Abstract
There has recently been increasing interest in the phenomenon of circadian rhythmicity. We have used circadian rhythms as a means to understanding the regulation of glucose absorption in the intestine. We and others have previously demonstrated rhythmicity in intestinal glucose uptake, mediated by rhythmicity in expression of the sodium glucose co-transporter SGLT1. Rhythmicity of clock gene expression was subsequently confirmed in the intestine, a phenomenon also demonstrated in other viscera. Clock genes have since been shown via a combination of in vitro and in vivo techniques to play a role in the transcriptional regulation of key absorptive proteins.
This review article highlights the importance of circadian rhythmicity in mammalian physiology and explores the role of clock genes in the regulation of intestinal function. We describe the studies that have examined clock gene-mediated regulation of gene transcription as well as the contribution of knockout mouse models in understanding the importance of circadian rhythmicity in intestinal function.
Understanding the physiological regulation of nutrient uptake has profound implications for the modulation of intestinal absorption as a therapeutic option for conditions such as diabetes and obesity.
Keywords: Clock genes, SGLT1, circadian, intestine
Intestinal absorption of nutrients
The major role of the intestine in absorption of luminal nutrients into the bloodstream [1] has driven evolution of its physiology and anatomy to meet this function. Prominent examples are its villous structure to increase surface area and the co-localization of hydrolases and transporters to facilitate absorption. Ultimately, epithelial flux of nutrients, fluid and electrolytes is proportional to intestinal length[1]. Therefore, the loss of an intestinal segment, most often caused by massive small bowel resection, significantly compromises intestinal function and results in its failure to meet nutritional needs, a phenomenon known as short bowel syndrome[2]. The limited range of therapeutic options with their many associated side-effects merits the continued search for more effective alternatives[2, 3]. Development of improved treatments will likely be realised through a better understanding of intestinal absorption and, in particular, its regulation.
A remarkable observation has been a dramatic circadian increase in intestinal absorptive capacity. Because the daily changes in intestinal absorptive capacity are quite dramatic, many researchers have begun to investigate the mechanisms underlying this rhythmicity in intestinal function to gain new insights that might have therapeutic benefit. This review describes the circadian rhythmicity in intestinal function, with particular emphasis on the rhythmicity of clock genes in the intestine and the postulated role of this molecular clockwork in the regulation of intestinal absorption.
Circadian rhythmicity
Circadian rhythmicity is the innate ability of organisms to detect and adapt to external stimuli, evidenced by the adaptation to the 24-hour cyclic pattern of light and the daily and seasonal patterns of food availability and temperature[4]. Circadian rhythmicity has been found in organisms from the unicellular to the complex, such as plants and humans[4]. Physiological and pathological examples of circadian rhythms in visceral function include circadian rhythms in heart rate[5], blood pressure[6], intestinal function[7], nocturnal worsening of asthma [8] and gout[9–12]. The characteristics of circadian rhythms are similar among most species, including flies, mice and humans. First, synchronization occurs by environmental stimuli (zeitgebers – “time-givers”)[13], the most potent of which is light in most mammals. Second, circadian rhythmicity persists in the absence of cycling zeitgebers, i.e. under constant conditions, such as constant darkness[14]. This persistent rhythm is controlled by an internal cellular clock as discussed below.
The circadian clockwork
Circadian rhythms are controlled at a cellular level by a set of genes collectively known as “clock genes” - transcriptional regulators that maintain 24-h rhythmicity via two interacting transcription loops. Physiological output rhythms are subsequently generated by transcriptional regulation of key “clock-controlled genes” (CCG) that are responsive to the clock gene transcription factors. Cells in most tissues express circadian clock genes, with the central clock in the suprachiasmatic nucleus (SCN) responsive to environmental stimuli and subsidiary clocks in peripheral tissues that serve to regulate local functions and are cued to the central clock.
Central circadian clock
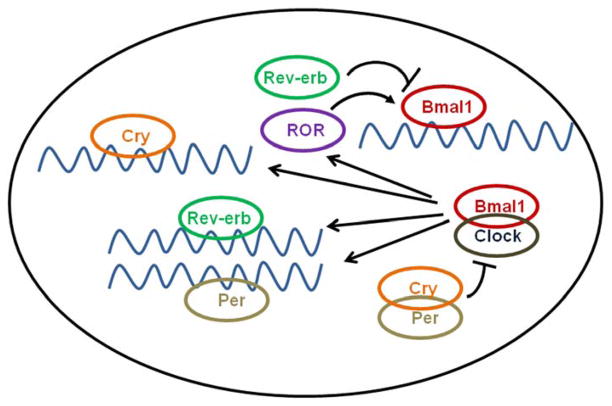
In mammals, the master clock resides in the SCN and maintains a 24-h periodicity entrained by light, but persists in the absence of light/dark cycles[15]. This periodicity is regulated via opposing positive and negative molecular feedback loops involving the clock genes [16, 17]. The negative-feedback loop involves the rhythmic induction of three Period genes (designated Per1–3) and two Cryptochrome genes (Cry1 and Cry2) by heterodimerization of the transcription factors CLOCK and BMAL1 (Figure 1)[18–21]. CLOCK and BMAL1 contain the functionally important basic helix-loop-helix (bHLH)–PER-ARNT-SIM (PAS) protein dimerization domain[22, 23], which binds to and activates E-boxes (sequences of CAnnTG) on the promoters of the PER and CRY genes[23, 24]. PER and CRY subsequently form multimers that translocate to the nucleus, where they directly interact with CLOCK and/or BMAL1 to inhibit transcription[18, 20]. This in turn induces rhythmicity in the transcription of Bmal1, with a phase opposite that of Per or Cry, resulting in a positive-feedback loop[20, 25].
Figure 1.
Schematic showing the molecular feedback loops of circadian clock genes. Black pointed arrows indicate stimulation of one clock gene component by another, while the truncated arrows indicate repression.
More recently three further genes have been identified – Retinoic acid-receptor-related orphan receptor (ROR) -α and Rev-erbA and B[26]. These orphan nuclear receptors have been identified as key regulators linking the positive and negative limbs of the circadian oscillator, with Reverb and ROR-α transcription driven by BMAL1/CLOCK and in turn suppressing and activating Bmal1 expression respectively, acting as an accessory loop to improve the amplitude and stability of the core clock regulatory loops[26, 27].
Clock gene expression in the SCN is cued by the detection of light by the retina, which is transmitted to the SCN by several convergent pathways, the most important of which is the retinohypothalamic tract[28–30] which arises from the retinal ganglion cells and transmits the light signal to the SCN using the neurotransmitter glutamate[31]. The persistence of circadian rhythms in blind humans and their ability to respond to light with circadian phase changes suggests a separate non-visual light detection system, independent of rods and cones, which mediates phase setting by the circadian clock[32]. Indirect pathways such as the geniculohypothalamic tract also play a role, as do neurohumoral signals such as the pineal hormone melatonin[17].
Output genes
Clock genes subsequently regulate downstream events via rhythmically expressed CCGs that are regulated (directly or indirectly) by the core feedback loops[33, 34]. Examples of CCGs directly regulated via CLOCK:BMAL1 heterodimers acting on E-box enhancers include arginine vasopressin (AVP)[33] and albumin D-element-binding protein (DBP)[33, 34].
Clock genes are clearly important transcriptional regulators[35–37]. Rhythmic expression of several proteins has been found to be regulated by clock and clock-controlled genes; for instance CLOCK/BMAL1 heterodimers bind directly to the promoter of the Na+/H+ exchanger Nhe3 in the kidney to induce circadian rhythmicity of Nhe3 mRNA in the rat[35]. In the intestine, the clock-controlled output gene, DBP has been noted to bind to and activate the promoter of the oligopeptide transporter Pept1 [36], while similarly clock-controlled output genes hepatic leukemia factor (HLF) and E4 promoter binding protein-4 (E4BP4) regulated transcription of the multidrug resistance 1 (Mdr1a) gene via a reciprocating mechanism involving competition for the same Mdr1a promoter DNA binding site[37].
Peripheral circadian oscillators
Circadian rhythmicity exists in many tissues besides the SCN[38–42] and may be mediated by rhythmically expressed clock genes in these tissues. Although cued by the SCN under normal physiological conditions, the ability of peripheral oscillators to be entrained by other external stimuli such as nutrients[43–45] allows the organism to optimally adapt to match environmental challenges, for example to alter liver function to match the timing of delivery of nutrients and metabolites[43, 44]. The presence of peripheral oscillators likely provides increased flexibility to allow the organism to adapt to a greater variety of stimuli. Peripheral oscillators have now been characterized in a number of other tissues, including the heart, lung, kidney, peripheral blood cells and liver, with a 3–9h phase delay in oscillatory rhythms relative to the SCN [39–42].
The molecular machinery driving rhythmicity of the circadian clock in peripheral tissues appears to exhibit some significant differences from that in the SCN. Firstly, peripheral clock gene rhythms exhibit a phase delay relative to that in the SCN[43, 44]. Secondly, in mice lacking a functional CLOCK protein (Clock−/− mutants), Bmal1 levels are elevated in peripheral tissues but blunted and expressed at reduced levels in the SCN[46], suggesting that negative autoregulation by CLOCK/BMAL1 heterodimers affects Bmal1 transcription in the periphery but not in the SCN. Thirdly, unlike in vitro cultures of SCN cells, which maintain their rhythms indefinitely[47], cultures derived from peripheral tissues exhibit circadian rhythms which dampen after 2–7 cycles[48]. These findings suggest that peripheral oscillators may involve a population of asynchronous cells requiring intermittent synchronizing stimuli, either from the SCN or external entraining cues such as feeding. This is further corroborated by the ability of a serum shock consisting of 50% serum to induce rhythmicity in clock gene transcription in cell lines including fibroblast derived cell lines in vitro[49].
The intestine is a prime example of a tissue exhibiting peripheral clocks. Clock genes are expressed in all regions of the gut in rodents, from the stomach to the caecum and predominantly in enterocytes[50]. Clock, Bmal1, Per1, Per2, Cry1, Cry2, ReverbA and ReverbB have all been found to exhibit diurnal rhythmicity in rodent jejunal mucosa[51] with a similar phase compared to the liver but a phase delay of approximately 6 hours compared to the SCN[41, 43, 52, 53]. Several studies have subsequently confirmed that the transcriptional rhythmicity of some of these genes is matched by rhythmicity in protein expression[45, 54, 55].
Peripheral clocks are primarily cued by the SCN master clock - destruction of the SCN flattens oscillatory rhythms in peripheral tissues such as the liver[56]. Circadian transcripts continued to oscillate following liver-specific deletion of Clock in mice in the presence of an intact SCN clock[57, 58], possibly as a result of persistent rhythmicity in feeding behaviour. The nature of the signal from SCN to the peripheral oscillators is not known.
Although light, the predominant Zeitgeber for the SCN, may indirectly initiate rhythmicity of peripheral tissues, other stimuli can also regulate peripheral oscillators and dissociate rhythms of the peripheral clock from that of the SCN[43, 44, 56, 59–62]. These include adrenergic stimulation[56], glucocorticoid exposure[59–62] and one of the strongest cues – nutrient availability[43, 44]. Nutrient availability is a powerful Zeitgeber - restricted feeding was able to reinstate circadian rhythmicity of hundreds of genes which had been blunted in Cry1−/−Cry2−/− mice and in otherwise arrhythmic SCN-lesioned mice[63]. Restricted feeding also reset the peripheral clocks in the liver, kidney, heart and pancreas within one week with no change in the phase of the SCN clock[43, 44, 64]. Glucocorticoid signalling has been postulated as a potential regulator of peripheral clocks, specifically as mediators of the effects of nutrient-induced phase shifts in clock gene expression[59–62]. However recent studies have demonstrated that the phase shifts in rat liver seen in restricted feeding could not be induced by daily corticosterone injections to simulate the endogenous peak of these hormones in animals subjected to restricted feeding[43]. Le Minh et al developed these findings further by showing that rather than facilitating phase shift, glucocorticoids act to inhibit phase shifts of peripheral oscillators to daytime feeding and may in fact exist to prevent a rapid uncoupling of peripheral oscillators from the SCN[65].
Peripheral rhythms can persist in the absence of photic entrainment, presumably by the free-running rhythmic behaviour maintained under constant conditions. Specifically, mice held in constant darkness continued to exhibit rhythmic feeding and 15% of their liver transcripts continued to exhibit circadian rhythmicity despite a 24-hour dark-dark cycle[66]. This study demonstrated that the liver clock does not need a light dark cycle for operation.
Circadian rhythmicity in nutrient absorption
Early studies
Furuya and Yugari first demonstrated in 1974 that intestinal absorption exhibits a diurnal rhythm in rats, with a peak in histidine absorption during their normal nocturnal feeding period[67, 68]. This work was extended by Fisher and Gardner in 1976, who confirmed peak absorption of glucose during the nocturnal period, but further demonstrated that rhythmicity of sugar absorption was dependent on nutrient availability by showing that peak absorption could be shifted to daytime by restricting the feeding to the light phase[7]. These findings preceded the discovery and characterization of specific glucose transporters, hence these experiments measured total glucose uptake using isolated segments of intestine at diurnal timepoints. Other groups have also demonstrated rhythmicity in intestinal function besides rhythms in absorption. Saito demonstrated diurnal rhythms in the activities of maltase and leucylnaphthylamidase, with peak activity occurring during feeding periods[69]. Subsequently circadian rhythmicity in intestinal absorption was confirmed and further developed in a series of experiments by Stevenson et al, who demonstrated circadian rhythmicity in the activity of sucrase, lactase, trehalase, γ-glutamyltransferase and as previously shown, maltase and leucylnaphthylamidase[70].
Sugar uptake
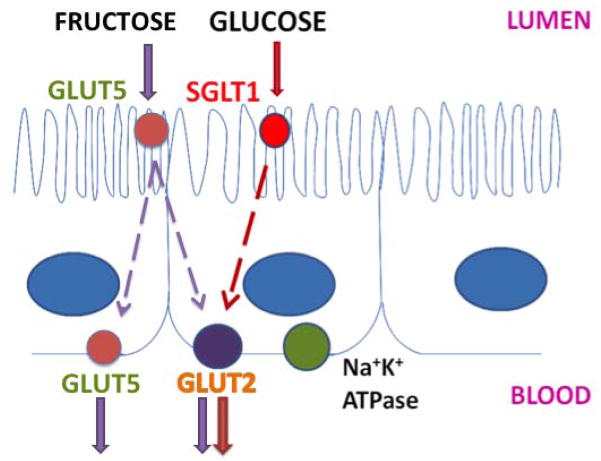
While circadian clock genes have been shown to regulate rhythms of intestinal ion, drug and peptide transporters[35–37], their role in the rhythmicity of glucose absorption is less well understood. The sodium glucose co-transporter SGLT1 is responsible for all active glucose uptake in the intestine; subsequent movement of glucose across the basolateral membrane occurs down a diffusion gradient (Figure 2)[71, 72]. Thus, SGLT1 is an essential component of intestinal glucose absorption and understanding its regulation is of paramount importance. The initial findings of diurnal rhythmicity in intestinal glucose absorption had preceded the characterization of specific intestinal transporters. Subsequent studies from our laboratory and others demonstrated diurnal rhythmicity of Sglt1 in rats with peak mRNA expression in the afternoon and evening, in anticipation of and during the time of peak nutrient uptake[73–75]. These studies used phloridzin as an SGLT1-specific inhibitor and identified complete loss of rhythmicity following SGLT1 inhibition, indicating that SGLT1-mediated glucose uptake was entirely responsible for the diurnal rhythmicity of glucose absorption[73, 75].
Figure 2.
Schematic showing the localization of glucose and fructose transporters in enterocytes. SGLT1 (red circle) is situated on the apical border of the enterocyte while GLUT2 (purple circle) is on the basolateral membrane. Fructose is transported by GLUT5 on the apical membrane and by both GLUT5 and GLUT2 on the basolateral membrane. The Na+K+ ATPase (green circle), also localized to the basolateral membrane, provides the Na+ gradient necessary for SGLT1 activity.
SGLT1 rhythmicity has also been documented in non-human primates[74], with a 12 hour difference in the time of peak expression consistent with the nocturnal rat versus the diurnal Rhesus monkey. This highlights the relevance of circadian rhythmicity across species and suggests that the circadian rhythmicity noted in the rat can be extrapolated to primates.
Two other intestinal transporters cooperate with SGLT1 in the intestinal absorption of glucose – GLUT2 and GLUT5[76–78]. While SGLT1 transports glucose from the lumen into the cytoplasm of the enterocyte, the high-capacity low-affinity transporter GLUT2 resides on the basolateral membrane of enterocytes and transports glucose, fructose, galactose and mannose out of the cell[76, 77] (Figure 2). The facilitated glucose transporter GLUT5 is responsible for the uptake of fructose at the brush border membrane of the intestine[78]. Fructose is then transported out of the enterocyte by both GLUT 5 and GLUT2 at the basolateral membrane[78, 79]. Rhythmicity of GLUT2 and GLUT5 mRNA and protein expression has also been demonstrated[80].
Peptide uptake
The transporter PEPT1 is the only oligopeptide transporter at the intestinal brush-border, responsible for the transport of 400 dipeptides and 8000 tripeptides as well as a wide range of peptide-like drugs, such as β-lactam antibiotics and angiotensin-converting enzyme inhibitors[81]. PEPT1 is a H+-peptide cotransporter that relies on a proton gradient for the uphill transport of peptides, which are subsequently transported across the basolateral membrane by the facilitative basolateral peptide transporter[82]. Peptide absorption has been shown to display diurnal rhythmicity with peak in absorption of L-histidine occurring during the dark phase in nocturnal animals[67], coincident with higher levels of PEPT1 protein expression during the dark phase than the light phase[83].
Recent studies have shown that certain drug transporters also exhibit rhythmicity of expression in the intestine[84]. These transporters act as efflux proteins in the intestine to facilitate the excretion of the metabolic products of many drugs including digoxin[85] and methotrexate[84. Five drug transporters (Mdr1, Mct1, Mrp2, Bcrp and the peptide transporter Pept1, which also transports β-lactam drugs[86]) are diurnally rhythmic in the intestine at a transcriptional level[84]. A 2–5 fold change in expression was observed between peak and trough times and peak expression varied from morning to late afternoon[84]. This observation of circadian rhythmicity of drug transporters highlights the opportunity for chemotherapy regimens targeted to match peak transporter expression to optimise therapeutic efficacy[87].
Lipid uptake
Rhythmicity of intestinal absorption also applies to the absorption of lipids; mice have been shown to absorb significantly higher amounts of triglyceride and cholesterol during the night than during the day, corresponding with rhythmicity in expression of two proteins involved in lipid absorption, intestinal microsomal triglyceride transfer protein (MTP) and apoB[50].
Lessons from Clock gene knockout mice
As noted above, all regions of the rodent gut express circadian clocks[45]. The effects of clock genes on the rhythmicity of intestinal function were further studied by Pan et al in a murine Clock mutant model[50]. Rhythmicity of circadian clock gene expression was dampened or absent in the intestines of Clock, mutant mice compared to wild-type (WT) mice. In addition, Clock mutants demonstrated overall higher rates of monosaccharide and lipid absorption, coincident with increased basal expression of the hexose transporters SGLT1, GLUT2 and GLUT5. In contrast PEPT1 expression and peptide absorption were lower in mutant mice, who also displayed loss of rhythmicity of SGLT1, GLUT2, GLUT5 and PEPT1 expression as well as loss of rhythmicity of monosaccharide, peptide and lipid absorption compared to WT mice. Clock mutants also failed to entrain to restricted feeding regimes unlike WT mice, suggesting that Clock is necessary in nutrient-mediated entrainment of the peripheral clock.
Cues regulating peripheral circadian rhythms
Nutrient availability vs. light cycle
The majority of research performed on circadian rhythmicity in nutrient absorption has been performed in the rats, nocturnal feeders. The association of nutrient consumption with the dark phase in the rat[73] has necessitated further experiments to isolate the period of nutrient availability from light cycle. The first experiments on this were performed by Fisher and Gardner[7], who demonstrated that restricting feeding to only a few hours during the daytime resulted in an initial reduction in food consumption and weight loss, but this normalized within one week. Rates of absorption were similar toward the end of the feeding period regardless of whether animals were fed during the lights on or lights off period[7]. Restricted feeding produced a coordinated phase shift in the temporal expression of clock genes as well as SGLT1; peak expression of these genes was shifted forwards by 8 hours in rats fed during the lights-on period compared to those fed during the lights-off period, suggesting nutrients to be a strong cue of rhythmicity in the intestine, that over rides the central clock[51]. A more recent study by Polidarova et al showed partial preservation of circadian rhythmicity of gene expression in rats rendered behaviourally arrhythmic by prolonged exposure to constant lights-on conditions[88]. Differential preservation of rhythmicity was noted dependent on the section of the GI tract; the greatest degree of preservation was maintained in the duodenum compared to the liver and colon. Similarly the introduction of a restricted feeding regimen under constant lights-on conditions resynchronized the peripheral clock of the duodenum and liver to a greater extent than that of the colon. Resynchronization of the colon was more robust under 12:12 light-dark conditions[55], suggesting that despite its response to nutrient cues the colon may be more susceptible than the duodenum to environmental factors such as constant lighting.
Glucocorticoids
Glucocorticoids are secreted by the adrenal gland[1] with a circadian rhythm[89], reaching a peak around or just before the onset of activity in both diurnal and nocturnal mammals and have been implicated in the regulation of circadian function as the rhythmicity in their expression is able to rapidly adapt to alterations in activity or feeding schedule[65]. Administration of glucocorticoids was able to synchronize asynchronous fibroblasts in vitro to a common circadian phase[49, 90] and transiently induce the expression of mPer1 in the mouse liver in vivo[61]. The effects of glucocorticoids are dependent on a functional glucocorticoid receptor, which is absent in the suprachiasmatic nucleus (SCN)[61, 91], hence glucocorticoid administration was unable to shift SCN clock gene expression. Glucocorticoids are also known to affect glucose absorption in rats by stimulating transcription of glucose transporters[92]. The specific role of glucocorticoids in the regulation of the circadian rhythm of intestinal glucose absorption has not been studied.
Recent studies
Several groups have attempted to better delineate the mechanisms governing circadian regulation of intestinal function. A recent study by Iwashina et al demonstrated circadian rhythmicity of the binding of BMAL1 to regions at −400 and +0 on the Sglt1 promoter relative to the Sglt1 transcription start site in mice, suggesting a role for the importance of the proximal promoter region in the regulation of Sglt1 rhythmicity[93]. Functionality of BMAL1 on the E-boxes was not specifically assesed in their study and is yet to be determined[93]. As yet there are no data on the exact relationship of specific clock genes to the basolateral glucose transporter GLUT2.
The role of clock-mediated regulation of lipid absorption is also a topic of current interest. Douris et al demonstrated that the rhythmically expressed clock-regulated deadenylase nocturnin plays a key role in the optimization of dietary lipid absorption by enterocytes via regulation of chylomicron transit[94]. These findings may explain the increased absorption of triglyerides and cholesterol during the lights-off period, which has been noted to occur independent of the timing of nutrient intake[50].
Conclusions
The intestine exhibits striking rhythms in function, cued by nutrient availability and coincident with rhythmic expression of circadian clock genes as well as key intestinal transporters. Clock genes are transcriptional regulators of other circadian genes and have been implicated in the regulation of intestinal transporters however the exact molecular mechanisms behind this remain unclear. Greater insights into the role of clock genes in the regulation of rhythmicity in intestinal function may facilitate the development of new therapies to boost absorptive and other functions in malabsorptive conditions, thereby improving the management of intestinal failure and short bowel syndrome.
Abbreviations
- AVP
arginine vasopressin
- Bmal1
brain muscle Arnt-like 1
- CCG
Clock controlled genes
- Cry
cryptochrome
- DBP
albumin D-element-binding protein
- E4BP4
E4 promoter binding protein-4
- HLF
hepatic leukemia factor
- Mdr1
multidrug resistance 1
- mRNA
messenger ribonucleic acid
- Nhe3
Na+/H+ exchanger
- Per
period
- SCN
suprachiasmatic nucleus
- SGLT1
sodium glucose cotransporter 1
- WT
wild-type
Footnotes
Disclosures:
All authors confirm no conflicts of interest.
Copyright licence statement:
The authors attest that this article is not currently under consideration by any other journal.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sinnathamby C. Last’s Anatomy. 10. Churchill Livingstone: Elsevier; 2005. [Google Scholar]
- 2.Buchman AL. Etiology and initial management of short bowel syndrome. Gastroenterology. 2006;130(2 Suppl 1):S5–S15. doi: 10.1053/j.gastro.2005.07.063. [DOI] [PubMed] [Google Scholar]
- 3.Tavakkolizadeh A, Whang EE. Understanding and augmenting human intestinal adaptation: a call for more clinical research. JPEN J Parenter Enteral Nutr. 2002;26(4):251–5. doi: 10.1177/0148607102026004251. [DOI] [PubMed] [Google Scholar]
- 4.Gehring W, Rosbash M. The coevolution of blue-light photoreception and circadian rhythms. J Mol Evol. 2003;57(Suppl 1):S286–9. doi: 10.1007/s00239-003-0038-8. [DOI] [PubMed] [Google Scholar]
- 5.Struthius J. Ars Sphymica. Basel: Ludovici Konigs; 1602. [Google Scholar]
- 6.Zadek J. Die Messung des Blutdrucks des Menschen mittels des Basch’schen Apparates. Z Klin Med. 1881;2:509–551. [Google Scholar]
- 7.Fisher RB, Gardner ML. A diurnal rhythm in the absorption of glucose and water by isolated rat small intestine. J Physiol. 1976;254(3):821–5. doi: 10.1113/jphysiol.1976.sp011262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aurelianus C. JC Amman, recensuit, emaculavit, notulasque adjecit. Amsterdam: 1722. De morbis acutis & chronicis; p. 429.p. 527. [Google Scholar]
- 9.Sydenham T. The second edition corrected from the original Latin by John Perchey. Richard Wellington; London: 1697. The Whole Works of the Excellent Practical Physician Dr. Thomas Sydenham. [Google Scholar]
- 10.Lemmer B. Discoveries of rhythms in human biological functions: a historical review. Chronobiol Int. 2009;26(6):1019–68. doi: 10.3109/07420520903237984. [DOI] [PubMed] [Google Scholar]
- 11.Cannon CP, et al. Circadian variation in the onset of unstable angina and non-Q-wave acute myocardial infarction (the TIMI III Registry and TIMI IIIB) Am J Cardiol. 1997;79(3):253–8. doi: 10.1016/s0002-9149(97)00743-1. [DOI] [PubMed] [Google Scholar]
- 12.Krantz DS, et al. Circadian variation of ambulatory myocardial ischemia. Triggering by daily activities and evidence for an endogenous circadian component. Circulation. 1996;93(7):1364–71. doi: 10.1161/01.cir.93.7.1364. [DOI] [PubMed] [Google Scholar]
- 13.Mills JN. Human circadian rhythms. Physiol Rev. 1966;46(1):128–71. doi: 10.1152/physrev.1966.46.1.128. [DOI] [PubMed] [Google Scholar]
- 14.Halberg F, et al. 24-Hour rhythms at several levels of integration in mice on different lighting regimens. Proc Soc Exp Biol Med. 1958;97(4):897–900. doi: 10.3181/00379727-97-23915. [DOI] [PubMed] [Google Scholar]
- 15.Hastings MH. Circadian clocks. Curr Biol. 1997;7(11):R670–2. doi: 10.1016/s0960-9822(06)00350-2. [DOI] [PubMed] [Google Scholar]
- 16.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 17.Reppert SM, Weaver DR. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–76. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 18.Kume K, et al. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98(2):193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 19.Vitaterna MH, et al. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A. 1999;96(21):12114–9. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shearman LP, et al. Interacting molecular loops in the mammalian circadian clock. Science. 2000;288(5468):1013–9. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 21.Bae K, et al. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30(2):525–36. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 22.King DP, et al. Positional cloning of the mouse circadian clock gene. Cell. 1997;89(4):641–53. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hogenesch JB, et al. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A. 1998;95(10):5474–9. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gekakis N, et al. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280(5369):1564–9. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 25.Yu W, Nomura M, Ikeda M. Interactivating feedback loops within the mammalian clock: BMAL1 is negatively autoregulated and upregulated by CRY1, CRY2, and PER2. Biochem Biophys Res Commun. 2002;290(3):933–41. doi: 10.1006/bbrc.2001.6300. [DOI] [PubMed] [Google Scholar]
- 26.Preitner N, et al. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–60. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 27.Ueda HR, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418(6897):534–9. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 28.Johnson RF, Moore RY, Morin LP. Loss of entrainment and anatomical plasticity after lesions of the hamster retinohypothalamic tract. Brain Res. 1988;460(2):297–313. doi: 10.1016/0006-8993(88)90374-5. [DOI] [PubMed] [Google Scholar]
- 29.Lucas RJ, et al. Identifying the photoreceptive inputs to the mammalian circadian system using transgenic and retinally degenerate mice. Behav Brain Res. 2001;125(1–2):97–102. doi: 10.1016/s0166-4328(01)00274-1. [DOI] [PubMed] [Google Scholar]
- 30.Wright KP, Jr, Czeisler CA. Absence of circadian phase resetting in response to bright light behind the knees. Science. 2002;297(5581):571. doi: 10.1126/science.1071697. [DOI] [PubMed] [Google Scholar]
- 31.Ding JM, et al. Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science. 1994;266(5191):1713–7. doi: 10.1126/science.7527589. [DOI] [PubMed] [Google Scholar]
- 32.Van Gelder RN. Circadian rhythms: eyes of the clock. Curr Biol. 1998;8(22):R798–801. doi: 10.1016/s0960-9822(07)00503-9. [DOI] [PubMed] [Google Scholar]
- 33.Jin X, et al. A molecular mechanism regulating rhythmic output from the suprachiasmatic circadian clock. Cell. 1999;96(1):57–68. doi: 10.1016/s0092-8674(00)80959-9. [DOI] [PubMed] [Google Scholar]
- 34.Ripperger JA, et al. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14(6):679–89. [PMC free article] [PubMed] [Google Scholar]
- 35.Rohman SM, Emoto N, Nonaka H, Okura R, Nishimura M, Yagita K, van der Horst GT, Matsuo M, Okamura H, Yokoyama M. Circadian clock genes directly regulate expression of the Na(+)/H(+) exchanger NHE3 in the kidney. Kidney International. 2005;67:1410–1419. doi: 10.1111/j.1523-1755.2005.00218.x. [DOI] [PubMed] [Google Scholar]
- 36.Saito H, et al. Regulatory mechanism governing the diurnal rhythm of intestinal H+/peptide cotransporter 1 (PEPT1) Am J Physiol Gastrointest Liver Physiol. 2008;295(2):G395–402. doi: 10.1152/ajpgi.90317.2008. [DOI] [PubMed] [Google Scholar]
- 37.Murakami Y, et al. Circadian clock-controlled intestinal expression of the multidrug-resistance gene mdr1a in mice. Gastroenterology. 2008;135(5):1636–1644. e3. doi: 10.1053/j.gastro.2008.07.073. [DOI] [PubMed] [Google Scholar]
- 38.Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272(5260):419–21. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 39.Oishi K, et al. Humoral signals mediate the circadian expression of rat period homologue (rPer2) mRNA in peripheral tissues. Neurosci Lett. 1998;256(2):117–9. doi: 10.1016/s0304-3940(98)00765-4. [DOI] [PubMed] [Google Scholar]
- 40.Lau NC, et al. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 41.Takata M, et al. Daily expression of mRNAs for the mammalian Clock genes Per2 and clock in mouse suprachiasmatic nuclei and liver and human peripheral blood mononuclear cells. Jpn J Pharmacol. 2002;90(3):263–9. doi: 10.1254/jjp.90.263. [DOI] [PubMed] [Google Scholar]
- 42.Sakamoto K, et al. Circadian expression of clock genes during ontogeny in the rat heart. Neuroreport. 2002;13(10):1239–42. doi: 10.1097/00001756-200207190-00003. [DOI] [PubMed] [Google Scholar]
- 43.Stokkan KA, et al. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291(5503):490–3. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 44.Damiola F, et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14(23):2950–61. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoogerwerf WA, et al. Clock gene expression in the murine gastrointestinal tract: endogenous rhythmicity and effects of a feeding regimen. Gastroenterology. 2007;133(4):1250–60. doi: 10.1053/j.gastro.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 46.Oishi K, Fukui H, Ishida N. Rhythmic expression of BMAL1 mRNA is altered in Clock mutant mice: differential regulation in the suprachiasmatic nucleus and peripheral tissues. Biochem Biophys Res Commun. 2000;268(1):164–71. doi: 10.1006/bbrc.1999.2054. [DOI] [PubMed] [Google Scholar]
- 47.Welsh DK, et al. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14(4):697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 48.Yamazaki S, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288(5466):682–5. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 49.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93(6):929–37. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 50.Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. J Lipid Res. 2009;50(9):1800–13. doi: 10.1194/jlr.M900085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Balakrishnan A, et al. Restricted feeding phase shifts clock gene and sodium glucose cotransporter 1 (SGLT1) expression in rats. J Nutr. 2010;140(5):908–14. doi: 10.3945/jn.109.116749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Froy O, Chapnik N, Miskin R. Mouse intestinal cryptdins exhibit circadian oscillation. Faseb J. 2005;19(13):1920–2. doi: 10.1096/fj.05-4216fje. [DOI] [PubMed] [Google Scholar]
- 53.Zylka MJ, et al. Three period homologs in mammals: differential light responses in the suprachiasmatic circadian clock and oscillating transcripts outside of brain. Neuron. 1998;20(6):1103–10. doi: 10.1016/s0896-6273(00)80492-4. [DOI] [PubMed] [Google Scholar]
- 54.Pan X, Hussain MM. Clock is important for food and circadian regulation of macronutrient absorption in mice. J Lipid Res. 2009 doi: 10.1194/jlr.M900085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sladek M, et al. Insight into the circadian clock within rat colonic epithelial cells. Gastroenterology. 2007;133(4):1240–9. doi: 10.1053/j.gastro.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 56.Terazono H, et al. Adrenergic regulation of clock gene expression in mouse liver. Proc Natl Acad Sci U S A. 2003;100(11):6795–800. doi: 10.1073/pnas.0936797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kornmann B, et al. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5(2):e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105(39):15172–7. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamamoto T, et al. Acute physical stress elevates mouse period1 mRNA expression in mouse peripheral tissues via a glucocorticoid-responsive element. J Biol Chem. 2005;280(51):42036–43. doi: 10.1074/jbc.M509600200. [DOI] [PubMed] [Google Scholar]
- 60.Torra IP, et al. Circadian and glucocorticoid regulation of Rev-erbalpha expression in liver. Endocrinology. 2000;141(10):3799–806. doi: 10.1210/endo.141.10.7708. [DOI] [PubMed] [Google Scholar]
- 61.Balsalobre A, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289(5488):2344–7. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 62.Stearns AT, BA, Abolmaali K, Rhoads DB, Ashley SW, Tavakkolizadeh A. Corticosteroids Phase-Shift Intestinal Clocks and Diurnal Functional Rhythms. Gastroenterology. 2009;136(5):A-96. [Google Scholar]
- 63.Hara R, et al. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. Genes Cells. 2001;6(3):269–78. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- 64.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 65.Le Minh N, et al. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. Embo J. 2001;20(24):7128–36. doi: 10.1093/emboj/20.24.7128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–41. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Furuya S, Yugari Y. Daily rhythmic change of L-histidine and glucose absorptions in rat small intestine in vivo. Biochim Biophys Acta. 1974;343(3):558–64. doi: 10.1016/0304-4165(74)90274-8. [DOI] [PubMed] [Google Scholar]
- 68.Furuya S, Yugari Y. Daily rhythmic change in the transport of histidine by everted sacs of rat small intestine. Biochim Biophys Acta. 1971;241(1):245–8. doi: 10.1016/0005-2736(71)90321-x. [DOI] [PubMed] [Google Scholar]
- 69.Saito M. Daily rhythmic changes in brush border enzymes of the small intestine and kidney in rat. Biochim Biophys Acta. 1972;286(1):212–5. doi: 10.1016/0304-4165(72)90108-0. [DOI] [PubMed] [Google Scholar]
- 70.Stevenson NR, et al. Effect of changes in feeding schedule on the diurnal rhythms and daily activity levels of intestinal brush border enzymes and transport systems. Biochim Biophys Acta. 1975;406(1):131–45. doi: 10.1016/0005-2736(75)90048-6. [DOI] [PubMed] [Google Scholar]
- 71.Crane RK. Na+-dependent transport in the intestine and other animal tissues. Fed Proc. 1965;24(5):1000–6. [PubMed] [Google Scholar]
- 72.Wright EM, Martin MG, Turk E. Intestinal absorption in health and disease--sugars. Best Pract Res Clin Gastroenterol. 2003;17(6):943–56. doi: 10.1016/s1521-6918(03)00107-0. [DOI] [PubMed] [Google Scholar]
- 73.Tavakkolizadeh A, et al. Diurnal rhythmicity in intestinal SGLT-1 function, V(max), and mRNA expression topography. Am J Physiol Gastrointest Liver Physiol. 2001;280(2):G209–15. doi: 10.1152/ajpgi.2001.280.2.G209. [DOI] [PubMed] [Google Scholar]
- 74.Rhoads DB, et al. Circadian periodicity of intestinal Na+/glucose cotransporter 1 mRNA levels is transcriptionally regulated. J Biol Chem. 1998;273(16):9510–6. doi: 10.1074/jbc.273.16.9510. [DOI] [PubMed] [Google Scholar]
- 75.Balakrishnan A, et al. Diurnal rhythmicity in glucose uptake is mediated by temporal periodicity in the expression of the sodium-glucose cotransporter (SGLT1) Surgery. 2008;143(6):813–8. doi: 10.1016/j.surg.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thorens B, et al. Cloning and functional expression in bacteria of a novel glucose transporter present in liver, intestine, kidney, and beta-pancreatic islet cells. Cell. 1988;55(2):281–90. doi: 10.1016/0092-8674(88)90051-7. [DOI] [PubMed] [Google Scholar]
- 77.Cheeseman CI. GLUT2 is the transporter for fructose across the rat intestinal basolateral membrane. Gastroenterology. 1993;105(4):1050–6. doi: 10.1016/0016-5085(93)90948-c. [DOI] [PubMed] [Google Scholar]
- 78.Douard V, Ferraris RP. Regulation of the fructose transporter GLUT5 in health and disease. Am J Physiol Endocrinol Metab. 2008;295(2):E227–37. doi: 10.1152/ajpendo.90245.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jones HF, Butler RN, Brooks DA. Intestinal fructose transport and malabsorption in humans. Am J Physiol Gastrointest Liver Physiol. 2011;300(2):G202–6. doi: 10.1152/ajpgi.00457.2010. [DOI] [PubMed] [Google Scholar]
- 80.Fatima J, et al. Hexose transporter expression and function in mouse small intestine: role of diurnal rhythm. J Gastrointest Surg. 2009;13(4):634–41. doi: 10.1007/s11605-008-0776-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Adibi SA. The oligopeptide transporter (Pept-1) in human intestine: biology and function. Gastroenterology. 1997;113(1):332–40. doi: 10.1016/s0016-5085(97)70112-4. [DOI] [PubMed] [Google Scholar]
- 82.Terada T, Inui K. Peptide transporters: structure, function, regulation and application for drug delivery. Curr Drug Metab. 2004;5(1):85–94. doi: 10.2174/1389200043489153. [DOI] [PubMed] [Google Scholar]
- 83.Pan X, et al. Diurnal rhythm of H+-peptide cotransporter in rat small intestine. Am J Physiol Gastrointest Liver Physiol. 2002;283(1):G57–64. doi: 10.1152/ajpgi.00545.2001. [DOI] [PubMed] [Google Scholar]
- 84.Stearns AT, et al. Diurnal rhythmicity in the transcription of jejunal drug transporters. J Pharmacol Sci. 2008;108(1):144–8. doi: 10.1254/jphs.08100sc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Drescher S, et al. P-glycoprotein-mediated intestinal and biliary digoxin transport in humans. Clin Pharmacol Ther. 2003;73(3):223–31. doi: 10.1067/mcp.2003.27. [DOI] [PubMed] [Google Scholar]
- 86.Boll M, et al. Expression cloning of a cDNA from rabbit small intestine related to proton-coupled transport of peptides, beta-lactam antibiotics and ACE-inhibitors. Pflugers Arch. 1994;429(1):146–9. doi: 10.1007/BF02584043. [DOI] [PubMed] [Google Scholar]
- 87.Abolmaali K, et al. Circadian variation in intestinal dihydropyrimidine dehydrogenase (DPD) expression: a potential mechanism for benefits of 5FU chrono-chemotherapy. Surgery. 2009;146(2):269–73. doi: 10.1016/j.surg.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 88.Polidarova L, et al. Hepatic, duodenal, and colonic circadian clocks differ in their persistence under conditions of constant light and in their entrainment by restricted feeding. Chronobiol Int. 2011;28(3):204–15. doi: 10.3109/07420528.2010.548615. [DOI] [PubMed] [Google Scholar]
- 89.Moore RY, V, Eichler B. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42(1):201–6. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- 90.Nagoshi E, et al. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119(5):693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 91.Rosenfeld P, et al. Ontogeny of the type 2 glucocorticoid receptor in discrete rat brain regions: an immunocytochemical study. Brain Res. 1988;470(1):119–27. doi: 10.1016/0165-3806(88)90207-6. [DOI] [PubMed] [Google Scholar]
- 92.Thiesen A, et al. The locally acting glucocorticosteroid budesonide enhances intestinal sugar uptake following intestinal resection in rats. Gut. 2003;52(2):252–9. doi: 10.1136/gut.52.2.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Iwashina I, et al. Clock genes regulate the feeding schedule-dependent diurnal rhythm changes in hexose transporter gene expressions through the binding of BMAL1 to the promoter/enhancer and transcribed regions. J Nutr Biochem. 22(4):334–43. doi: 10.1016/j.jnutbio.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 94.Douris N, et al. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Curr Biol. 2011;21(16):1347–55. doi: 10.1016/j.cub.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]