SUMMARY
The regulatory networks of differentiation programs have been partly characterized; however, the molecular mechanisms of lineage-specific gene regulation by highly similar transcription factors remain largely unknown. Here we compare the genome-wide binding and transcription profiles of NEUROD2-mediated neurogenesis with MYOD-mediated myogenesis. We demonstrate that NEUROD2 and MYOD bind a shared CAGCTG E-box motif and E-box motifs specific for each factor: CAGGTG for MYOD and CAGATG for NEUROD2. Binding at factor-specific motifs is associated with gene transcription, whereas binding at shared sites is associated with regional epigenetic modifications but not as strongly associated with gene transcription. Binding is largely constrained to E-boxes pre-set in an accessible chromatin context that determines the set of target genes activated in each cell type. These findings demonstrate that the differentiation program is genetically determined by E-box sequence whereas cell lineage epigenetically determines the availability of E-boxes for each differentiation program.
INTRODUCTION
The family of basic-helix-loop-helix (bHLH) transcription factors regulates differentiation programs in numerous cell types. For example, the Neurod family regulates neuronal differentiation, the Myod family regulates skeletal muscle differentiation, and the E-proteins regulate B and T-cell differentiation and also function as heterodimer partners for both Myod and Neurod family members (Berkes and Tapscott, 2005; Chae et al., 2004; Murre, 2005). Each of these factors binds to a motif containing the core CANNTG sequence, termed an E-box. Although some additional sequence preferences have been described for each factor (Blackwell and Weintraub, 1990; Seo et al., 2007), the degrees of shared and specific binding relative to differentiation programs have not been described. Since the bHLH family evolved by duplication and divergence, the maintenance of a common core E-box sequence suggests the possibility of shared sites among family members that might have a functional role in gene regulation and/or other aspects of differentiation. However, the large differences in the transcriptional programs of neurons, muscles, and T-cells strongly indicates that each factor has evolved mechanisms for factor-specific gene regulation.
Other studies of transcription factor families suggest several mechanisms of achieving factor-specific gene regulation within a family of highly related transcription factors. In several cases, transcriptional activity and site-specific binding are regulated separately. For instance, members of the NF-κB family were shown to bind at the same sequences, but demonstrated sequence specific transcriptional activity (Leung et al., 2004). Similarly, the glucocorticoid receptor (GR) will bind to many similar sequences but transcriptional activation depends on a DNA sequence-specific allosteric activation of the bound GR (Meijsing et al., 2009). Therefore, hormone receptor families and NF-κB families of transcription factors might achieve gene-specific regulation by regulated activity at binding sites shared by multiple family members. The ETS family of transcription factors also has sites bound by many different family members; however, factor-specific transcription appears to be achieved by a subset of binding sites with sequence variations that favor a specific family member, as well as binding of co-factors specific to the transcriptional program (Hollenhorst et al., 2011; Hollenhorst et al., 2007). In the case of the ETS family, the binding sites shared by multiple family members are associated with constitutively expressed genes, suggesting that factor binding corresponds to local gene activation for both the shared and factor-specific binding sites.
In addition to binding site sequence, several recent studies indicate that chromatin structure limits access for factor binding to subsets of potential sites. For the GR receptor, the activator protein 1 (AP1) facilitates open chromatin and GR receptor binding, essentially presetting the chromatin context for response to GR activation (Biddie et al., 2011; Hakim et al., 2011; John et al., 2011). Factor binding can also be prevented at accessible sites by the competitive binding of factors, such as CBF1 occluding accessible sites and preventing the binding of PHO4 in yeast (Zhou and O’Shea, 2011). The role of chromatin accessibility can also be demonstrated in a developmental context in Drosophila, where it limits the sites accessible to transcription factors and correlates with local gene transcription (Li et al., 2011; Thomas et al., 2011).
Within this context, we have examined the genome-wide binding and transcriptional activity of NEUROD2 in P19 cells and MYOD in mouse embryonic fibroblasts (MEFs). NEUROD2 expression converts P19 cells to neurons and MYOD expression converts MEFs to skeletal muscle cells; whereas NEUROD2 does not induce neurogenesis in MEFs, nor does MYOD induce myogenesis in P19 cells. We determined that NEUROD2 and MYOD bind a shared E-box motif, RRCAGCTG, and E-boxes with motifs specific to each factor: CAGATG for NEUROD2 and CAGGTG for MYOD. Binding at the NEUROD2-specific motif was associated with transcription of the neuronal differentiation program, whereas binding at the shared sites was associated with regional epigenetic modifications but not regional gene transcription, and a similar trend was observed for MYOD-specific motifs. In each cell type, binding is largely constrained to E-boxes pre-set in an accessible chromatin context and lineage-restricted differentiation reflects differences in E-box accessibility. These findings demonstrate that the differentiation program is genetically encoded by the location of the factor-specific E-boxes and that cell lineage establishes the set of E-boxes in an accessible chromatin context.
RESULTS
NEUROD2 ChIP-Seq Demonstrates Genome Wide Binding and Regional Histone Acetylation
We have previously shown that the pluripotent mouse cell line P19 can be converted to neurons by the exogenous expression of NEUROD2 (Farah et al., 2000). Transduction of P19 cells with a NEUROD2 expressing lentivirus achieved nearly complete conversion to neurons (Figure 1A). Expression array analysis identified the up-regulation of 532 genes and down-regulation of 278 genes (Table S1). Gene set enrichment analysis (GSEA) showed that up-regulated genes were associated with Gene Ontology (GO) categories involving neuron development and differentiation (Table 1).
Figure 1. NEUROD2 binds genome-wide in P19 cells differentiated into neurons.
(A) Differentiation of P19 cells by expression of NEUROD2. Immunostaining of P19 cells before (top panels) and 72 hours after (bottom panels) transduction with NEUROD2 lentivirus (green: Tuj1 antibody; red: NEUROD2 antibody; blue: DAPI).
(B) NEUROD2 antibody is specific for NEUROD2. Immunoprecipitation of 35S-labeled in vitro translated bHLH proteins relative to 10% input (lane 1) with either non-specific IgG (lane 2) or 2 different NEUROD2 antibodies (lanes 3 and 4).
(C) NEUROD2 binding is associated with, but does not reliably predict, gene up-regulation. NEUROD2 ChIP-Seq and microarrays were performed in P19 cells before and 72 hours after transduction with NEUROD2 lentivirus. NEUROD2 peak height (Y-axis, square root transformation) of binding sites located within the CTCF domain of gene TSSs is plotted against the log-2 fold change in mRNA expression (X-axis) in smooth scatter plot (left) and boxplot (right) binned by level of activation. The blue trend line in the scatter plot was computed using the loess local regression method; in the boxplot, the vertical bounds represent the 25th and 75th percentile, the width represents the size of the dataset, the dot is the median value, and the whisker extends to the extreme value (minimum or maximum), bounded by 1.5 times IQR (25th and 75th interquartile range) from the box
See also Figure S1.
Table 1.
GO categories of NEUROD2 up-regulated genes identified by expression array.
| GO ID | P-value | Odds Ratio | Count | Size | Term |
|---|---|---|---|---|---|
| GO:0007275 | 3.2E-30 | 4.14 | 120 | 1,141 | multicellular organismal development |
| GO:0048858 | 8.6E-19 | 7.25 | 39 | 192 | cell projection morphogenesis |
| GO:0031175 | 5.3E-18 | 7.86 | 35 | 163 | neuron projection development |
| GO:0045202 | 5.8E-15 | 6.01 | 35 | 200 | synapse |
| GO:0023046 | 1.6E-13 | 2.44 | 109 | 1,476 | signaling process |
| GO:0007411 | 2.8E-12 | 12.77 | 17 | 54 | axon guidance |
| GO:0005509 | 3.6E-12 | 3.18 | 57 | 561 | calcium ion binding |
| GO:0048856 | 5.0E-12 | 6.03 | 27 | 176 | anatomical structure development |
| GO:0007155 | 5.5E-12 | 5.18 | 31 | 202 | cell adhesion |
| GO:0009653 | 8.8E-12 | 5.07 | 31 | 214 | anatomical structure morphogenesis |
| GO:0007268 | 4.6E-11 | 6.95 | 22 | 110 | synaptic transmission |
| GO:0030424 | 7.2E-10 | 7.61 | 18 | 83 | axon |
| GO:0007399 | 5.2E-09 | 6.99 | 17 | 95 | nervous system development |
| GO:0030054 | 1.0E-08 | 4.24 | 26 | 196 | cell junction |
To identify NEUROD2 binding sites, we used two different rabbit polyclonal antibodies that specifically pull down NEUROD2 (Figure 1B) and performed chromatin immunoprecipitations in P19 cells transduced with the NEUROD2 lentivirus followed by high throughput sequencing (ChIP-Seq). Reads with a unique match in the mouse genome were extended to a total length of 200 nucleotides (nt), which was the estimated average fragment size, and the number of overlapping reads at each position in the genome was computed to generate peak heights for NEUROD2 or the control ChIP samples (P19 cells ChIPed with pre-immune serum) (see Experimental Procedures). The two individual antisera were highly concordant with a Pearson correlation coefficient of 0.9 (Figure S1A) and the data from the two antisera were pooled for further analysis. The small number of regions enriched in the pre-immune ChIP control were subtracted from our analyses.
To identify NEUROD2 binding sites, we compared the false discovery rate (FDR) at various read coverage cutoffs in anti-NEUROD2 and pre-immune samples (Figure S1B). Using a conservative cutoff of a p-value < 10−10 corresponding to ~23 reads or more (FDR < 2.4 x 10−8), 35,042 peaks were identified in the anti-NEUROD2 samples (Table S2). Although the region +/−2kb around a transcription start site (TSS) contained the highest density of binding regions (Figure S1C), the majority of peaks were located in introns and intergenic regions, the latter defined as regions more than 10 kb away from any known transcripts (Table S2).
We used three different approaches to assign a peak to a gene, or genes: (1) binding within the immediate region of the gene, defined as 2 kb upstream from the TSS to 2 kb downstream of the polyadenylation site; (2) binding within the domain established by the two CTCF binding regions (SRX 000540, from the NCBI Sequence Read Archive) that flank the TSS for each gene; and (3) binding +/−2 kb from a TSS. Using these approaches, 40% of 18,054 annotated genes were bound by NEUROD2; 48% of the annotated genes had at least one TSS with a NEUROD2 bound region(s) within the flanking CTCF domain; and 19% of genes had a TSS with NEUROD2 bound in the +/−2 kb region. GSEA on all annotated genes possessing NEUROD2 peaks +/−2kb from the TSS demonstrated enrichment for GO categories including neurogenesis and neuronal differentiation and development (Table 2). However, while there is a trend for genes up-regulated by NEUROD2 to be bound by NEUROD2 either within 2 kb of the TSS (data not shown) or within the CTCF domain flanking the TSS (Figure 1C), the majority of genes bound by NEUROD2 do not change in expression. Therefore, NEUROD2 binding does not reliably predict transcriptional regulation of the closest TSS.
Table 2.
GO categories of all annotated genes possessing NEUROD2 peaks within +/− 2kb of the TSS.
| GO ID | P-value | Odds Ratio | Count | Size | Term |
|---|---|---|---|---|---|
| GO:0031175 | 2.3E-10 | 2.88 | 70 | 158 | neuron projection development |
| GO:0048858 | 3.6E-10 | 2.57 | 82 | 197 | cell projection morphogenesis |
| GO:0048856 | 4.7E-10 | 2.65 | 76 | 185 | anatomical structure development |
| GO:0007399 | 9.3E-09 | 3.06 | 52 | 115 | nervous system development |
| GO:0030424 | 1.3E-08 | 4.39 | 34 | 62 | axon |
| GO:0003779 | 3.9E-07 | 2.07 | 82 | 224 | actin binding |
| GO:0030036 | 2.8E-06 | 3.89 | 26 | 50 | actin cytoskeleton organization |
| GO:0007050 | 4.1E-06 | 4.08 | 24 | 45 | cell cycle arrest |
| GO:0022603 | 5.2E-06 | 2.59 | 42 | 100 | regulation of anatomical structure morphogenesis |
| GO:0051960 | 5.4E-06 | 2.37 | 49 | 123 | regulation of nervous system development |
| GO:0007242 | 1.3E-05 | 2.09 | 58 | 159 | intracellular signaling cascade |
| GO:0016055 | 5.4E-05 | 2.54 | 34 | 82 | Wnt receptor signaling pathway |
| GO:0007411 | 5.4E-05 | 2.87 | 28 | 63 | axon guidance |
| GO:0004725 | 5.9E-05 | 2.52 | 34 | 82 | protein tyrosine phosphatase activity |
Characteristics of NEUROD2 Binding Sites
To determine the sequence characteristics of NEUROD2 binding sites, we examined the E-box sequences found in the 200 nt region centered under the NEUROD2 peak summit. Within these regions, 98% of peaks contained at least one canonical E-box (CANNTG), and 80% of peaks contained an E-box within 20 nt of the peak summit. A strong sequence preference was observed for the central dinucleotides within these E-boxes, with a high frequency of GA and GC E-boxes at NEUROD2 peaks (Figure S2A). Focusing on the nearest E-box to the peak summit, 47% contained the motif GA, and 33% were GC. Within the entire 200 nt window, 65% of peaks contained a GA, and 50% contained a GC E-box. Further refinement of the motif model indicates that the sequence preference also extended to the flanking nucleotides, with a G or A at the −2 and −1 positions, a strong absence of T at the −1 position, and a preference for G at the +1 position, yielding a consensus NEUROD2 binding site of RRCAGMTGG (Figure 2A, top panels). ChIP of the endogenous NEUROD2 in P19 cells differentiated to neurons by treatment with retinoic acid followed by quantitative PCR at ten loci confirmed that the endogenous NEUROD2 binds ChIP-Seq identified sites with either CAGCTG or CAGATG E-boxes (Figure S2B).
Figure 2. NEUROD2 and MYOD possess shared and private binding sites.
(A) E-box motif enrichment at NEUROD2 and MYOD peaks (a and d) demonstrates central dinucleotide and flanking sequence preferences that consist of a factor-specific motif (b and e) and a shared motif (c and f).
(B) Motifs enriched (see Experimental Procedures) under NEUROD2 peaks in P19 cells compared to background. All motifs posses z-values > 5 based on a logistic regression model, with an approximate p-value of < 10−7 (ratio: enriched/depleted ratio of motifs; fg.frac, bg.frac: fraction of foreground/background sequences that contain at least one motif occurrence).
(C) NEUROD2 and MYOD bind with higher affinity to their private E-box sequences. Top: EMSA using translated NEUROD2 or MYOD and E12 mixed with probes containing identical flanking sequences and either a MYOD-preferred (probe A), or NEUROD2-preferred (probe B) E-box and competed with cold A or B probe as shown above each lane. * indicates E12 homodimer. Bottom: EMSA using probes containing either a MYOD-preferred (probe C) or NEUROD2-preferred (probe D) E-box with flanking sequence from a natural site.
(D) Comparison of the top 30,000 peaks (30K) bound by NEUROD2 in P19 cells (X-axis) and MYOD in MEFs (Y-axis) demonstrates ~20% overlap of binding sites. From the origin, bins represent the top 3K peaks, then the top 6K peaks, etc, as determined by peak height rank. Colors represent the proportion of sites bound by both NEUROD2 and MYOD (see Experimental Procedures).
(E) NEUROD2 and MYOD shared E-box sequence correlates with shared binding sites. Comparison of binding site overlap between NEUROD2 in P19 cells and MYOD in MEFs restricted to the top 30K peaks centered on a GC E-box demonstrates ~40% overlap.
See also Figure S2.
To determine other potential factors influencing NEUROD2 binding, we performed a de novo motif search for all motifs enriched within an area spanning 200 nt around the peak summit and compared this to randomly selected regions in the genome of similar GC content. In addition to enrichment for E-boxes, we also observed an increased frequency of the AATCAAT PBX motif (Figure 2B). PBX proteins have previously been demonstrated to be important during retinoic acid-mediated neuronal differentiation of P19 cells (Qin et al., 2004). We also observed enrichment for other homeobox-like motifs with the consensus sequences DGATTA, TAATKA and CAATTA. Numerous homeobox proteins expressed in P19 cells, such as LHX2, PITX2, OTX2, and EN2, have roles in neuronal development and neural lineage specification (Acampora et al., 1999; Evans and Gage, 2005; Koenig et al., 2010; Subramanian et al., 2011), and these homeobox motifs also represent potential binding sites for POU domain factors, one of which, Brn2, has recently been described to assist in the direct conversion of fibroblasts to neurons (Pang et al., 2011; Vierbuchen et al., 2010).
NEUROD2 and MYOD Bind to Shared and Private E-boxes That Correlate With Common and Distinct Genomic Binding Sites
The preferred E-box for NEUROD2 (CAG[C/A]TG) overlaps with the preferred E-box for MYOD (CAG[C/G]TG), derived from a similar analysis of MYOD binding sites in mouse muscle cells and embryonic fibroblasts converted to muscle by transduction with MYOD (Cao et al., 2010)(Figure 2A). This suggested that there might be a common set of binding sites with a GC core as well as a set of NEUROD2-specific (GA core) and MYOD-specific (GG core) sites. To evaluate this further, we estimated the E-box binding affinities of NEUROD2 and MYOD with in vitro gel shift competition assays. While both bHLH proteins are capable of binding each of these E-boxes in vitro, there is a clear binding preference of NEUROD2 for the GA E-box and of MYOD for the GG E-box (Figure 2C). Conversely, we observed an approximately equivalent affinity between NEUROD2 and MYOD for the GC E-box (data not shown).
To determine if these shared (GC) and private (GA for NEUROD2 and GG for MYOD) E-box motifs correlated with shared and private binding, we compared ChIP-Seq peaks from NEUROD2 in P19 cells and MYOD in cells differentiated into skeletal muscle. For the MYOD binding profile, we used MEFs converted to skeletal muscle by lentiviral expression of MYOD. This binding profile was similar to both the previously published profile in differentiated C2C12 mouse myoblasts and primary muscle cells (manuscript in preparation).
The total number of NEUROD2 peaks in P19 cells ranged from ~35,000 (p < 10−10) to ~72,000 (p < 10−5), and the number of MYOD peaks in MEFs ranged from ~67,000 to ~124,000 at the same p-value thresholds (Table S2). To evaluate the degree of peak overlap, we organized peaks into bins based on their rank according to peak p-values, and plotted the degree of overlap within corresponding bins representing the top 30,000 peaks for both NEUROD2 and MYOD. Overall, there was ~20% overlap of the top 30,000 NEUROD2 and MYOD peaks (Figure 2D). As anticipated, we observed very little overlap between NEUROD2 and MYOD binding at peaks centered on a GA or GG E-boxes: 8.8% of GA peaks bound by NEUROD2 were also bound by MYOD, whereas 12.5% of GG peaks bound by MYOD were also bound by NEUROD2. In contrast, ~40% of GC peaks bound by NEUROD2 were also bound by MYOD (Figure 2E), indicating that the majority of shared NEUROD2 and MYOD binding occurs at GC E-boxes, while binding at private E-boxes is not shared.
NEUROD2 and MYOD Private Sites are Associated with Differentiation Programs
The presence of shared and private NEUROD2 and MYOD peaks suggested the possibility of shared and private gene expression profiles. While NEUROD2 and MYOD are presumed to control neuron and muscle specific genes, respectively, a direct comparison of their regulated genes has not yet been performed, to our knowledge. Based on our expression array data, there were 990 genes up-regulated by MYOD in MEFs and 532 genes up-regulated by NEUROD2 in P19 cells, with 67 genes up-regulated by both factors (all compared to the same cell-type not expressing MYOD or NEUROD2, with a log 2-fold change cutoff).
We determined whether these private and shared transcription programs correlated with private or shared binding by assessing the presence of private or shared ChIP-Seq peaks within the promoter regions (+/− 2 kb of the TSS) of genes expressed in neurons, muscle, or both (Figure 3A). The private genes specifically activated by NEUROD2 and not MYOD were enriched for NEUROD2 private peaks (i.e., bound by NEUROD2 and not MYOD). The differential binding was significantly correlated with the presence of a NEUROD2 preferred PWM (color coded in Figure 3A). Similarly, the genes activated by MYOD and not NEUROD2 were enriched for MYOD private peaks bound to a MYOD preferred PWM.
Figure 3. The neurogenic and myogenic programs correlate with NEUROD2 and MYOD binding to private sites.
(A) Scatter plot of private and shared NEUROD2 and MYOD peaks within the promoter regions (+/− 2kb from the TSS) of genes up-regulated by NEUROD2 in P19 cells (ND.P19), MYOD in MEFs (MD.MM), or up-regulated by both (shared). The number of reads is represented in square root transformation. Sites are further characterized by PWM score (see Experimental Procedures): green, NEUROD2 private site; red, MYOD private site; black, shared site. Genes with multiple TSS were excluded.
(B) Sites occurring within 2 kb of a TSS plotted against the fold change in gene activation for (a) NEUROD2 and (b) MYOD. (Y-axis: square root transformation of peak height; X-axis: log-2 fold change in gene expression). (c) box plot of fold change in gene activation (log 2) comparing private and shared sites for NEUROD2 and MYOD. There is greater gene activation associated with private sites based on a Wilcoxon Rank Sum test for NEUROD2 (p < 10−6) and for MYOD (p = 0.027). Using a threshold of 2-fold change in expression, 21.1% of genes associated with NEUROD2 private sites have fold change >/= 2, compared to 5.4% of genes associated with shared sites (p = 8.2e-9 per Fisher’s exact test). For MYOD, 25.1% of genes associated with private peaks have fold change >/= 2, and 23.5% for shared peaks (p = 0.67), whereas 11.8% of genes associated with private sites have a fold-change >/= 8 compared to 6.3% of genes associated with a shared site (p = 0.03).
(C) Reporter constructs containing paired E-boxes with the indicated central nucleotides were transfected into P19 cells with NEUROD2 (top) or MEFs with MYOD (bottom). *p-value < 0.05 by t-test compared to vector without E-box insertion (pGL3); error bars represent 1 standard deviation.
(D) Scatter plot of peak height derived from native ChIP-Seq for acetyl-histone 4 in P19 cells prior to (X-axis) and after (Y-axis) transduction with NEUROD2. Shared, NEUROD2-induced change in acetylation at sites shared sites; ND.private, acetylation at NEUROD2 private sites; MD.private, acetylation at sites not bound by NEUROD2 in P19 cells. Number of reads are shown in square root transformation.
(E) Y-axis represents the number of raw reads from native ChIP-Seq for acetyl-histone 4, divided by strand (blue: + strand, red: - strand). X-axis represents nucleotide position centered on the E-box closest to the summit of either the private (left half) or shared (right half) NEUROD2 peaks. There is little histone acetylation in P19 cells at baseline (bottom panels), and a significant increase in histone acetylation after differentiation with NEUROD2 (top panels).
See also Table S3.
The shared genes that were activated by both MYOD and NEUROD2 were associated with shared peaks (i.e., MYOD and NEUROD2 peaks in the same location as demonstrated by their distribution along the 45 degree axis of the scatter plot comparing MYOD and NEUROD2 binding within the promoter regions of these genes, Figure 3A ‘shared’ panel). While in many instances this shared binding appeared to be associated with NEUROD2 or MYOD private E-boxes (note the green and red points along the 45 degree axis), assessment of individual promoters demonstrated the presence of both shared GC E-boxes and private E-boxes within close proximity of the peak summits (Figure S2C). The set of genes regulated by both MYOD and NEUROD2 included many factors involved in signaling, vesicle transport and other components of cell differentiation (Table S3). It is notable that nearly 10% of this set of genes regulates the activity of bHLH factors, either by directly interacting with bHLH factors (Id1, Id2, Hes6, Cbfa2t3), or by binding to E-box sequences (Znf238, Zeb1). In addition, the shared program was enriched in Notch signaling pathway genes (5/67=7%: Notch1, Dll1, Dner, Hes6, and Megf10) and genes critical for cell differentiation (Cdk5r1, Pou4f1, Mllt11, and Rb1). Therefore, both MYOD and NEUROD2 bind a common set of sites and regulate a shared program of cell differentiation that includes critical genes in the Notch, cell cycle, and differentiation pathways.
To further assess the relative importance of the private and shared E-box motifs for gene regulation, we asked whether binding at a private or shared motif better correlated with regional gene expression. For this analysis we defined shared and private sites by the presence of a shared or private peak that also contained a shared or private motif with a high PWM score and assigned these sites to a gene if they were within 2 kb of a TSS. Genes bound by NEUROD2 at private sites showed greater up-regulation than genes bound by NEUROD2 at shared sites (Figure 3B), despite similar peak heights in both groups (data not shown). Genes bound by MYOD at private sites also showed a trend toward higher activation compared to genes bound by MYOD at shared sites, although not as significant as for NEUROD2 (Figure 3B). GSEA analysis on the genes associated with the NEUROD2 or MYOD private sites, whether transcriptionally regulated or not, demonstrated enrichment for categories associated with neurogenic and myogenic development, respectively, while the genes associated with shared sites were associated with general cellular and metabolic processes (Table S3). Together, this suggests that the private NEUROD2 and MYOD sites are more important for the regulation of lineage specific genes.
To determine whether the apparently greater transcriptional activity of the NEUROD2 private sites can be partly attributed to the private E-box motif, we tested the ability of NEUROD2 to activate paired E-box reporter constructs that differed only in the core dinucleotides of the E-box sequence. NEUROD2 preferentially activated a reporter construct driven by paired NEUROD2 private E-boxes (GA) compared to MYOD private E-boxes (GG) or shared E-boxes (GC) (Figure 3C). Many bHLH factors, such as MYOD and NEUROD2, require paired binding sites for transcriptional activation (or an E-box paired with another factor binding site) because cooperative interactions stabilize the weak binding to an isolated E-box (Weintraub et al., 1990). Combining one GA E-box with an E-box that NEUROD2 will not bind (CG core that is bound by the MYC family but not NEUROD2 or MYOD) or binds relatively weakly (GG core) resulted in significantly decreased activity compared to the paired GA E-boxes; whereas pairing a single GA E-box with a GC E-box had substantially more activity than the paired GC E-boxes, although less than the paired GA E-boxes. Therefore, it appears that a GC E-box can facilitate the activity of a GA E-box, possibly by facilitating cooperative binding. Although MYOD activated the reporter with MYOD private E-boxes more than with shared E-boxes, the difference was not as dramatic as for NEUROD2, and MYOD also activated a reporter with the NEUROD2 private E-boxes (Figure 3C, lower panel). Although speculative, it will be interesting to determine whether additional flanking motifs at MYOD sites will confer a greater distinction in the activities of private and shared sites.
Previously, we demonstrated that MYOD binding was associated with regional histone 4 acetylation (Cao et al., 2010). To further understand the role of the shared binding sites, we compared histone acetylation changes specifically at shared and private sites. We observed enhanced acetylation occurring equivalently at both the private GA and the shared GC NEUROD2 binding sites (Figure 3D). In addition, binding of NEUROD2 at both of these sets of sites induces a bi-modal histone distribution that has been suggested to indicate a functional binding site (Hoffman et al., 2010) (Figure 3E). Therefore, although relatively few genes are commonly regulated by both MYOD and NEUROD2, their shared genome-wide binding at GC E-boxes results in widespread alterations of nucleosome positioning and modification.
NEUROD2 and MYOD Binding is Determined by Chromatin Accessibility and Co-factor Motifs
While there was ~40% overlap between the GC binding profiles of NEUROD2 in P19 and MYOD in MEF, a significant proportion of these E-boxes were not shared. To assess chromatin accessibility at all GC E-boxes, we exposed nuclei from P19 cells and MEFs to PvuII, which cleaves CAGCTG sites, and sequenced the cleaved sites, a modification of NA-Seq (Gargiulo et al., 2009) (see Experimental Procedures and Figure S3A). We ranked GC E-boxes based on their relative accessibilities to PvuII prior to introduction of NEUROD2 or MYOD.
P19 cells and MEFs had ~50% overlap of the top 100,000 accessible GC E-boxes (Figure S3B), indicating cell-type differences in chromatin accessibility. To investigate the extent to which chromatin accessibility determines factor binding, we restricted our analysis to GC E-boxes with a high PWM score for MYOD and NEUROD2 in their flanking nucleotides, and thus good binding motifs for either MYOD or NEUROD2. Notably, E-boxes with very low nuclease accessibility in P19 cells or MEFs had very few NEUROD2 or MYOD peaks, respectively, indicating that nuclease inaccessible E-boxes were also inaccessible to these factors (Figure 4A). E-boxes that were accessible to the nuclease, on the other hand, showed a broad range of binding.
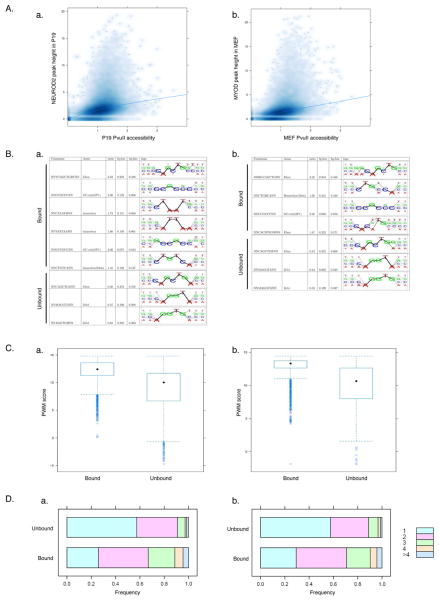
Figure 4. Chromatin accessibility is necessary but not sufficient for NEUROD2 and MYOD binding.
(A) Scatter plots comparing (a) NEUROD2 binding sites in P19 cells and (b) MYOD binding sites in MEFs with PvuII nuclease accessibility at these sites. NEUROD2 and MYOD peak height (Y-axis) and the normalized accessibility of PvuII sites (X-axis, see Experimental Procedures for calculation) are represented in square root transformation. Only PvuII sites in the context of good MYOD and NEUROD2 motif matches with PWM scores >/= 14 are included. Blue line is the fitted loess curve.
(B) Motif enrichment analysis comparing bound and unbound sites within PvuII accessible areas for (a) NEUROD2 in P19 cells and (b) MYOD in MEFs (ratio: enriched/depleted ratio of motifs; fg.frac, bg.frac: fraction of foreground/background sequences that contain at least one motif occurrence).
(C) Plot of E-box PWM (Y-axis) for (a) NEUROD2 and (b) MYOD bound and unbound sites within PvuII accessible regions demonstrates a higher average PWM at bound regions.
(D) Plot of the number of E-boxes at PvuII accessible regions either bound or unbound by (a) NEUROD2 in P19 cells or (b) MYOD in MEFs. Colors represent the number of E-boxes located within the 200bp window of a PvuII accessible site. X-axis is the frequency of sites containing the depicted number of E-boxes.
See also Figure S3.
We looked for motifs that might distinguish bound sites from unbound sites within nuclease accessible regions. Compared to accessible but unbound sites, NEUROD2 bound sites in P19 cells were enriched 1.5-fold for the MEIS motif and 1.7 to 1.8-fold for motifs of other homeodomain factors (Figure 4B). Accessible sites bound by MYOD in MEFs were enriched 1.4-fold for the MEIS motif. In addition to these factor motifs, the sites that were accessible and bound were enriched for good consensus E-boxes with a higher average PWM compared to accessible and unbound sites (Figure 4C). There was also a higher average total number of E-boxes at NEUROD2 and MYOD bound sites compared to unbound sites within PvuII accessible areas (Figure 4D). Together, these results indicate that, in addition to chromatin accessibility, good PWM E-boxes, additional adjacent E-boxes, and motifs for other potential cooperative factors modulate the binding of NEUROD2 in P19 cells and MYOD in MEFs. These data are consistent with prior studies showing that a MEIS-containing complex cooperates with MYOD binding at the Myogenin promoter (Berkes et al., 2004).
For both MYOD and NEUROD2, accessible but unbound sites were enriched for the ZEB1 motif. As noted above, both MYOD and NEUROD2 activate the expression of Zeb1 and Znf238, both factors that bind E-boxes and suppress activity of MYOD and/or NEUROD2 (Postigo and Dean, 1997; Yokoyama et al., 2009). Therefore, both MYOD and NEUROD2 activate the expression of factors that might prevent their access to a subset of E-boxes.
Cell Lineage Determines Binding and Gene Regulation
Our results suggest that private sites correlate with, and likely determine, the private genes activated by NEUROD2 and, to a lesser extent, by MYOD. To determine whether cell lineage constrains differentiation potential by site accessibility, we compared gene expression and binding profiles for MYOD expressed in P19 cells and NEUROD2 expressed in MEFs. Neither expression of MYOD in P19 cells nor NEUROD2 in MEFs by lentiviral delivery resulted in myogenesis or neurogenesis. This is consistent with prior studies showing that <3% of P19 cells transfected with MYOD differentiate into muscle, and NEUROD2 expression alone is insufficient to convert fibroblasts to neurons (Skerjanc et al., 1994; Yoo et al., 2011). However, despite the absence of differentiation, there remained a significant degree of genome wide binding and gene regulation by these factors.
There were 51,004 NEUROD2 peaks in MEFs and 21,695 MYOD peaks in P19 cells at a threshold p-value of 10−10 (Table S2). Comparing NEUROD2 and MYOD peaks across cell types, we observed ~30% overlap for the top 30,000 NEUROD2 peaks between P19 cells and MEFs, and a similar overlap between the top 30,000 MYOD peaks in both cell types (Figure 5D). 605 genes were up-regulated by NEUROD2 in MEFs and only 83 genes overlapped with genes up-regulated by NEUROD2 in P19 cells. The majority of genes uniquely up-regulated by NEUROD2 in MEFs were associated with GO categories not involved in neural development, but rather extracellular and membrane components (Table S1). For MYOD, 134 genes were up-regulated in P19 cells and 68 overlapped with genes up-regulated by MYOD in MEFs. In contrast to NEUROD2 in MEFs, these genes were associated with a number of GO terms related to muscle development, potentially representing a partial activation of the myogenic program (Table S1). Overall, however, the majority of the transcriptional programs of neurogenesis and myogenesis were not activated when MYOD or NEUROD2 was expressed in the opposite cell type, and this correlated with decreased binding at promoter-proximal sites near these genes (Figure S3C). A motif analysis again identified the PBX motif (ATCAAT) as enriched in genes up-regulated by NEUROD2 and not MYOD in P19 cells, and RUNX (ACCACA) at genes up-regulated by MYOD and not NEUROD2 in MEFs (Figure S3D), again implicating PBX and RUNX as cell-type specific co-factors. However, further analysis of ChIP-Seq peaks did not demonstrate a preferential enrichment for these motifs at peaks near regulated genes compared to peaks near unchanged genes (data not shown), suggesting these factors might primarily influence binding but not transcriptional activation.
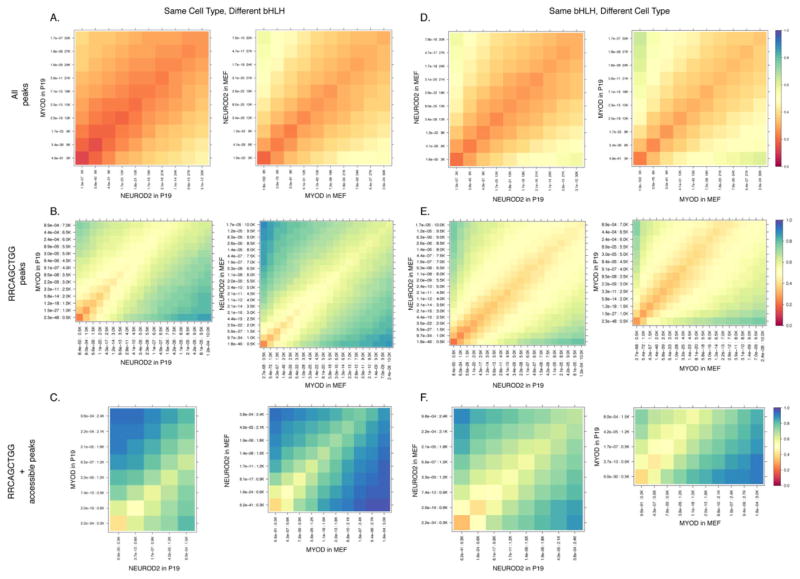
Figure 5. NEUROD2 and MYOD binding between cell types is strongly determined by chromatin accessibility.
(A) Comparison of the top 30,000 peaks (30K) bound by NEUROD2 and MYOD in the same cell type (left: P19 cells, right: MEFs) demonstrates a ~30% overlap of peaks. From the origin, bins represent the top 3K peaks, then the top 6K peaks, etc, as determined by peak height rank. Color scale represents the percentage of peaks bound by both NEUROD2 and MYOD (see Experimental Procedures). Bins are presented with their corresponding p-values for peak height.
(B) Restriction of the comparison in (A) to the top 7–10,000 peaks (7–10K) containing a RRCAGCTGG E-box.
(C) Further restriction of the comparison in (B) to the top 1,500–3,000 peaks (1.5–3K) containing a RRCAGCTGG E-box with a high nuclease accessibility score (normalized value > 2).
(D) Comparison of the top 30,000 peaks (30K) bound by NEUROD2 in both P19 and MEFs (left) or MYOD in both P19 and MEFs (right).
(E) Restriction of the comparison in (D) to the top 7–10,000 peaks (7–10K) containing a RRCAGCTGG E-box.
(F) Further restriction of the comparison in (E) to the top 1,500–3,000 peaks (1.5–3K), and containing a RRCAGCTGG E-box with a high nuclease accessibility score.
Chromatin Accessibility Is the Major Determinant of Lineage-Specific Binding
To determine whether chromatin accessibility in each cell type is the major determinant of binding pattern for each factor, we compared the binding profile of NEUROD2 in P19 and MEFs and the profile of MYOD in P19 and MEFs at all sites or at accessible sites. When expressed in the same cell type, where accessibility is the same for both factors, there was ~30% overlap between MYOD and NEUROD2 when comparing the top 30,000 peaks grouped by rank (Figure 5A), which included both private and shared sites. Restricting this comparison to the top 10,000 peaks containing a good consensus MYOD and NEUROD2 shared E-box (RRCAGCTGG), however, significantly increased the overlap to ~70% for MYOD and NEUROD2 peaks within the same cell type (Figure 5B). Further restricting the analysis to the shared E-box peaks with high accessibility by the PvuII assay showed a nearly complete overlap of MYOD and NEUROD2 binding within the same cell type (Figure 5C).
As stated previously, when comparing NEUROD2 or MYOD binding profiles between different cell types, there was ~30% overlap for all sites (see Figure 5D). This degree of overlap increased only modestly to ~40% upon restricting the analysis to the top 10,000 peaks with a consensus shared E-box (RRCAGCTGG, Figure 5E). However, further restriction of the analysis to consensus E-boxes with high nuclease accessibility scores in both cell types increased the overlap to ~80–90% (Figure 5F). These findings indicate that the pre-existing chromatin structure and associated binding site accessibility are major determinants of MYOD and NEUROD2 binding in the different cell types.
DISCUSSION
Our results are consistent with prior studies on the specific activity of individual members of a family of transcription factors and suggest an emerging model of how related transcription factors maintain some common functions and yet achieve specific transcriptional activity. Similar to studies on the ETS family of factors, MYOD and NEUROD2 bind to a shared E-box motif and each has its own distinct private E-box motif. Binding at the NEUROD2 private sites, and to a lesser extent at the MYOD private sites, is correlated with transcriptional activation of their respective differentiation programs, which is similar to the reported association of factor-specific binding sites with genes regulated by individual members of the ETS family. In contrast, binding at the NEUROD2 or MYOD shared sites does not show the same degree of regional gene activation. In addition, NEUROD2 showed stronger transcriptional activation of a reporter driven by its private E-boxes compared to the shared E-box motifs, and MYOD showed the same trend. This does not appear secondary to affinity, since peak height was similar at private and shared sites (data not shown). These findings indicate that motif sequence might confer a level of transcriptional activity on the bound NEUROD2 or MYOD, similar to the sequence-specific allosteric activation described for the GR receptor (Meijsing et al., 2009).
The E-box motif for NEUROD2 is similar to the consensus binding site identified for the related neurogenic bHLH factor ATOH1 (RMCAKMTGKY) in a ChIP-Seq study from mouse cerebellum (Klisch et al., 2011). The central dinucleotide preferences are similar to NEUROD2, whereas ATOH1 appears to have a palindromic flanking nucleotide preference different from NEUROD2, although this might result from the motif algorithm method used. Interestingly, a subset of flanking nucleotides are enriched at ATOH1 E-boxes in enhancers of genes expressed in dorsal interneurons (AMCAGMTG) (Lai et al., 2011), suggesting E-box specificity might have a role in neuronal subtype gene regulation; however, functional differences were not observed in this study.
The biological role of the NEUROD2 and MYOD shared sites remains unclear. Although we do not yet know the biological significance of these shared sites, the induction of a bimodal H4 acetylation signal is similar to the criteria developed for biologically functional binding sites for several transcription factors (FOXA2, PDX1, HNF4A) in liver development (Hoffman et al., 2010), and it is interesting to speculate that the alteration of histone modifications at many thousands of sites genome-wide might have a yet unknown biological function that is distinct from regional transcription, perhaps related to nuclear compartments and/or architecture (Lieberman-Aiden et al., 2009).
It is interesting that MYOD and NEUROD2 both induce the expression of Znf238 and Zeb1. ZNF238 binds to a consensus sequence that includes the CAGATG E-box, whereas the ZEB1 site includes the CAGGTG E-box. In skeletal muscle cells, ZNF238 has been shown to inhibit the expression of the Id genes and its binding appears to prevent MYOD activity at the same region (Yokoyama et al., 2009). Similarly, ZEB has been shown to bind the E-box in the IgH enhancer and prevent its activation in non-B cells (Genetta et al., 1994). Therefore, MYOD and NEUROD2 initiate the expression of factors that can suppress their activities at a subset of E-boxes, possibly limiting the genes regulated by each factor. This is consistent with the transient activation of Id genes by MYOD and might be a general method of suppressing the early programs initiated by MYOD and NEUROD2.
It is interesting that NEUROD2 activates approximately the same number of genes in MEFs as in P19 cells but there is very little overlap in the set of regulated genes. Similarly, MYOD activated different sets of genes with partial overlap in P19 cells and MEFs. Therefore, both are active transcription factors in both cell types, but the cell-type determines the target genes that will be activated. Our nuclease access studies indicate that chromatin structure is a major determinant of binding site accessibility in the different cell lineages. This is consistent with the studies showing that nuclease accessibility predicts GR binding (Biddie et al., 2011; John et al., 2011). However, accessibility is not the only determinant of binding at a particular site. Motif analysis determined that additional E-boxes were associated with both NEUROD2 and MYOD peaks and PBX and homeobox-like motifs with NEUROD2 peaks. This study together with our prior MYOD ChIP-Seq study (Cao et al., 2010) identified MEIS and RUNX motifs with MYOD peaks. Therefore, accessibility is important for the spectrum of sites available for MYOD and NEUROD2, whereas other factor motifs may influence the degree of binding at particular accessible sites. Although associated with NEUROD2 or MYOD binding, we did not find an association of these motifs specifically with regulated genes (data not shown), suggesting a role in binding rather than transcriptional activation. This is in contrast to the strong association of RUNX1 motifs near TAL1 binding sites in T-cells (Palii et al., 2011), where RUNX appears to play a direct role in TAL1 binding and gene regulation. It is also important to note that in our study we are identifying associated motifs and have not directly identified the factors binding at these motfis.
In this study and in our prior MYOD ChIP-Seq study (Cao et al., 2010) we identified tens of thousands of bound sites. In both studies, neither peak height nor p-value accurately predicted peaks that were associated with a regulated gene. An important consideration in this study is that we have forced the expression of both MYOD and NEUROD2 by lentiviral transduction. Our previous publication on endogenous MYOD binding in C2C12 mouse muscle cells and MEFs virally transduced with MYOD showed a 90% similarity in peak location. In addition, comparison of the lentiviral MYOD binding in MEFs with endogenous MYOD in C2C12 cells and primary mouse myotubes shows a similar level of concordance (Z. Yao, manuscript in preparation), indicating that the lentiviral transduction produces an accurate representation of the binding of endogenous MYOD, possibly because of limiting amounts of the endogenous E-protein dimerization partner, which would also be true for NEUROD2.
In summary, both NEUROD2 and MYOD bind to tens of thousands of sites genome-wide. Factor-specific transcriptional programs appear to be encoded, at least in part, by private E-boxes that drive the transcriptional programs of neurons and muscles in P19 cells and MEFs, respectively; whereas many thousands of shared sites are associated with histone acetylation but not as strongly associated with regional gene transcription, particularly for NEUROD2. Cell lineage determines the accessibility of the sites and constrains the transcriptional response by each factor. The fact that NEUROD2 and MYOD activate the expression of large numbers of genes that are not normally a part of their differentiation program when expressed in a different lineage (i.e., NEUROD2 in MEFs and MYOD in P19 cells) indicates that lineage transitions, such as epithelial-to-mesenchymal transition, could profoundly alter the transcriptional program of these, or other, transcription factors.
EXPERIMENTAL PROCEDURES
Microarray and GO analysis
Total RNA samples were collected in triplicate from undifferentiated and differentiated P19 cells and MEFs and labeled cDNA was made per Affymetrix protocol. Samples were hybridized on Affymetrix Mouse 430 2.0 Expression Arrays. The microarrays were analyzed using Bioconductor simpleaffy and limma package. Differentially expressed genes were chosen with fdr cutoff 0.05 and fold change cutoff of 2. The trend line in Figure 1C was computed using a loess local regression method. GO analysis was performed using the Bioconductor GOstats package. Association studies of peak binding affinity and gene expression were performed as previously described (Cao et al., 2010).
ChIP-Seq
ChIP was performed as previously described (Cao et al., 2010). Briefly, ~108 cells were fixed in 1% formaldehyde for 11 minutes, quenched with glycine, lysed, and then sonicated to generate final DNA fragments of 150–600 bp. The soluble chromatin was diluted 1:10 and pre-cleared with a 1:1 Protein A:G slurry for 2 hours at 4°C. Chromatin was mixed with antibody overnight at 4°C, then Protein A:G beads for 2 hours. Beads were washed and de-crosslinked overnight for ~16 hours in 1%SDS, 0.1M NaHCO3 and 70 μg Proteinase K. ChIP samples were validated by qPCR and prepared for sequencing per the Illumina Sample Preparation protocol, with two modifications: (1) DNA fragments of 150–300 bp were selected at the gel-selection step; (2) 21 cycles of PCR were performed at the amplification step instead of 18. For the controls, we used P19 cells ChIPed with pre-immune serum and MEFs ChIPed with MYOD antibody. We performed native ChIP with micrococcal nuclease digestion per a published protocol (Brand et al., 2008). Mononucleosomes were isolated at the final gel-selection step. All samples were sequenced with the Illumina Genome Analyzer II and IIx platforms.
PvuII endonuclease accessibility assay
5x106 cells were trypsinized and washed once in reticulocyte suspension buffer (RSB: 10mM Tris pH 7.4, 10mM NaCl, 5mM MgCl2), followed by resuspension in lysis buffer (RSB + 0.1% NP-40) at a final concentration of 1.5x106 cells/mL and incubation on ice for 10 minutes. Nuclei were pelleted and washed in lysis buffer, followed by resuspension in 200μl of 1X NEB buffer 2, addition of 40 units of PvuII (NEB) per 106 nuclei, and incubation at 37°C for 30 minutes. 200μl of STOP buffer (0.6M NaCl, 20mM Tris pH 7.4,10mM EDTA, 1% SDS, 2 mg/mL proteinase K) was added and the reaction was incubated at 37°C overnight. Genomic DNA was isolated using Qiagen DNeasy spin columns. 5μg of DNA was used for labeling, beginning with addition of an ‘A’ tail to the blunt ends generated by PvuII digestion using Klenow 3–5′ exo− (NEB). After purification through MinElute columns (Qiagen), custom designed biotinylated adapters with a ‘T’ overhang and an EcoRV site immediately upstream of the ‘T’ (purchased from IDT) were ligated onto the ‘A’-tailed ends using Quick Ligase (NEB). After purification (Qiagen), DNA was fragmented to 150–350bp using a Diagenode Bioruptor (low amplitude, 30 seconds/cycle, 30 cycles). Biotinylated fragments were enriched with Streptavidin-conjugated Dynabeads (Invitrogen), and DNA was released from the beads by digestion with EcoRV for 1 hour at 37°C. Fragments were subsequently purified and labeled for sequencing as above.
ChIP-Seq peak calling and significance inference
Sequences were extracted using the GApipeline software. Reads mapping to the X and Y chromosomes were excluded from our analysis. Reads were aligned using MAQ and BWA to the mouse genome (mm9). Duplicate sequences were discarded to minimize affects of PCR amplification. Each read was extended in the sequencing orientation to a total of 200 bases to infer the coverage at each genomic position. We performed peak calling by an in-house developed R package “peakSig”(pending submission to Bioconductor), which models background reads by a negative binomial distribution. The negative binomial distribution can be viewed as a continuous mixture of Poisson distribution where the mixing distribution of the Poisson rate is modeled as a Gamma prior. This prior distribution is used to capture the variation of background read density across the genome. The parameters of the negative binomial distribution were estimated by fitting the truncated distribution on the number of nucleotides with coverage 1–3, to avoid the problem of inferring effective genome size excluding the non-mappable regions, and to eliminate contamination of any foreground signals in the high coverage regions. We also fit separate model parameters based on the binned GC content of the flanking sequence, which based on our observations heavily correlates with background read density. Therefore, the significance of the peaks is determined not only by peak height, but also by the GC content of the flanking sequence. We used control ChIP-Seq samples (pre-immune serum in P19 cells, MYOD antibody in MEFs) to eliminate statistically significant peaks likely due to artifact. We removed all peaks that overlap with the peaks in the control sample at p-value cutoff of 10^−5, and required all remaining peaks to have a much more significant p-value (10^−3) than in the control sample.
Motif analysis
We used a discriminative de-novo motif discovery tool described previously (Cao et al., 2010; Palii et al., 2011) to find motifs that distinguish foreground and background sequence datasets. To find motifs enriched under ChIP-Seq peaks, we selected background sequences using random genomic regions sampled with similar GC content and distance to TSS. The motif z-values follow a normal distribution if there is no distinction between foreground and background sequences. To learn a positional weight matrix (PWM) model, we used the output motif instances from the motif discovery tool as the seed to initialize the iterative expectation-maximization (EM) refinement process, which is essentially the same as MEME. In some cases, the motifs are extended iteratively as long as there is sequence preference in the flanking region, and refined in the same EM process.
ChIP-Seq sample comparison
Cross cell-type comparison is difficult, as it is unclear how to set up a fair comparison baseline due to the differences in the sample preparation protocol, total number of reads, foreground/background reads distribution, and in some cases, even the underlying genome sequences. Here, we adopt a rank-based paradigm to compare ChIP-Seq samples of different transcription factors and cell types, while still taking the peak p-value significance into account. We rank all peaks by their p-values and group ranks into bins of 3000 (i.e., the top 3K peaks, then the top 6K peaks, etc). Then we compute the fraction of top x peaks in one sample that overlap with the top y peaks in another sample, where x and y vary from 3K to 30K, and y is equal to or greater than x. For the top 20,000 peaks overlapping between NEUROD2 in P19 and MYOD in MEFs, the average degree of overlap was 529bp (69% of peak width), with 90% of peaks overlapping more than 369bp. For comparison of overlap at specific E-boxes, we estimate the overlap of peaks containing a GC E-box underneath the summit. The same procedure is then used, except that the peaks are ranked among all GC-containing subset.
PvuII and histone 4 acetylation data analysis
The reads at a typical PvuII site with GC E-box can be divided into four categories, based on whether they are from the 5′ end or 3′ end of the fragments, whether they are at the cleaved ends of the fragments, or the random sonicated ends. We define the accessibility by combining reads both at the cleaved ends and the at the sonicated ends within 200bp from the cleavage site, and normalizing this value by dividing it with the median value of the reads at all PvuII sites. These two components are comparable for the bulk of the data. For the histone acetylation data, we used 500bp sliding windows across the genome, and used the number of reads falling into each window to assess the genome-wide acetylation pattern. Then we evaluated the histone acetylation within the 500bp window centered at the NEUROD2 or MYOD binding sites to study the association between the two. To assess the degree of change in chromatin state in MEFs and P19 cells with or without NEUROD2 or MYOD, we used the DESEQ Bioconductor package to detect changes in accessibility or histone acetylation under two conditions. For the PvuII nuclease accessibility data, we used replicates for a better estimate of variance. For the histone acetylation data without replicates, we used the pooled variance estimates.
Supplementary Material
HIGHLIGHTS.
MYOD and NEUROD2 bind to shared and factor-specific E-boxes
Binding at factor-specific motifs is associated with gene transcription.
MYOD and NEUROD2 binding is constrained to E-boxes in accessible chromatin
Lineage determines accessibility of genetically determined differentiation programs
Acknowledgments
S.J.T. was supported by NIH NIAMS R01AR045113; A.P.F was supported by a grant from the University of Washington Child Health Research Center, NIH U5K12HD043376-08; Z.Y. was supported by an Interdisciplinary Training Program grant, T32 CA080416.
Footnotes
ACCESSION NUMBERS
Microarray and ChIP-Seq data have been deposited in Gene expression Omnibus (GEO) under accession number GSE34908 (subseries GSE34906 and GSE34907). The MYOD ChIP-Seq control data is in the Short Read Archive study ID:SRP001761, Experiment ID:SRX016194.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acampora D, Gulisano M, Simeone A. Otx genes and the genetic control of brain morphogenesis. Mol Cell Neurosci. 1999;13:1–8. doi: 10.1006/mcne.1998.0730. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Bergstrom DA, Penn BH, Seaver KJ, Knoepfler PS, Tapscott SJ. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Molecular cell. 2004;14:465–477. doi: 10.1016/s1097-2765(04)00260-6. [DOI] [PubMed] [Google Scholar]
- Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, et al. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Molecular cell. 2011;43:145–155. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc. 2008;3:398–409. doi: 10.1038/nprot.2008.8. [DOI] [PubMed] [Google Scholar]
- Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez GJ, Parker MH, MacQuarrie KL, Davison J, Morgan MT, Ruzzo WL, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae JH, Stein GH, Lee JE. NeuroD: the predicted and the surprising. Mol Cells. 2004;18:271–288. [PubMed] [Google Scholar]
- Evans AL, Gage PJ. Expression of the homeobox gene Pitx2 in neural crest is required for optic stalk and ocular anterior segment development. Hum Mol Genet. 2005;14:3347–3359. doi: 10.1093/hmg/ddi365. [DOI] [PubMed] [Google Scholar]
- Farah MH, Olson JM, Sucic HB, Hume RI, Tapscott SJ, Turner DL. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Gargiulo G, Levy S, Bucci G, Romanenghi M, Fornasari L, Beeson KY, Goldberg SM, Cesaroni M, Ballarini M, Santoro F, et al. NA-Seq: a discovery tool for the analysis of chromatin structure and dynamics during differentiation. Dev Cell. 2009;16:466–481. doi: 10.1016/j.devcel.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakim O, Sung MH, Voss TC, Splinter E, John S, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, de Laat W, Hager GL. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res. 2011;21:697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman BG, Robertson G, Zavaglia B, Beach M, Cullum R, Lee S, Soukhatcheva G, Li L, Wederell ED, Thiessen N, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res. 2010;20:1037–1051. doi: 10.1101/gr.104356.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, McIntosh LP, Graves BJ. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem. 2011;80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nature genetics. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klisch TJ, Xi Y, Flora A, Wang L, Li W, Zoghbi HY. In vivo Atoh1 targetome reveals how a proneural transcription factor regulates cerebellar development. Proc Natl Acad Sci U S A. 2011;108:3288–3293. doi: 10.1073/pnas.1100230108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig SF, Brentle S, Hamdi K, Fichtner D, Wedlich D, Gradl D. En2, Pax2/5 and Tcf-4 transcription factors cooperate in patterning the Xenopus brain. Dev Biol. 2010;340:318–328. doi: 10.1016/j.ydbio.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Lai HC, Klisch TJ, Roberts R, Zoghbi HY, Johnson JE. In vivo neuronal subtype-specific targets of Atoh1 (Math1) in dorsal spinal cord. J Neurosci. 2011;31:10859–10871. doi: 10.1523/JNEUROSCI.0445-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung TH, Hoffmann A, Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Li XY, Thomas S, Sabo PJ, Eisen MB, Stamatoyannopoulos JA, Biggin MD. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome biology. 2011;12:R34. doi: 10.1186/gb-2011-12-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murre C. Helix-loop-helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- Palii CG, Perez-Iratxeta C, Yao Z, Cao Y, Dai F, Davison J, Atkins H, Allan D, Dilworth FJ, Gentleman R, et al. Differential genomic targeting of the transcription factor TAL1 in alternate haematopoietic lineages. Embo J. 2011;30:494–509. doi: 10.1038/emboj.2010.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, et al. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo AA, Dean DC. ZEB, a vertebrate homolog of Drosophila Zfh-1, is a negative regulator of muscle differentiation. Embo J. 1997;16:3935–3943. doi: 10.1093/emboj/16.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P, Haberbusch JM, Zhang Z, Soprano KJ, Soprano DR. Pre-B cell leukemia transcription factor (PBX) proteins are important mediators for retinoic acid-dependent endodermal and neuronal differentiation of mouse embryonal carcinoma P19 cells. J Biol Chem. 2004;279:16263–16271. doi: 10.1074/jbc.M313938200. [DOI] [PubMed] [Google Scholar]
- Seo S, Lim JW, Yellajoshyula D, Chang LW, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. Embo J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skerjanc IS, Slack RS, McBurney MW. Cellular aggregation enhances MyoD-directed skeletal myogenesis in embryonal carcinoma cells. Mol Cell Biol. 1994;14:8451–8459. doi: 10.1128/mcb.14.12.8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian L, Sarkar A, Shetty AS, Muralidharan B, Padmanabhan H, Piper M, Monuki ES, Bach I, Gronostajski RM, Richards LJ, et al. Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus. Proc Natl Acad Sci U S A. 2011;108:E265–274. doi: 10.1073/pnas.1101109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Li XY, Sabo PJ, Sandstrom R, Thurman RE, Canfield TK, Giste E, Fisher W, Hammonds A, Celniker SE, et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome biology. 2011;12:R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Davis R, Lockshon D, Lassar A. MyoD binds cooperatively to two sites in a target enhancer sequence: occupancy of two sites is required for activation. Proc Natl Acad Sci U S A. 1990;87:5623–5627. doi: 10.1073/pnas.87.15.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Ito Y, Ueno-Kudoh H, Shimizu H, Uchibe K, Albini S, Mitsuoka K, Miyaki S, Kiso M, Nagai A, et al. A systems approach reveals that the myogenesis genome network is regulated by the transcriptional repressor RP58. Dev Cell. 2009;17:836–848. doi: 10.1016/j.devcel.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, O’Shea EK. Integrated approaches reveal determinants of genome-wide binding and function of the transcription factor Pho4. Molecular cell. 2011;42:826–836. doi: 10.1016/j.molcel.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.