37. doi: 10.1242/dev.121.11.3529. [DOI] [PubMed] [Google Scholar]
- 106.Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–38. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 107.Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, et al. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–87. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]