Abstract
KIR2DL4 is unique among human KIR genes in expression, cellular localization, structure, and function, yet the transcription factors required for its expression have not been identified. Using mutagenesis, electrophoretic mobility shift assay, and co-transfection assays, we identified two redundant Runx binding sites in the 2DL4 promoter as essential for constitutive 2DL4 transcription, with contributions by a CRE site and initiator elements. IL-2-and IL-15-stimulated human NK cell lines increased 2DL4 promoter activity, which required functional Runx, CRE, and Ets sites. Chromatin immunoprecipitation experiments show that Runx3 and Ets1 bind the 2DL4 promoter in situ. 2DL4 promoter activity had similar transcription factor requirements in T cells. Runx, CRE, and Ets binding motifs are present in 2DL4 promoters from across primate species, but other postulated transcription factor binding sites are not preserved. Differences between 2DL4 and clonally-restricted KIR promoters suggest a model that explains the unique 2DL4 expression pattern in human NK cells.
Keywords: Human, Natural Killer Cells, Cell Activation and Differentiation, Gene Regulation, Transcription Factors
INTRODUCTION
NK lymphocytes use MHC class I-specific clonally-restricted KIR (crKIR) receptors to distinguish healthy and aberrant cells in immunity to intracellular pathogens and in cancer surveillance (1). Inhibitory crKIR have cytoplasmic tails that contain ITIM motifs, which recruit SHP-1 and SHP-2. These proteins dephosphorylate Vav1 and disrupt actin polymerization needed for signaling from a variety of activating receptors (2). Activating crKIR are missing the ITIM motif because they have shortened cytoplasmic tails, but have a transmembrane region that contain a basic residue that recruits ITAM-containing proteins, which generate activating signals (1). In contrast to crKIR, the function of KIR2DL4 is less clear. However, 2DL4 is likely very important because of all KIR, 2DL4 is the only KIR gene with orthologs present in all hominid species and in old world monkeys, such as the rhesus monkey (3).
Compared with crKIR, 2DL4 has unusual expression, cellular localization, structure, and function. Although both crKIR and 2DL4 expression is controlled by DNA methylation, crKIR expression is allele-specific while 2DL4 is bi-allelically expressed by all NK cells (4–9). crKIR expression is largely absent from immature CD56bright NK cells and is found on more mature CD56dim NK cells. By comparison, KIR2DL4 is expressed by CD56bright NK cells and expression declines with further NK cell maturation (10–12). Unlike most crKIR, 2DL4 is poorly expressed on the cell surface, but is strongly expressed in early endosomes (13). KIR2DL4 has both a cytoplasmic ITIM motif and a transmembrane arginine that associates with the activating effector molecule, FcεR1γ (14). Despite these dual activating and inhibitory structural features, 2DL4 usually transduces activating signals (10, 15). Distinct from activating crKIR, 2DL4 ligation weakly stimulates cytotoxicity (10, 16). Instead, 2DL4 ligation stimulates NK cells to produce a unique repertoire of inflammatory and angiogenic cytokines and chemokines, including IFN-γ, IL-6, IL-8, and IL-23 (13). The classical MHC class I (HLA-A, B, C) ligands for crKIR are widely expressed. In contrast, 2DL4 recognizes HLA-G, a non-classical MHC class I protein that is expressed by invasive fetal trophoblast cells, but few other cells (13, 17–19).
NK cells dominate the uterine leukocyte population during the early stages of pregnancy, where they may be involved in uterine tissue remodeling (20). Fetal trophoblast cells invade the uterine mucosa where they encounter maternal NK cells. These uterine NK cells secrete inflammatory and angiogenic factors that encourage trophoblast cell growth, differentiation, and migration, and spiral artery remodeling, all of which are important functions for successful pregnancy (20). Recent data suggests that 2DL4 may play a role in this process. Ligation of decidual NK cell 2DL4 with mAb or HLA-G induced production of IL-1β, IL-6, TNF-α, and IL-8 inflammatory cytokines and chemokine, which have been hypothesized to promote uterine remodeling (19). KIR2DL4 expression is not absolutely required for successful pregnancy, perhaps because other leukocyte receptors fill redundant functions (19).
In addition to sense transcription from the proximal promoter, KIR transcription may be regulated by more complex mechanisms. Several KIR genes are transcribed from distal promoters that are upstream of the more proximal promoters, including a promoter ~ 10 kb from the 2DL4 translation start site (21). Furthermore, both crKIR and 2DL4 proximal promoters are transcribed in both sense and antisense directions (22). However, the importance of these transcripts in KIR gene expression regulation has not been firmly established. Focusing on the proximal promoter, we reported that sense transcription of the 3DL1 crKIR gene depends on five different transcription factors, each of which make a small contribution to full promoter activity (23). The 2DL4 promoter has about 60% sequence identity to the crKIR promoters, including homologous transcription factor binding sites, suggesting both shared and distinct transcriptional control mechanisms (24). A previous report suggested that Runx factors inhibit 2DL4 expression (25), but stimulatory transcription factors have not been identified. Given the unique role of 2DL4 in NK cell biology, we performed a systematic, unbiased study of the 2DL4 promoter. We demonstrate that Runx transcription factors are absolutely required to stimulate 2DL4 expression. Our data also provide a plausible mechanism for differential 2DL4 and crKIR expression in NK cell development.
MATERIALS and METHODS
Cells and constructs
Human YT-HY cells (hereafter referred to as YT) were cultured as described (23). Human NKL cells were grown in suspension in RPMI 1640 (Invitrogen, Lonza) + 10% FBS (Hyclone) + 2 mM glutamine (Invitrogen) + 1 mM pyruvate (Invitrogen) + 200 U/mL IL-2 (NIH). Mouse LNK cells were grown as monolayers in RPMI 1640 + 10% FBS + 50 μM 2-mercaptoethanol (Invitrogen), nonessential amino acids (Invitrogen), + 20 mM HEPES (Invitrogen) + 200 U/mL IL-2. Human Jurkat, Hut-78, and CEM-T4 cells were grown in RPMI 1640 + 10% FBS. Human NK 92.26.5 cells were grown as previously described (26). Primary NK cells were enriched from peripheral blood by incubating them with antibody complexes bispecific for erythrocyte glycophorin A and leukocyte CD3, CD4, CD19, CD36, and CD66b (RosetteSep™; StemCell Technologies Inc.). Erythrocyte-leukocyte rosettes were removed by density gradient centrifugation through Lymphoprep™ (Axis-Shield); flow cytometry indicated that > 90% of enriched cells were NK cells. The 2DL4 promoter pGL3 reporter plasmid has been described (24) and includes bases −7 to −269 bp upstream of the 2DL4 ATG translational start site. We performed PCR-based mutagenesis on this plasmid essentially as described, including substitution of 24 consecutive 10 bp segments beginning 10 bp upstream of the ATG site (23). Putative transcription factor binding sites were identified using the TESS computer algorithm (http://www.cbil.upenn.edu/cgi-bin/tess/tess). Human Ets1, Ets2 expression constructs and the pSG5 parent vector were generous gifts of Dennis Watson (Medical University of South Carolina, Charleston, SC, USA). Human Runx1, Runx2, Runx3, and the mouse CBF beta expressing constructs in the pEF-Bos vector were generous gifts of Yoshiaki Ito (National University of Singapore). Surface 2DL4 was detected in flow cytometry with a PE-labeled mAb (347005 PE, Biolegend).
EMSA
YT cell nuclear extracts were prepared and EMSA was carried out as described (23). The Ab used for supershift studies were from Santa Cruz Biotechnology, CREB (186X), ATF-1 (270X), and Runx1 (8584X), or from Active Motif, Runx 2 (AML-3, 39302), Runx3 (AML-2, 39301).
Transient transfection
YT and HeLa cells were transfected as described (23). Other cells were transfected by electroporation using plasmid “midipreps” (Bio-Rad), including LNK (3 × 106 trypsinized cells, 5.0 μg luciferase plasmid). All other electroporation conditions included 20.0 μg luciferase plasmid with the indicated number of cells: NKL (20 × 106); Jurkat, Hut-78 and CEM-T4 (7.5 × 106). All electroporations included 100 ng CMV-Renilla or SV40-Renilla control plasmids and were conducted in a Bio-Rad Gene Pulser II (250 V, 600 μF) with a 2 mmgap cuvette (USA Scientific) in 400 μl conditioned media (including serum), with the exception of CEM-T4 cells, which were transfected in fresh RPMI 1640 without serum. After electroporation, cells were incubated in 50% fresh/50% conditioned media. Cell lysates were prepared ~40 h after electroporation and luciferase and Renilla activity was measured as described (23). Where indicated, at 24 and 12 h before harvest, IL-2 (40 and 20 U/ml, respectively) or IL-15 (10 ng/ml each time) was added to YT transfectants. Luciferase activity was divided by Renilla activity and this value was normalized to the value for the wild type promoter vector.
ChIP
Chromatin isolation and transcription factor immunoprecipitation was carried out using the EZ ChIP Protocol (Millipore) with modifications. 80 × 106 NK 92.26.5 cells [or 15 – 20 × 106 primary NK cells—values within brackets below refer to primary NK cells] were resuspended in 40 [10] mL NK-92 media (26) and treated with 1% formaldehyde (freshly prepared as an 18.5% stock solution from paraformaldehyde (Fisher) using the instructions provided in the kit) for 10 min at room temperature; the reaction was terminated by incubating on ice for 5 min after adjustment to 0.125 M glycine, 1 × EDTA-free Roche complete protease inhibitor cocktail (PIC), and 0.4% IPA saturated with PMSF (Sigma). All subsequent steps were carried out at 4 °C: Cells were washed twice with 40 [10] mL of PBS containing 0.5 [1.0] × PIC and 0.4 % IPA/PMSF. The cell pellet was next resuspended in 1.6 [1.0] mL of Swelling Buffer (100 mM Tris, 10 mM KCl, 15 mM MgCl2, 0.5% NP-40, pH 8.0) containing 1 × PIC and 0.4% IPA/PMSF. After 10 min, cells were sheared with 5 strokes from a glass Dounce homogenizer, centrifuged at 2500 × g for 5 min, and the supernatant was removed. The cell pellet was then resuspended in 600 [200] μl of Lysis Buffer (EZ-ChIP kit) with 1 × PIC and 0.4% IPA/PMSF, and 200 [50] μl of glass beads (106 microns, Sigma-Aldrich) was added. Chromatin was fragmented to an average size of 500 bp by sonicating (Branson 450 Sonifier) for 15 pulses (10 s each at 30% of the microtip limit with 1 min breaks), and then centrifuged at 14,000 × g for 1 h. 600 [200] μl supernatant was resuspended to a final volume of 6.0 [2.0] mL in ChIP Dilution Buffer (EZ-ChIP kit) with 1 × PIC and 0.4% IPA/PMSF. 600 [150] μl of Protein A agarose/salmon sperm DNA solution (Millipore) was added and the mixture was gently rocked for 2.5 h. The resin was removed by centrifugation and the chromatin was split into eight 720 μl [two 950 μl] aliquots, with extra chromatin saved as Input. 3.0 [2.0] μg of Ab were added to individual aliquots, including mouse anti-Runx3 (clone 5G4, D235-3 and clone 6E9, D234-3, from MBL), and several Ab from Santa Cruz Biotech: mouse anti-FLAG (G-8, 166384), rabbit anti-Ets1 (C-20, 350X), rabbit anti-FLAG (D-8, 807), and normal rabbit IgG (2027). After overnight incubation with rotation, 9.0 [6.0] μg of rabbit anti-mouse H + L chain secondary Ab (Millipore, m-06-371) was added to the aliquots with mouse Ab, and incubated with rotation for 1.5 h. The samples were centrifuged at 13,000 × g for 30 min to remove non-specific precipitates, and 60 [45] μl of protein A agarose was added to the supernatants and incubated with rotation for 1 h. The samples were centrifuged for 15 min at 6000 × g to pellet the resin, and the resin was washed once with 1 mL of buffers of increasing ionic strength, for 5 min: Low Salt Immune Complex Buffer, High Salt Immune Complex Buffer, LiCl Immune complex Buffer (EZ ChIP kit), and then 2 washes of TE Buffer. Chromatin bound to the resin was eluted with two 100 μl washes of room temperature elution buffer (0.1 M sodium bicarbonate + 1.0% SDS, pH 9.0). Crosslinks were released overnight at 65 °C. Protein was digested as described in the protocol and DNA was purified using QIAquick Spin (Qiagen) columns. Quantitative PCR was carried out using 1× SYBR Green JumpStart Taq Ready mix (Sigma) or SensiFAST SYBR Hi-Rox Kit (Bioline) on an ABI 7000 qPCR system or a Bio-Rad CFX96 Real-Time System. The primer sequences used were: 2DL4 promoter, sense: AATACATCAAATTTCCTCATGTGA, antisense: TCTGCTGCCAGGACGCAGTGA at 200 nM final concentration; 2DL4 Intron 4, sense: GTCACAGGTGAGGAAAGCCAAT, antisense: CCATGCTGCATCTTCTATCCA (200 nM), KIR3DL1F, GTGAAGGACGC GAGGTGTCAATTCTAGTGACAG and KIR3DL1R, ACCTCTAGGC CCATATCTTTACCTCCAAGT (250 nM, underline bases indicate mismatches designed to improve specificity). For the 2DL4 promoter and intron 4, an initial denaturation step at 94 °C for 2 min, was followed by cycles of 15 s at 94 °C, 1 min at 63 °C, 1 min at 72 °C. For the 3DL1 promoter, the program was 95 °C for 2 min, and then cycles of 10 s at 95 °C, 10 s at 63 °C, 40 s at 72 °C The quantity of DNA in the samples was calculated from a 4 or 5 point standard curve generated from dilution from the Input sample; this standard curve typically indicated a ~ 100% amplification efficiency with r2 > 0.95. In each case, agarose gels indicated a single product of the expected size.
Statistical testing
F-statistics indicated that luciferase and ChIP data showed equivalent variances after logarithmic transformation, allowing parametric testing. Data shown in Figures 2, 3A, 4A, 7, and 8 were tested using paired, two-tailed t tests. Data shown in figure 6 were tested using unpaired, two-tailed t tests. P values are listed in the figures and figure legends. All error bars represent SEM.
Figure 2.
A. YT cells express high levels of 2DL4. YT cells were treated either with a PE-labeled mAb specific for 2DL4 (solid line) or with an isotype control (shaded) and surface staining was detected by flow cytometry. B. Linker-scanning mutagenesis identifies few 2DL4 promoter cis-acting sites. YT cells were transfected with the indicated 2DL4 substitution, numbered as in Fig. 1. WT, wild type. B, background level produced by the empty pGL3-basic vector. Values represent averages from tests of 3–8 different plasmid preparations (each measured in duplicate). Asterisks indicate significant differences from WT (p < 0.05).
Figure 3.
An unmethylated CRE site contributes to 2DL4 promoter activity. A. 2DL4 promoter activity is reduced by mutations affecting the CRE site. Shown is 2DL4 promoter activity without (WT) and with CRE site point mutations (CRE1 and CRE2, see Table II). Promoter activity was measured as described in Fig. 2, and asterisks indicate significant differences from WT (p < 0.01). B. CpG methylation diminishes 2DL4 promoter DNA binding to CREB and ATF-1 in EMSA. YT nuclear extract was incubated with an unmethylated (Wt CRE, lanes 1–6) or a CpG methylated (Me CRE, lanes 7–12) probe encompassing the CRE site, either alone (lanes 1, 7) or with 150-fold excess of self-competitor (Wt, lanes 2, 8) or competitor with a mutated CRE site (M (CREm2, Table I), lanes 3, 9). Alternatively, nuclear extracts were pre-incubated with non-specific rabbit IgG (Ig, lanes 4, 10), or with Ab to CREB (Cb, lanes 5, 11) or to ATF-1 (A, lanes 6, 12) as indicated. The arrows indicate supershifted bands. The experiment shown is representative of three independent experiments with similar results.
Figure 4.
2DL4 promoter activity depends on redundant activating sites. A. The activities of single site promoter mutations in Ets, distal Runx (dRx1, dRx2, dRx3), and proximal Runx (pRx) sites (described in Table II), and combinations of these mutations are shown, along with empty vector pGL3 Basic background activity (B). Promoter activity was measured as described in Fig. 2. Asterisks indicate significant differences from WT (*, p < 0.005; ** p < 0.0005). B. The 2DL4 Ets site is functional. HeLa cells were transfected with 1 ng of the control SV-40 Renilla luciferase construct, either 0, 200, or 400 ng Ets1 (solid square) or Ets2 (open square) expression plasmids and 1.5 ug 2DL4 promoter reporter that was either wild type (solid line) or mutated at the Ets site (dashed line). Empty pSG5 plasmid DNA was added as needed to equalize the total amount of transfected DNA. Each point represents the average of five tests; each test had a separate reporter plasmid preparation and one of three different Ets expression plasmid preparations. Shown is one representative experiment of three with similar results. Error bars (not always visible) show within-experiment SEM.
Figure 7.
CRE, Ets, and Runx sites are required for full 2DL4 promoter activity in NK and T cells. YT (black), LNK (dark gray), NKL (light gray), and Hut-78 (white) cell lines were transfected with 2DL4 reporter plasmids with mutations to either the CRE (CRE2), Ets (Ets), proximal Runx (pRx), distal Runx (dRx1), or combined distal and proximal Runx mutations (dRx1 + pRx, mutations described in Table II), and were cultured with IL-2 (with the exception of Hut-78) as described in Methods. Promoter activity was measured as described in Fig. 2. With the exception of CRE2 and dRx1 for NKL, all mutations had significant declines in activity compared to wild type (p < 0.05). WT and B are described in Fig. 2.
Figure 8.
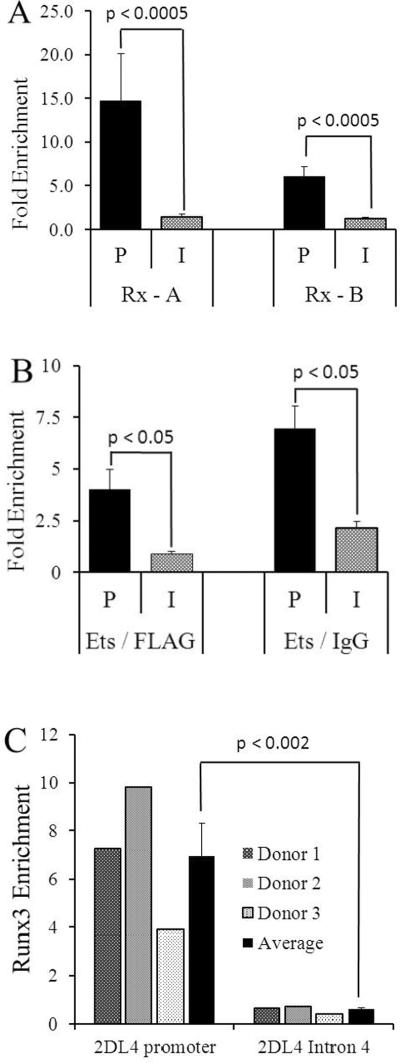
Runx3 and Ets1 specifically bind to the endogenous 2DL4 promoter. Cross-linked chromatin was purified from IL-2-cultured NK92.26.5 (A, B) or freshly-isolated primary NK (C) cells and immunoprecipitated with Ab to Runx3, Ets1, or with negative control Ab. DNA from immunoprecipitates was purified and qPCR amplified using primers specific either to the 2DL4 promoter (P) or 2DL4 intron 4 (I), as indicated. A. ChIP was performed with anti-Runx3 mAb 5G4 (Rx-A) and 6E9 (Rx-B), and negative control mAb (anti-FLAG). Values represent the average enrichment of DNA bound to specific Ab (Runx/FLAG) from tests of five different chromatin preparations. B. ChIP was performed with anti-Ets1 Ab and negative control Ab, either rabbit anti-FLAG or normal rabbit IgG. Values represent the average enrichment of DNA bound to Ab (Ets/FLAG or Ets/IgG) from tests of at least three different chromatin preparations. C. ChIP was performed with anti-Runx3 mAb 5G4 and a negative control mAb (anti-FLAG). Values represent the enrichment of DNA bound to specific Ab (Runx 5G4/FLAG) using chromatin preparations from three different human donors. Averages and p values are shown.
Figure 6.
Distinct 2DL4 promoter transcription factor requirements in constitutive and cytokine-treated cells. A. IL-2 treatment activates Segment 5 and 6 sites. YT cells transfected with 2DL4 segment substitution mutant reporter plasmids were either IL-2 treated or not treated (Nil), as described in Methods. Promoter activity was measured as described in Fig. 2. Asterisks indicate significant differences between the indicated groups (*, p < 0.0005). WT and B are described in Fig. B. IL-2 and IL-15 specifically activate the compound Ets/Runx site. YT cells transfected with 2DL4 promoter reporter plasmids with mutations in either the proximal Runx (pRx), Ets (Ets), proximal Runx and Ets site (pRx + Ets), or in the distal Runx site (dRx1, dRx2, dRx3, mutations described in Table II) were treated with IL-2, IL-15, or not treated (Nil) as described in Methods. Promoter activity was measured as described in Fig. 2. Asterisks indicate significant differences between the indicated groups (*, p < 0.05; ** p < 0.0005). WT and B are described in Fig. 2.
RESULTS
Systematic dissection of the 2DL4 promoter reveals few important cis-acting elements
KIR3DL1, a model crKIR gene, contains five promoter elements that are required for optimal transcription in NK cells: CRE, Ets, Runx, and Sp1 sites and an overlapping STAT/Ets/YY1 site (Fig. 1). Each of these elements made a relatively weak contribution to 3DL1 promoter activity, but mutation of all five sites abrogated 3DL1 promoter activity (23). Of these elements, only the Ets and Runx sites are also present at homologous 2DL4 locations (Fig. 1). Juxtaposed to the Ets site in the 2DL4 promoter is a second Runx site that is not found in crKIR promoters. Although the 2DL4 promoter does not contain the CRE site that is found in crKIR promoters, an inverted CRE site is present nearby (Fig. 1). This suggested that 2DL4 and crKIR promoters are activated by both shared and distinct transcription factors, as predicted by DNase I footprinting (24).
Figure 1.
3DL1 and 2DL4 promoters share overlapping but distinct sets of potential cis-acting elements. Shown are aligned 3DL1 (top) and 2DL4 (bottom) promoter sequences. Twenty-four contiguous 2DL4 10-bp segments (denoted S1–S24) were replaced with the linker sequence, GCAGATCCGC. Putative cis-acting elements are denoted by boxes. The sequences end at the ATG translational start site.
Because there has been limited testing of the transcription factors that drive 2DL4 expression, we directly tested the 2DL4 promoter. Using the unbiased linker scanning mutagenesis method, we replaced twenty-four 10-bp segments with a sequence that does not contain a known transcription factor binding motif (27). In contrast to deletions, substitutions do not affect promoter activity via phasing changes of transcription factor binding sites on the DNA helix (28, 29). We tested mutants in the YT NK cell line, which expresses endogenous 2DL4 (Fig. 2A and (30)) and in which expression of the 2DL4 promoter reporter is more than 20-fold greater than the reporter plasmid without insert (pGL3-Basic, Fig. 2B). Substitution of Segments 7, 10 and 12 increased promoter activity (Fig. 2B). The increased promoter activity could be due to removal of repressive elements or creation of new activating sites at the junction of the linker and the 2DL4 sequence (data not shown). In contrast, substitutions of Segments 1, 3 and 13 each significantly decreased 2DL4 promoter activity (about 50% for Segments 1 and 13). Because activating factors have not yet been identified for the 2DL4 promoter, we focused our investigation on the regions where substitution reduced promoter activity.
A CRE element is required for optimal 2DL4 promoter activity
Because 2DL4 Segment 13 contained a predicted CRE site (Fig. 1), we targeted this site with two point mutations. Both CRE mutations substantially decreased 2DL4 promoter activity (Fig. 3A). To further test the importance of the CRE site, we performed EMSA. YT NK cell nuclear factors formed several bands with the 2DL4 CRE probe and were specifically competed by unlabeled self oligonucleotide, but not by self oligonucleotide that contained a mutated CRE site (Fig. 3B). Furthermore, the nuclear complexes were supershifted by Ab to CRE-binding transcription factors, CREB and ATF-1. Thus, mutagenesis and EMSA studies indicate that optimal 2DL4 transcription, like crKIR transcription, requires a CRE site. Because 2DL4 and crKIR expression is controlled by DNA methylation (4–9), we tested CREB and ATF binding to a methylated 2DL4 probe. As expected from other gene studies (31), CREB and ATF-containing nuclear complexes poorly bound to the methylated 2DL4 probe. This result suggests that DNA methylation inhibits 2DL4 transcription, in part, by inhibiting CREB and ATF binding.
2DL4 core promoter elements
Substitution of Segment 1 significantly inhibited 2DL4 promoter activity. Because the TESS transcription factor binding program did not predict any high probability transcription factor binding sites in this region (32) and because 2DL4 transcription start sites are upstream of Segment 1 (24, 33), it was possible that Segment 1 contained elements of the core 2DL4 promoter. Core promoters can be classified into distinct types, depending on the sequence recognized by the general transcription factor TFIID of the RNA polymerase II holoenzyme complex, including TATA and Initiator (Inr) sequences. Most (8 of 9) crKIR promoters contain a TFIIB recognition element (BRE) and an adjacent A-rich sequence (AAATAAC) that has been postulated to act as a variant TATA element ((24), Fig. 1, and data not shown). The 2DL4 promoter does not contain the BRE motif or the A-rich sequence (Fig. 1). Instead, 2DL4 transcription often initiates at or near potential Inr sequences in Segments 2–4 (Table III), with putative initiating adenine residues at −28, −32, −37, and −42 relative to the ATG translation start site (24, 33). The weak effect of substitutions in Segments 2, 3, and 4 is consistent with multiple Inr sites, each contributing to 2DL4 activity (Fig. 2). To test this hypothesis, we made point mutations in the putative Inr elements. 2DL4 promoter activity was decreased by 47% by the InrA mutation, which replaced initiator adenines of the four putative Inr sites (Table III). Mutation of adenine in a consensus Inr sequence may not reduce promoter function because alternative sites can substitute for mutated Inr sites (34, 35), and so we mutated additional bp surrounding the initiating adenines (InrB). This mutation decreased 2DL4 promoter activity by 60% (Table III). Statistically significant and substantial reduction, but not elimination, of 2DL4 promoter activity by the InrA and InrB mutations is consistent with an important role for Inr elements, but suggest involvement of other initiating sites. Indeed, 2DL4 transcription often initiates from sites distinct from the Inr elements (24, 33). We conclude that 2DL4 has functional Inr elements that contribute to promoter activity.
Table III.
Inr Mutations Decrease 2DL4 Promoter Activity
| Construct | Sequence | Activity (SEM) |
|---|---|---|
| WT | tcAgtcGagccgagtcActGc | 1.0 |
| InrA | tcGgtcgGgccgGgtcGctgc | 0.53 (0.045)* |
| InrB | tcGgCGgTgccgagtcGcGgc | 0.40 (0.021)* |
Shown are 2DL4 promoter sequences −44 to −24 with initiating adenine residues of putative Inr sites (YYANWYY motif) underlined and major transcription start sites identified by Radeloff et al (33) are shown in capital letters in the wild type (WT) sequence. The InrA and InrB substitutions are indicated by capital letters. The relative promoter activity represents averages from tests of at least 10 different plasmid preparations (with SEM).
Significantly different from WT, p <1 × 10−5.
To investigate the hypothesis that crKIR promoters contain a functional TATA box, we purified recombinant human TBP and tested whether it would shift oligonucleotides containing either the 2DL4 or 3DL1 core promoter sequence, as visualized by EMSA. Although purified human TBP shifted the well-characterized adenovirus major late promoter TBP positive control (AdMLP, Table I), TBP did not shift a TBP negative control probe (36) or either of the KIR probes tested (Supplemental Fig. S1A). Addition of human TFIIB to the incubation also failed to result in a shift (Supplemental Fig. S1A). Although the 2DL4 and crKIR core promoters are distinct, our results do not support the hypothesis that crKIR promoters contain a functional TATA box.
Table I.
2DL4 EMSA Probes and Competitors
| Reagent | Oligonucleotide Sequence |
|---|---|
| Wt CRE | GGA CCT CAT ATG ACG TAG AAG AAG CC |
| Me CRE | GGA CCT CAT ATG ACG TAG AAG AAG CC |
| CRE m2 | GGA CCT CAT ATA ATG TAG AAG AAG CC |
| 3DL1 A | GGC TCC CAT GAT GTG GTC AAC ATG TAA |
| Mut 3DL1 A | GGC TCC CAT GAA CTA GTC AAC ATG TAA |
| 2DL4 A | GGT TCA CAT GTT GTG GTC AAT GTG TCA A |
| Mut 2DL4 A | GGT TCA CAT GTA CTT GTC AAT GTG TCA A |
| 3DL1 B | GGC GCC AAA TAA CAT CCT GTG CGC TGC TG |
| 2DL4 B | GGC CCC TCA CCA CAT CCT CTG CAC CGG TC |
| Mut 2DL4 B | GGC CCC TCA CTA CAT CCT CTG CAC CGG TC |
| AdmLP | GGC TGA AGG GGG GCT ATA AAA GGG GGT GGG GG |
| TBP negative control | GGG CTG CGC CGG CTG TCA CGC CAG GCT GCG CC |
| 3DL1 core | GGG GGC AGG GCG CCA AAT AAC ATC CTG TGC GC |
| 2DL4 core | GGG ATC CGG GCC CCT CAC CAC ATC CTC TGC AC |
| 2DL4 BTE | GGG ATC CGG GCG CCA AAT AAC ATC CTC TGC AC |
Underling denotes mutated bases, bold lettering denotes 5-methylcytosine.
We considered whether the Segment 1 substitution decreased 2DL4 promoter activity by eliminating other putative elements of the core promoter, such as elements with sequence homology to a Motif 10 Element and a Downstream Core Element (35). However, mutagenesis studies did not show evidence for the function of these sites (Supplemental Fig. S1B) and the role of Segment 1 in 2DL4 gene expression remains to be defined.
Runx/Ets interaction in the 2DL4 promoter
3DL1 promoter activity was affected by mutations in the Ets and Runx sites (23, 37). Both of these sites are preserved at homologous locations in the 2DL4 promoter (Fig. 1), so we expected that they would be required for optimal 2DL4 transcription. Surprisingly, 2DL4 promoter activity was not decreased by substitutions of Segment 5, which contained the Ets site, or of Segment 9, which contained the distal Runx site, homologous to the 3DL1 site (Fig. 2). We then further investigated the importance of the Ets and the two 2DL4 Runx sites. A 2-bp substitution targeting the Ets site that had diminished 3DL1 promoter activity by more than 60% did not decrease 2DL4 promoter activity (Fig.4A). Similarly, three mutations of the distal Runx site that had diminished 3DL1 promoter activity by 35–45% produced little or no decrease in 2DL4 promoter activity. We also tested the Segment 5 proximal Runx site that did not have a homologous site in crKIR promoters. A 2-bp substitution that replaced a GG dinucleotide present in nearly all Runx sites and a second 1-bp Runx substitution did not significantly reduce promoter activity (Fig. 4A and data not shown). Therefore, with the exception of the CRE site, none of the putative transcription factor binding sites shared with crKIR promoters appeared to be critical for 2DL4 promoter activity. We hypothesized that the Ets site interacts with the juxtaposed proximal Runx site, and that mutation of both sites in the same construct maybe required to reveal an interaction. Combination of the Ets mutation and the proximal Runx mutation reduced 2DL4 promoter activity by 40% (Fig. 4A). The synergy between the mutations suggests that Runx and Ets family proteins interact at the compound proximal Runx/Ets site.
Given the ability of the Ets site to synergize with the proximal Runx site, we wished to further investigate whether Ets family members were involved. We had previously shown that YT NK nuclear complexes containing Ets family members, GABP and Elf1, bound the 3DL1 Ets site in EMSA (23). Although YT nuclear proteins bound the Ets probe and were competed by mutant, but not Ets-mutant competitor, neither GABP nor Elf1 binding was shown by Ab supershift (results not shown). This suggested that other Ets family members contribute to 2DL4 transcription. To investigate possible roles for Ets1 or Ets2, which are poorly detected in EMSA, we performed transactivation studies in HeLa cells, which are deficient in several lymphoid specific Ets factors (38, 39). We co-transfected HeLa cells with plasmids encoding either Ets1 or Ets2 together with the wild-type or Ets-site mutated 2DL4 promoter reporter plasmids. Both Ets1 and Ets2 increased transcription from the intact 2DL4 promoter relative to the Ets-mutated promoter (Fig. 4B). Ets1 and Ets2 transactivation of 2DL4 was at least as great as that of 3DL1 (data not shown). This indicated that the 2DL4 Ets site is functional and responsive to both Ets1 and Ets2, although it does not rule out a potential role for other Ets family members.
Runx transcription factors are critical for 2DL4 promoter activity
We tested possible redundancy of the two Runx sites in the 2DL4 promoter. Combined mutation of both proximal and distal Runx sites reduced luciferase activity nearly to that of the pGL3-Basic empty vector (Fig. 4A). These results suggest that Runx transcription factors, acting at either of the two promoter sites, are absolutely required for 2DL4 transcription in NK cells. To further explore a role for the two Runx sites, we tested the ability of YT NK cell nuclear proteins to bind to proximal and distal Runx probes, comparing 2DL4 probes to probes from homologous regions of the 3DL1 promoter (Fig. 5A). Several YT NK cell nuclear protein complexes bound the 2DL4 and 3DL1 distal Runx probes in EMSA (Fig. 5B) and were competed by unlabeled self oligonucleotides, but not by Runx-mutated oligonucleotides. Ab specific for Runx2 and Runx3, but not Runx1, supershifted both 2DL4 and 3DL1 (Fig. 5B). These results indicate that YT NK cell Runx2 and Runx3 proteins bind the homologous distal Runx sites in the 2DL4 and 3DL1 promoters, regardless of non-identical flanking sequences. Runx2 and Runx3 also bound the 2DL4 proximal Runx probe (Fig. 5C). As expected, Runx proteins did not bind a homologous 3DL1 probe that did not contain a Runx motif (Fig. 5C). Therefore, the 2DL4 promoter differs from crKIR promoters in having two functional Runx sites. To test which Runx transcription factors can activate the 2DL4 promoter, we investigated 2DL4 promoter activity in HeLa cells, which do not express endogenous Runx factors (40). We co-transfected intact and Runx-mutated 2DL4 promoter plasmids with expression plasmids for Runx1, Runx2, or Runx3, together with CBFβ, the Runx heterodimer partner protein (41). All three Runx factors greatly stimulated transcription from the 2DL4 promoter, and Runx transactivation was substantially lower with Runx-mutated 2DL4 promoters (Fig. 5D). These results show that Runx factors act directly on the 2DL4 promoter. Interestingly, mutation of either the proximal or distal Runx site significantly decreased Runx transactivation, although mutation of both sites was even more destructive. These results reinforce mutation and EMSA findings, showing that Runx transcription factors are critical for 2DL4 promoter activity.
Figure 5.
Runx transcription factor family members bind the 2DL4 promoter at two functional sites. A. Shown are sequences surrounding the 2DL4 distal (left) and proximal (right) Runx sites and the aligned 3DL1 promoter sequences. Boxes denote Runx motifs and the sequences shown denote EMSA probes. B. Runx2 and Runx3 bind to 2DL4 and 3DL1 distal Runx sites. YT nuclear extract was incubated with either 3DL1 (3DL1 A, lanes 1–7) or 2DL4 (2DL4 A, lanes 8–14) probes encompassing the distal Runx site either alone (lanes 1, 8) or in the presence of 150-fold excess of self-competitor (Wt, lanes 2, 9) or competitor with a mutated Runx site (M, lanes 3 and 10 (mut 3DL1 A, mut 2DL4 A, Table I)). Alternatively, probe was added to nuclear extracts that had been pre-incubated with non-specific rabbit IgG (Ig, lanes 4, 11), or with Ab to Runx1 (1, lanes 5, 12), Runx2 (2, lanes 6, 13) or Runx3 (3, lanes 7, 14) as indicated. Arrows indicate supershifted bands. C. Runx2 and Runx3 bind to the 2DL4 proximal Runx site but not to the 3DL1 aligned region. YT nuclear extract was incubated with either a 3DL1 (3DL1 B, lanes 1–6) or 2DL4 (2DL4 B, lanes 7–13) probe encompassing the proximal Runx site (2DL4) or the aligned 3DL1 sequence alone (lanes 1, 7) or in the presence of 150-fold excess of self-competitor (Wt, lanes 2, 8) or a competitor with a mutated proximal Runx site (M (mut 2DL4 B, Table I) lane 9). Alternatively, nuclear extracts were pre-incubated with IgG (Ig, lanes 3, 10), or with Ab to Runx1 (1, lanes 4, 11), Runx2 (2, lanes 5, 12) or Runx3 (3, lanes 6, 13) as indicated. For B. and C., the experiments shown are representative of three independent experiments with similar results. D. Both proximal and distal Runx sites are functional. HeLa cells were transfected with 1 ng of a control SV-40 Renilla luciferase construct, 500 ng CBFβ expression plasmid, 500 ng Runx1, Runx2, or Runx3 expression plasmids as indicated, and 1.5 ug 2DL4 promoter reporter that was either wild type (WT), mutated at the proximal Runx site (pRx), at the distal Runx site (dRx1), or at both Runx sites (pRx + dRx1, mutations described in Table II). Each group represents the average of five tests; each test had a separate reporter plasmid preparation and one of three different Runx1, Runx2 or Runx3 expression plasmid preparations. Error bars denote within-experiment SEM. Shown is one representative experiment of two with similar results.
DNase I protection experiments (24) and our own TESS search had suggested additional 2DL4 promoter cis-acting sites: CP2, myogenic differentiation antigen 1, AP-1, interferon regulatory factor 2, and GATA-3 sites located in Segments 4, 4, 16, 21, and 21, respectively. We found that mutation of these specific sites, produced little or no reduction in 2DL4 promoter activity (Supplemental Fig. S1C).
Distinct transcription factors act on the 2DL4 promoter in IL-2/15-treated lymphocytes
NK cells require IL-15 signals for maturation, proliferation and survival. Moreover, 2DL4 expression is relatively high in immature CD56bright NK cells that have recently developed from CD122+ NK precursor cells under the influence of IL-2 and IL-15 in the lymph node and the bone marrow, respectively, and NK cell 2DL4 levels increase with cytokine stimulation (10–12, 42). To test whether the 2DL4 promoter uses distinct transcription factors in the presence of cytokines, we used YT NK cells that respond to both IL-2 and IL-15 ((43) and data not shown). The 2DL4 promoter was 1.55-fold more active with IL-2 stimulation (data not shown). Comparing wild type and mutated KIR promoters under the same cytokine condition, the 24 3DL1 segment substitutions showed little or no relative change in promoter activity in the presence vs. the absence of IL-2 (Supplemental Fig. S1D). For the 2DL4 promoter, 22 of 24 segment substitutions showed little or no IL-2 effect, including Segment 9 (distal Runx) and Segment 13 (CRE). However, substitutions of Segment 5 (Ets) and Segment 6 (proximal Runx) significantly reduced relative transcription only when tested in IL-2-treated YT cells (Fig. 6A).
To confirm that the compound Ets and proximal Runx site is important in the context of cytokine treatment, we tested point mutations in either IL-2- or IL-15-treated YT cells. Consistent with the Segment 5 substitution data, the Ets mutation strongly reduced 2DL4 promoter activity only in the presence of IL-2 or IL-15 treatment (Fig. 6B). This indicates that Ets transcription factors are much more important for 2DL4 transcription in the presence of IL-2/15 than constitutive 2DL4 transcription in YT NK cells. Mutation of the proximal Runx site also reduced 2DL4 promoter response to IL-15 (Fig. 6B). As before, the combination of the Ets mutation and the proximal Runx mutation reduced constitutive 2DL4 promoter activity, which was relatively more reduced in the presence of IL-2 or IL-15. As a control, we tested three Segment 9 distal Runx mutations under the same conditions. The relative response was similar in the presence or absence of cytokine stimulation (Fig. 6B). Therefore, although the distal and proximal Runx sites have identical sequence motifs, mutations directed at the distal Runx site did not prevent the 2DL4 promoter from responding to IL-2 or IL-15.
Use of 2DL4 promoter elements in T and NK cell lines
Because 2DL4 is expressed by both mature and immature NK cells and by a subset of T cells, we tested several NK and T cell lines. Mouse LNK cells are a model of immature NK cells and human NKL cells are models of mature cytotoxic NK cells (44, 45). Both NK cell lines require cytokine for growth. Hut-78 is a model of KIR-expressing CD4+ T cells. All cell lines, with the exception of Hut-78, were tested while being treated with IL-2 (Fig. 7). YT, LNK, NKL, and Hut-78 cells required the CRE site for optimal 2DL4 promoter activity, showing that CRE-binding elements contribute to 2DL4 transcription under all conditions tested. Similar to IL-2-treated YT cells, but distinct from untreated YT cells, the other NK and T cell lines required the Ets site and the proximal Runx site for optimal 2DL4 promoter activity (Fig. 7). Therefore, the compound proximal Runx/Ets site plays a unique and nonredundant role in cytokine-stimulated NK cells and in T cells. To further investigate a role for the distal Runx site, we mutated both Runx sites (Fig. 7). Combined mutation of both sites virtually eliminated 2DL4 promoter activity in all cell lines tested, showing that at least one 2DL4 promoter Runx site is required by NK and T cells. Similar results were seen in untreated CEM-T4 and Jurkat T cell lines (data not shown). We conclude that Runx transcription factors are absolutely required for 2DL4 transcription in both NK and T cell lines, in both resting and cytokine-stimulated conditions.
Runx3 and Ets1 bind to the 2DL4 promoter in situ
We carried out ChIP using Ab specific for Runx1 and Runx3, because these two isoforms are well expressed while Runx2 is poorly expressed in mature NK cells (46–48). First, we tested the IL-2-dependent NK92.26.5 cell line that expresses both 2DL4 and 3DL1. Runx3 associated with the 2DL4 promoter, as shown with two anti-Runx3 mAb (5G4 and 6E9), but not with anti-Runx1 Ab (Fig. 8A and data not shown). Ets1 also associated with the 2DL4 promoter in IL-2-treated NK92.26.5 cells (Figure 8B). Runx3 and Ets1 association was significant in comparison with a control genomic location (2DL4 intron 4, Fig. 8). We did not observe an association of Runx3 and Ets1 with the 3DL1 promoter in 3DL1-expressing NK92.26.5 cells (data not shown). We also tested whether Runx3 bound to the endogenous 2DL4 promoter in primary NK cells. To preserve chromatin structure, NK cells were purified from peripheral blood by buoyant density negative selection and fixed immediately after isolation. We found that Runx3 associated with the 2DL4 promoter, and that association was significant in comparison with a control genomic location (2DL4 intron 4, Fig. 8C).
DISCUSSION
Alignment of the 2DL4 and 3DL1 promoters (Fig. 1) suggests that while the Ets and distal Runx sites are conserved in human KIR promoters, many potential 2DL4 sites (proximal Runx, CRE, AP-1) are not found at homologous positions in crKIR promoters. We identified potential cis-elements and corresponding trans factors required for optimal 2DL4 promoter activity in a systematic and unbiased fashion using linker scanning mutagenesis. Tentatively identified factors were confirmed in point mutation, EMSA, co-transfection, and ChIP experiments. For optimal 2DL4 promoter activity, a Runx family member is absolutely required at one of two redundant Runx sites, the CRE site and Inr sites are important, and a compound proximal Runx/Ets site is important only in certain contexts, such as T cell and cytokine-treated NK cell lines. The roles of the 2DL4 Runx, CRE, and Ets sites are supported by an examination of orthologous 2DL4 promoter sequences in chimpanzee, orangutan, and the more distantly related rhesus monkey (Fig. 9). The distal Runx site and the Ets site are perfectly conserved in these primate 2DL4 promoters. Although a CRE site is not present in a homologous position in the rhesus 2DL4 promoter, a CRE site appears in the same position and having the same sequence as human crKIR promoters, which was shown to be functional (23). The rhesus 2DL4 promoter does not contain a proximal Runx site and we predict that Runx proteins stimulate 2DL4 transcription through the distal Runx site in rhesus monkey NK cells. Based on DNase I footprinting, GATA-3, LEF-1/TCF-2, IRF-2, and AP-1 sites were hypothesized to regulate 2DL4 transcription (24). With the exception of AP-1, these transcription factor consensus motifs are not well matched in the human 2DL4 promoter and diverge even more from consensus in the rhesus monkey 2DL4 promoter (Figure 9 and data not shown). This observation is consistent with our mutagenesis studies showing that these sites are not required for 2DL4 promoter activity (Supplemental Figure S1C). Thus, primate 2DL4 promoter alignment supports our conclusion that 2DL4 transcription requires Runx protein binding, along with a partial dependence on Ets and CRE sites. In addition, KIR2DL5 alleles that do not have a functional distal Runx promoter site are not expressed by NK cells (6). Hence, the distal Runx site is required for in vivo crKIR expression, possibly through an epigenetic mechanism (6).
Figure 9.
CRE, Ets, and distal Runx promoter sites are present in 2DL4 primate orthologs. Chimpanzee (C), orangutan (O), and rhesus monkey (R) promoter sequences were aligned to the human (H) sequence (−225 to +3) using CLUSTALW2. Putative cis-acting elements are denoted by boxes. Chimpanzee, orangutan, and rhesus monkey sequences had 98%, 95%, and 69% identity, respectively, to the human sequence.
The three Runx homologs (Runx1, 2, 3) act as activators or repressors, depending on the context in which they are bound. Runx proteins dimerize with CBFβ to achieve a high affinity DNA interaction and functional activity (41, 49, 50). Runx factors likely are required for NK cell development because reduction of CBFβ activity to 15% of WT levels in mouse fetal liver cells almost completely eliminated the production of immature and mature NK cells (47, 49). Furthermore, transgenic expression of a dominant negative Runx protein reduced mature NK cell numbers and reduced expression of proteins required for NK function (46, 49). During mouse and human NK cell development, Runx1 levels remain relatively constant while Runx3 levels increase steadily, so that Runx3 becomes the dominant Runx family member expressed in mature NK cells (46–49). Additionally, mature human NK cells express Runx3 from a distal promoter that is associated with robust protein translation (48). Despite the importance of Runx factors in NK cell development, very few NK cell genes are known to be controlled by Runx factors. Our report provides evidence that Runx3 binds to the KIR2DL4 promoter in human primary NK cells and activates its transcription.
Trompeter et al. reported that point mutation of the distal Runx site increased promoter activity 2-fold in NK3.3 cells and concluded that Runx represses 2DL4 transcription (25). Based on multiple lines of evidence, we found that Runx proteins activate 2DL4 transcription in several different settings. It is not clear why our findings differ because we tested the same mutation (dRx3) as reported by Trompeter et al. It should be noted that Trompeter et al. achieved 2DL4 promoter activity that was no more than 3-fold greater than that of the pGL3 Basic empty vector. In contrast, 2DL4 promoter activity was 10–35-fold above that of pGL3 Basic vector in the IL-2-treated NK cell lines that we tested. We conclude that Runx proteins are required for 2DL4 transcription.
KIR2DL4 gene expression differs significantly from that of 3DL1 and other crKIR genes. 2DL4 is expressed earlier in development and may be a prerequisite for crKIR expression in NK cells and CD8 T cells. Although both 2DL4 and crKIR transcription are controlled by promoter DNA methylation, 2DL4 is bi-allelically expressed in all NK cells, but crKIR have variegate expression that is typically mono-allelic (4–9). Our analysis of the 2DL4 promoter shows important differences with crKIR promoters and may explain how expression is differentially regulated. Despite overall sequence conservation of human KIR promoters, Runx elements are absolutely required for 2DL4 promoter activity, whereas Runx transcription factors make only a small contribution to 3DL1 promoter activity (23). This may be related to the observation that crKIR promoters, but not the 2DL4 promoter, contains an active Sp1 site and an overlapping STAT/Ets/YY1 site. Alternatively, the strong importance of Runx for the 2DL4 promoter may be related to the fact that 2DL4, unlike crKIR promoters, contains redundant Runx sites. Given that the redundant Runx sites are critical elements required for 2DL4 promoter activity, we propose that Runx3 is a major driver of 2DL4 transcription in mature NK cells. In a second contrast with crKIR, 2DL4 promoter activity is increased by IL-2 and IL-15 cytokine treatment, an increase that depends upon the 2DL4 compound proximal Runx/Ets site that is not found in crKIR promoters. We propose that the compound site serves multiple roles. In mature NK cells, cytokine-stimulated Ets1 binding increases Runx3 driven expression in an additive manner (our findings). The site also has the potential to promote Runx1/Ets1 synergy (50). Therefore, the compound Runx/Ets site may be especially important for initiating 2DL4 transcription in pre-NK cells or early NK cells, when Runx1 is relatively more abundant (46, 47, 49). In parallel with the TCRα enhancer and the TCR Vβ 8.1 promoter (51–53), Runx1/Ets1 protein-protein contacts and synergistic DNA binding, in cooperation with CREB or ATF, may allow assembly of a three-dimensional multiprotein-DNA complex at the 2DL4 promoter that is not possible at crKIR promoters during early NK cell development, because the crKIR promoters lack a compound Ets-Runx site. In contrast to these TCR elements, however, there is much less cooperativity between CRE, Runx, and Ets factors at the 2DL4 promoter. Mutation of TCRα enhancer CRE, Runx, or Ets site alone nearly abrogated enhancer activity in transfected T cells (51), but mutation of 2DL4 CRE, Runx, or Ets site alone had much smaller effects in transfected NK cells. Therefore, although the TCRα enhancer, the TCRβ promoter, and the 2DL4 promoter share many cis-acting sites, their three-dimensional structures likely differ.
Based on our results that define the requirements for steady state and cytokine-induced transcription, we propose a model for initiation and maintenance of 2DL4 transcription. IL-15 and IL-2 signaling in bone marrow and lymph node, which are required for maturation of pre-NK cells into CD56bright NK cells, increase Ets1 levels and phosphorylate CREB (54, 55). We speculate that Runx1, which is expressed in both pre-NK cells and NK cells (46, 47, 49), forms complexes with Ets1 and CREB at the 2DL4 promoter via protein-protein interactions (50, 51, 56), compensating for CREB's poor affinity for methylated DNA (our findings and (31)). Transcription factor binding is known to block DNMT1 binding, which passively prevents methylation of replicating DNA in dividing cells and we propose a similar mechanism for 2DL4 (57). In our model, these transcription factors recruit CBP/p300, which acetylates and further stabilizes Runx (58–61). CBP/p300 also is known to acetylate histones, open chromatin, and induce transcription (62). Given that CD56bright NK cells express high affinity IL-2 receptors and are sensitive to low IL-2 concentrations (63), we propose that Ets1 and Runx protein cooperation at the 2DL4 promoter plays a role in cytokine-stimulated immature NK cells, and, in part, explains why CD56bright NK cells express relatively high levels of 2DL4 (10–12). In mature CD56dim NK cells that are less sensitive to IL-2 signaling, we propose that 2DL4 transcription is maintained at a relatively lower level, largely under the influence of Runx3 and possibly a CRE-binding transcription factor.
Supplementary Material
Table II.
KIR2DL4 mutations
| Site | Motifa | KIR Sequenceb | Mutated Sequencec |
|---|---|---|---|
| Segments 1–24 | GCAGATCCGC | ||
| CRE1 | TGACGTCA | TGACGTAG | TAACTTAG; −137, −134 |
| CRE2 | TGACGTCA | TGACGTAG | TAATGTAG; −137, −135 |
| MTE/DPE | CSARCSSAAC/RGWYVT | CTGGCAGCAGAAGC | CTGGTAGGAGTAGC; −16, −13 −10 |
| DCE1 | AGC (segment 1 of DCE) | AGC | TTT; −15 to −13 |
| DCE2 | AGC (segment 1 of DCE) | AGC | ACC; −14 |
| Ets | CMGGAWGYd | ACATCCTC | ACATAATC; −56, −55 |
| dR×1 | TGTGGT | TGTGGT | ACTTGT; −100, −99, −97 |
| dR×2 | TGTGGT | TGTGGT | TGTTGT; −97 |
| dR×3 | TGTGGT | TGTGGT | TGTAGT; −97 |
| pRx | TGTGGT | ACCACA | ATAACA; −62, −61 |
| pRx + Ets | CCACWTCCTd,e | CCACATCCT | TAACATATT; −62, −61, −56, −55 |
| CP2 | CNRG(N)5–6 CNRS | CCGGTCAGTCGAGCCGAG | CCGATCATTCGAGCCGAG; −45, −41 |
| MyoD | CANNTG | CAGTCG | TAGTCT; −43, −38 |
| AP1-A | TGASTCA | TGATTCA | GGCTTCA; −161, −159 |
| AP1-B | TGASTCA | TGACTGATTCA | TGTCGGCTGCA, −163, −161, −159, −157 |
| IRF-2 | AAANYGAAA | GAAGTGACAA | GAGGTGTCAA; −211, −207 |
| GATA-3 | WGATAR | TGATGT | TAACGT; −218, −216 |
Footnotes:
W = A, T; M = A, C; N = A, C, G, T; R = A, G; V =A, C, G; Y = C, T; S = C, G.
Underlining denotes mutation target
Underlining denotes new base at the position indicated
Reference (56)
Motif on antisense strand
ACKNOWLEDGEMENTS
The authors are grateful to Dennis Watson, John Trowsdale, Yoshiaki Ito, Frank Pugh, Nancy Speck, and Chung-Tsai Lee for reagents, and the National Cancer Institute for IL-2, Xiuqin Li for technical assistance, and Anthony Sinai, Jeffrey Ebersole, Luke Bradley, and Peter Nelson for equipment use. We thank Robert Coleman, Janice Telfer, Frank Pugh, Kerry Campbell, and Stephen Smale for helpful discussions.
This work was supported by National Institute of Health grant, R01 AI56506.
Abbreviations
- ATF-1
Activating Transcription Factor-1
- BRE
TFIIB Recognition Element
- CBFβ
Core Binding Factor β
- ChIP
chromatin immunoprecipitation
- CRE
cyclic AMP response element
- crKIR
clonally-restricted KIR
- Inr
initiator element
- IPA
isopropyl alcohol
- KIR
killer cell Ig-like receptor
- PIC
protease inhibitor cocktail
- TBP
TATA-binding protein
REFERENCES
- 1.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 2.Long EO. Negative signaling by inhibitory receptors: the NK cell paradigm. Immunol Rev. 2008;224:70–84. doi: 10.1111/j.1600-065X.2008.00660.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. 2004;172:356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- 4.Santourlidis S, Graffmann N, Christ J, Uhrberg M. Lineage-specific transition of histone signatures in the killer cell Ig-like receptor locus from hematopoietic progenitor to NK cells. J Immunol. 2008;180:418–425. doi: 10.4049/jimmunol.180.1.418. [DOI] [PubMed] [Google Scholar]
- 5.Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P, Uhrberg M. Crucial role of DNA methylation in determination of clonally distributed killer cell Ig-like receptor expression patterns in NK cells. J Immunol. 2002;169:4253–4261. doi: 10.4049/jimmunol.169.8.4253. [DOI] [PubMed] [Google Scholar]
- 6.Gómez-Lozano N, Trompeter H-I, de Pablo R, Estefanía E, Uhrberg M, Vilches C. Epigenetic silencing of potentially functional KIR2DL5 alleles: Implications for the acquisition of KIR repertoires by NK cells. Eur J Immunol. 2007;37:1954–1965. doi: 10.1002/eji.200737277. [DOI] [PubMed] [Google Scholar]
- 7.Chan H-W, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE, Carrington M, Trowsdale J, Lutz CT. DNA methylation maintains allele-specific KIR gene expression in human natural killer cells. J Exp Med. 2003;197:245–255. doi: 10.1084/jem.20021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan HW, Miller JS, Moore MB, Lutz CT. Epigenetic control of highly homologous killer Ig-like receptor gene alleles. J Immunol. 2005;175:5966–5974. doi: 10.4049/jimmunol.175.9.5966. [DOI] [PubMed] [Google Scholar]
- 9.Li G, Weyand CM, Goronzy JJ. Epigenetic mechanisms of age-dependent KIR2DL4 expression in T cells. J Leukoc Biol. 2008;84:824–834. doi: 10.1189/jlb.0807583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-γ production. J Immunol. 2003;171:3415–3425. doi: 10.4049/jimmunol.171.7.3415. [DOI] [PubMed] [Google Scholar]
- 11.Cooley S, Xiao F, Pitt M, Gleason M, McCullar V, Bergemann T, McQueen KL, Guethlein LA, Parham P, Miller JS. A subpopulation of human peripheral blood NK cells that lacks inhibitory receptors for self MHC is developmentally immature. Blood. 2007;110:578–586. doi: 10.1182/blood-2006-07-036228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodridge JP, Witt CS, Christiansen FT, Warren HS. KIR2DL4 (CD158d) genotype influences expression and function in NK cells. J Immunol. 2003;171:1768–1774. doi: 10.4049/jimmunol.171.4.1768. [DOI] [PubMed] [Google Scholar]
- 13.Rajagopalan S, Bryceson YT, Kuppusamy SP, Geraghty DE, van der Meer A, Joosten I, Long EO. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 2006;4:e9. doi: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kikuchi-Maki A, Catina TL, Campbell KS. Cutting edge: KIR2DL4 transduces signals into human NK cells through association with the Fc receptor γ protein. J Immunol. 2005;174:3859–3863. doi: 10.4049/jimmunol.174.7.3859. [DOI] [PubMed] [Google Scholar]
- 15.Faure M, Long EO. KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory potential. J Immunol. 2002;168:6208–6214. doi: 10.4049/jimmunol.168.12.6208. [DOI] [PubMed] [Google Scholar]
- 16.Rajagopalan S, Fu J, Long EO. Cutting edge: Induction of IFN-γ production but not cytotoxicity by the killer cell Ig-like receptor KIR2DL4 (CD158d) in resting NK cells. J Immunol. 2001;167:1877–1881. doi: 10.4049/jimmunol.167.4.1877. [DOI] [PubMed] [Google Scholar]
- 17.Apps R, Gardner L, Moffett A. A critical look at HLA-G. Trends Immunol. 2008;29:313–321. doi: 10.1016/j.it.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J Exp Med. 1999;189:1093–1100. doi: 10.1084/jem.189.7.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Houser BL, Nicotra ML, Strominger JL. HLA-G homodimer-induced cytokine secretion through HLA-G receptors on human decidual macrophages and natural killer cells. Proc Natl Acad Sci U S A. 2009;106:5767–5772. doi: 10.1073/pnas.0901173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vacca P, Moretta L, Moretta A, Mingari MC. Origin, phenotype and function of human natural killer cells in pregnancy. Trends Immunol. 2011;32:517–523. doi: 10.1016/j.it.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Stulberg MJ, Wright PW, Dang H, Hanson RJ, Miller JS, Anderson SK. Identification of distal KIR promoters and transcripts. Genes Immun. 2006;8:124–130. doi: 10.1038/sj.gene.6364363. [DOI] [PubMed] [Google Scholar]
- 22.Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS, Anderson SK. Identification of bidirectional promoters in the human KIR genes. Genes Immun. 2007;8:245–253. doi: 10.1038/sj.gene.6364381. [DOI] [PubMed] [Google Scholar]
- 23.Presnell SR, Zhang L, Ramilo CA, Chan H-W, Lutz CT. Functional redundancy of transcription factor-binding sites in the killer cell Ig-like receptor (KIR) gene promoter. Int. Immunol. 2006;18:1221–1232. doi: 10.1093/intimm/dxl043. [DOI] [PubMed] [Google Scholar]
- 24.Stewart CA, Van Bergen J, Trowsdale J. Different and divergent regulation of the KIR2DL4 and KIR3DL1 promoters. J Immunol. 2003;170:6073–6081. doi: 10.4049/jimmunol.170.12.6073. [DOI] [PubMed] [Google Scholar]
- 25.Trompeter H-I, Gómez-Lozano N, Santourlidis S, Eisermann B, Wernet P, Vilches C, Uhrberg M. Three structurally and functionally divergent kinds of promoters regulate expression of clonally distributed killer cell Ig-like receptors (KIR), of KIR2DL4, and of KIR3DL3. J Immunol. 2005;174:4135–4143. doi: 10.4049/jimmunol.174.7.4135. [DOI] [PubMed] [Google Scholar]
- 26.Lutz CT, Kurago ZB. Human leukocyte antigen class I expression on squamous cell carcinoma cells regulates natural killer cell activity. Cancer Res. 1999;59:5793–5799. [PubMed] [Google Scholar]
- 27.Karantzoulis-Fegaras F, Antoniou H, Lai S-LM, Kulkarni G, D'Abreo C, Wong GKT, Miller TL, Chan Y, Atkins J, Wang Y, Marsden PA. Characterization of the human endothelial nitric-oxide synthase promoter. J Biol Chem. 1999;274:3076–3093. doi: 10.1074/jbc.274.5.3076. [DOI] [PubMed] [Google Scholar]
- 28.Barthel R, Tsytsykova AV, Barczak AK, Tsai EY, Dascher CC, Brenner MB, Goldfeld AE. Regulation of tumor necrosis factor alpha gene expression by mycobacteria involves the assembly of a unique enhanceosome dependent on the coactivator proteins CBP/p300. Mol Cell Biol. 2003;23:526–533. doi: 10.1128/MCB.23.2.526-533.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniatis T, Falvo JV, Kim TH, Kim TK, Lin CH, Parekh BS, Wathelet MG. Structure and function of the interferon-β enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 30.Selvakumar A, Steffens U, Dupont B. NK cell receptor gene of the KIR family with two IG domains but highest homology to KIR receptors with three IG domains. Tissue Antigens. 1996;48:285–295. doi: 10.1111/j.1399-0039.1996.tb02647.x. [DOI] [PubMed] [Google Scholar]
- 31.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 32.Schug J. Using TESS to predict transcription factor binding sites in DNA sequence. In: Baxevanis AD, Davison DB, Page RDM, Petsko GA, Stein LD, Stormo GD, editors. Current Protocols in Bioinformatics. Greene Publishing Associates and Wiley-Interscience; New York: 2003. pp. 2.6.1–2.6.7. [DOI] [PubMed] [Google Scholar]
- 33.Radeloff B, Nagler L, Zirra M, Ziegler A, Volz A. Specific amplification of cDNA ends (SPACE): A new tool for the analysis of rare transcripts and its application for the promoter analysis of killer cell receptor genes. DNA Seq. 2005;16:44–52. doi: 10.1080/10425170400028202. [DOI] [PubMed] [Google Scholar]
- 34.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale ST. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Juven-Gershon T, Hsu J-Y, Theisen JWM, Kadonaga JT. The RNA polymerase II core promoter -- the gateway to transcription. Curr Opin Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wiley SR, Kraus RJ, Mertz JE. Functional binding of the “TATA” box binding component of transcription factor TFIID to the −30 region of TATA-less promoters. Proc Natl Acad Sci U S A. 1992;89:5814–5818. doi: 10.1073/pnas.89.13.5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J, Vallejo AN, Jiang Y, Weyand CM, Goronzy JJ. Distinct transcriptional control mechanisms of killer immunoglobulin-like receptors in natural killer (NK) and in T cells. J Biol Chem. 2005;280:24277–24285. doi: 10.1074/jbc.M500727200. [DOI] [PubMed] [Google Scholar]
- 38.Arman M, Calvo J, Trojanowska ME, Cockerill PN, Santana M, Lopez Cabrera M, Vives J, Lozano F. Transcriptional regulation of human CD5: important role of Ets transcription factors in CD5 expression in T cells. J Immunol. 2004;172:7519–7529. doi: 10.4049/jimmunol.172.12.7519. [DOI] [PubMed] [Google Scholar]
- 39.Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armesilla AL, Calvo D, Vega MA. Structural and functional characterization of the human CD36 gene promoter: identification of a proximal PEBP2/CBF site. J Biol Chem. 1996;271:7781–7787. doi: 10.1074/jbc.271.13.7781. [DOI] [PubMed] [Google Scholar]
- 41.de Bruijn MFTR, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23:4238–4248. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- 42.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, Caligiuri MA. Evidence for discrete stages of human natural killer cell differentiation in vivo. J. Exp. Med. 2006;203:1033–1043. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.John S, Robbins CM, Leonard WJ. An IL-2 response element in the human IL-2 receptor alpha chain promoter is a composite element that binds Stat5, Elf-1, HMG-I(Y) and a GATA family protein. Embo J. 1996;15:5627–5635. [PMC free article] [PubMed] [Google Scholar]
- 44.Robertson MJ, Cochran KJ, Cameron C, Le J-M, Tantravahi R, Ritz J. Characterization of cell line, NKL, derived from an aggressive human natural killer cell leukemia. Exp Hematol. 1996;24:406–415. [PubMed] [Google Scholar]
- 45.Tsutsui H, Nakanishi K, Matsui K, Higashino K, Okamura H, Miyazawa Y, Kaneda K. IFN-gamma-inducing factor up-regulates Fas ligand-mediated cytotoxic activity of murine natural killer cell clones. J Immunol. 1996;157:3967–3973. [PubMed] [Google Scholar]
- 46.Ohno S.-i., Sato T, Kohu K, Takeda K, Okumura K, Satake M, Habu S. Runx proteins are involved in regulation of CD122, Ly49 family and IFN-γ expression during NK cell differentiation. Int. Immunol. 2008;20:71–79. doi: 10.1093/intimm/dxm120. [DOI] [PubMed] [Google Scholar]
- 47.Guo Y, Maillard I, Chakraborti S, Rothenberg EV, Speck NA. Core binding factors are necessary for natural killer cell development and cooperate with Notch signaling during T-cell specification. Blood. 2008;112:480–492. doi: 10.1182/blood-2007-10-120261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai CB, Mager DL. The role of runt-related transcription factor 3 (RUNX3) in transcription regulation of natural cytotoxicity receptor 1 (NCR1/NKp46), an activating NK cell receptor. J Biol Chem. 2012 doi: 10.1074/jbc.M111.306936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hesslein DG, Lanier LL. Transcriptional control of natural killer cell development and function. Adv Immunol. 2011;109:45–85. doi: 10.1016/B978-0-12-387664-5.00002-9. [DOI] [PubMed] [Google Scholar]
- 50.Ito Y. RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res. 2008;99:33–76. doi: 10.1016/S0065-230X(07)99002-8. [DOI] [PubMed] [Google Scholar]
- 51.Giese K, Kingsley C, Kirshner JR, Grosschedl R. Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein-protein interactions. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 52.Halle JP, Haus-Seuffert P, Woltering C, Stelzer G, Meisterernst M. A conserved tissue-specific structure at a human T-cell receptor beta-chain core promoter. Mol Cell Biol. 1997;17:4220–4229. doi: 10.1128/mcb.17.8.4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mayall TP, Sheridan PL, Montminy MR, Jones KA. Distinct roles for P-CREB and LEF-1 in TCRα enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 54.Ponti C, Gibellini D, Boin F, Melloni E, Manzoli FA, Cocco L, Zauli G, Vitale M. Role of CREB transcription factor in c-fos activation in natural killer cells. Eur J Immunol. 2002;32:3358–3365. doi: 10.1002/1521-4141(200212)32:12<3358::AID-IMMU3358>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 55.Grund EM, Spyropoulos DD, Watson DK, Muise Helmericks RC. Interleukins 2 and 15 regulate Ets1 expression via ERK1/2 and MNK1 in human natural killer cells. J Biol Chem. 2005;280:4772–4778. doi: 10.1074/jbc.M408356200. [DOI] [PubMed] [Google Scholar]
- 56.Hollenhorst PC, Chandler KJ, Poulsen RL, Johnson WE, Speck NA, Graves BJ. DNA specificity determinants associate with distinct transcription factor functions. PLoS Genet. 2009;5:e1000778. doi: 10.1371/journal.pgen.1000778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsieh CL. Evidence that protein binding specifies sites of DNA demethylation. Mol Cell Biol. 1999;19:46–56. doi: 10.1128/mcb.19.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol. Cell. Biol. 2004;24:10954–10964. doi: 10.1128/MCB.24.24.10954-10964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang C, Shapiro LH, Rivera M, Kumar A, Brindle PK. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin Y-H, Jeon E-J, Li Q-L, Lee YH, Choi J-K, Kim W-J, Lee K-Y, Bae S-C. Transforming growth factor-β stimulates p300-dependent RUNX3 acetylation, which inhibits ubiquitination-mediated degradation. J Biol Chem. 2004;279:29409–29417. doi: 10.1074/jbc.M313120200. [DOI] [PubMed] [Google Scholar]
- 61.Wheeler JC, Shigesada K, Gergen JP, Ito Y. Mechanisms of transcriptional regulation by Runt domain proteins. Semin Cell Dev Biol. 2000;11:369–375. doi: 10.1006/scdb.2000.0184. [DOI] [PubMed] [Google Scholar]
- 62.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 63.Caligiuri MA, Zmuidzinas A, Manley TJ, Levine H, Smith KA, Ritz J. Functional consequences of interleukin 2 receptor expression on resting human lymphocytes. Identification of a novel natural killer cell subset with high affinity receptors. J Exp Med. 1990;171:1509–1526. doi: 10.1084/jem.171.5.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.