Abstract
To study the process of morphogenesis, one often needs to collect and segment time-lapse images of living tissues to accurately track changing cellular morphology. This task typically involves segmenting and tracking tens to hundreds of individual cells over hundreds of image frames – a scale that would certainly benefit from automated routines; however, any automated routine would need to reliably handle a large number of sporadic, and yet typical problems (e.g., illumination inconsistency, photobleaching, rapid cell motions, drift of focus or of cells moving through the imaging plane). Here, we present a segmentation and cell tracking approach based on the premise that users know their data best – interpreting and using image features that are not accounted for in any a priori algorithm design. We have developed a program, SeedWater Segmenter (SWS), that combines a parameter-less and fast automated watershed algorithm with a suite of manual intervention tools that enables users with little to no specialized knowledge of image processing to efficiently segment images with near-perfect accuracy based on simple user interactions.
Keywords: image segmentation, computer-assisted image processing, tissue analysis, confocal microscopy, Drosophila, embryogenesis
INTRODUCTION
In studying embryonic development and morphogenesis, one often needs to track the morphological changes of individual cells in living tissues. This requires the collection of time-lapse movies of labeled cells, segmentation of each image frame into individual cells, and tracking cell identity across frames. Collecting the image sets using confocal or multiphoton fluorescence is now routine (1), but image segmentation and cell tracking represent substantial analysis bottlenecks. A number of algorithms and tools have been proposed for automated and/or manual segmentation and tracking of surface-labeled cells (2–12, reviewed in depth in 13–14), but few can segment and track tens to hundreds of close-packed cells over hundreds of image frames with an accuracy that correctly segments all cells distinguishable by the human eye. Automated methods, such as CellCognition and CellProfiler (2,4,5,7), are fast but do not attain the needed accuracy. Manual interactive tools like TrakEM2 and ITK-SNAP can attain the needed accuracy but are prohibitively slow (9–12). The only currently available tool that is both interactive and capable of segmenting and tracking packed cells in tissue is Packing Analyzer (3); unfortunately it is still quite slow.
Our approach to solving the segmentation and tracking problem is based on the premise that users know their data best; they may be able to interpret and use image features that are not accounted for in any a priori algorithm design. Thus, we have designed a system that combines a parameter-less and fast watershed algorithm with a suite of manual intervention tools that allows users with little to no specialized knowledge about image processing to efficiently segment images with near-perfect accuracy based on simple user interactions.
In general, the segmentation and tracking process can be broken into three steps: object identification, boundary generation, and object tracking (i.e., maintaining a consistent ID on the cell through time). Each step can be performed either manually or automatically. For example, object identification has been performed by manually clicking on an image to generate a “seed” for each cell (8–10) or by automatically finding such seeds using the minima of a Gaussian-filtered image or taking a threshold (4,11,12). Boundary generation has been performed manually by drawing a perimeter (4,9) or automatically via seed-based space-filling algorithms like a watershed or level set method (7,11–12). Likewise, object tracking can be applied as a manual post-processing step or as an automated postprocess technique, e.g., using maximal overlap of segmented regions in different frames to map ID's from one frame to the next (5,6,14). Object tracking has also been automated in-process by using the region centroids from one frame to generate seeds for space-filling algorithms applied to the next frame (7).
Our approach provides an initial automated guess at the seed positions (based on minima of a Gaussian-filtered image or the region centroids from segmentation of a previous frame) and uses an automated watershed algorithm to generate the region boundaries. Manual intervention comes through the use of in-process tools to add, delete, group and move seeds. As each change is made, the watershed algorithm automatically redraws the region boundaries. This process is repeated as necessary – automating the tedious process of finding the exact boundary locations, but allowing user control of object identification, segmentation, and tracking to any level of user-desired accuracy. Here we show applications of this method to time-lapse image sets of morphogenesis and wound healing in Drosophila embryos.
MATERIALS AND METHODS
Sample Preparation and Imaging
The primary strain of Drosophila melanogaster used in this study is ubi-DE-Cad-GFP (Kyoto Drosophila Genetic Resource Center), which ubiquitously expresses a cadherin-GFP fusion that labels epithelial cell junctions (15). Fly embryos were dechorionated and prepared for live imaging as described previously (16). Time-lapse image sets were captured on a Zeiss LSM410 laser-scanning confocal microscope (inverted) with a 40× 1.3 NA oil-immersion objective. Cellular ablations were performed with the 3rd harmonic (355 nm) of a Q-switched Nd:YAG laser (Continuum Minilite II, Santa Clara, CA) (16).
Segmentation Algorithm
Our segmentation and tracking system is based on a watershed approach. Initiation of watershed segmentation requires an initial set of starting pixels or seeds. Each seed has a unique identifier value that denotes the segmented region to which it will contribute. This allows multiple seeds for each region. The algorithm then fills the space by expanding the regions around each seed, starting with the darkest pixels first and slowly raising the “water” level. This process continues until the regions meet at boundaries and all pixels are assigned a value (17–19). We chose a watershed approach for three reasons: 1) it does a fairly good and consistent job of determining cell boundaries based on the bright lines of our GFP-cadherin fluorescence images; 2) it has the flexibility to use different numbers of seeds for each cell – one for most, but two or more for cells that are difficult to segment; and 3) it has no user-selectable parameters. This last point means that the user does not need previous image processing expertise to guide parameter selection.
To initialize the watershed segmentation and tracking procedure for an xyt (or xyz) image stack, we select seeds for the first xy-image based on the local minima after application of a Gaussian filter. This does require a user-selectable parameter – the width of the Gaussian kernel – but it is easy to manually vary this parameter until one obtains a reasonable compromise between under- and over-segmentation as in Figure 1. A Gaussian filter is not used to identify seeds for subsequent images. Instead, once the user is satisfied with the segmentation of image frame j, the centroid of each cell in frame j is used as a seed for frame j + 1 (using the same identifier value). This approach advantageously and automatically provides in-process cell tracking (7).
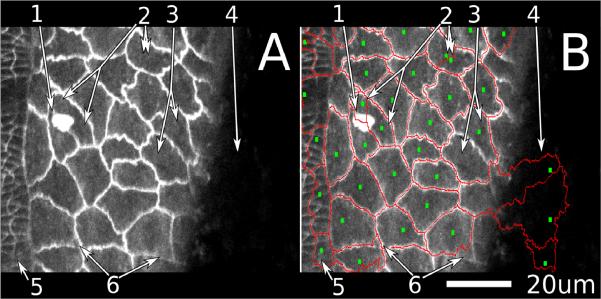
Figure 1.
Common segmentation difficulties in confocal images of living, cadherin-GFP stained fruit fly embryos. The large cells on the right of the image are amnioserosa cells; the smaller ones at the left are epidermal.
(A) is an unsegmented image and
(B) is the same image with an overlay of seeds (small green squares) generated automatically by application of a Gaussian filter (σ = 2.5 μm) and segment outlines (red lines) generated by a watershed algorithm.
The numbered arrows point to several common errors in automatic segmentation.
(1) An object obscures the view of the cell edge.
(2) A single cell is divided between two seeds, i.e., oversegmentation.
(3) Two cells share a single seed, i.e., undersegmentation.
(4) A region that should be part of the image background instead receives seeds and is assigned as cells.
(5) An area of epidermis cells that is very badly mis-segmented because the Gaussian filter is too large for these smaller cells. The user must decide if segmentation of this region should be completely reworked manually or skipped altogether. A smaller Gaussian filter (σ = 0.625 instead of 2.5) will effectively generate seeds for these smaller cells, but at the expense of severely oversegmenting the amnioserosa cells (into ~10 segments each, not shown).
(6) Sub-cellular regions are misassigned. One can often determine which cells these regions belong to based on other images in the time-lapse set.
Although the automated parts of this process yield generally reasonable results, there are obvious instances of incorrect segmentation. In fact, our time-lapse image sets contain frequent situations (such as sudden motion, movement of cells into or out of the imaging plane, or unusually bright regions internal to a cell) in which it is difficult or perhaps impossible for any automatic algorithm to properly segment the cells (Figure 1). We have thus chosen to develop a general-purpose framework and a suite of tools that enable a user to make these difficult decisions efficiently and directly. Although improved algorithms can tune segmentation for specific cases, our approach should be more flexible and more generally applicable.
Our novel approach to a hybrid of manual and automatic segmentation is not to adjust the final cell boundaries, but to directly adjust the seeds themselves. By doing so, we have a method that is robust, flexible, and easy to use. With a specified set of seeds, boundary generation by the watershed algorithm is fast and requires no user-selectable parameters, so incremental adjustment of the seeds and thus changes to the segmentation can be evaluated in real time. These features have also allowed us to create a simple save and load functionality that allows segmentation to be readjusted or completed at a later date. With this approach, if a user makes detailed changes to one section of an image, these changes will not have to be thrown out if coarse changes are later made in another section (as would be the case with some types of post-processing correction schemes based on manual image correction).
The seed manipulation tools we have developed are based on the ability to quickly add, delete, group and move seeds. There are one-click functions to “Add Seed”, “Delete Seed” and expand regions with “Extra Seeds”. This last feature allows the user to add secondary seeds for a cell, which the watershed algorithm then uses to expand that cell's boundaries. These extra seeds do not normally propagate in the tracking system, but normal tracking can be bypassed to copy all seeds (including extras) directly from the previous frame. This can be very useful with strangely shaped objects like thin curves or rings. There are also simple mouse/keyboard combinations to “Change Value” or “Swap Values” that change the identifier value associated with each cell. Finally, there is a “Lasso” tool that provides the ability to select individual seeds or groups of seeds (regardless of identifier value) which can then be moved or deleted en masse.
Technical Implementation
For the core watershed algorithm, we used the function cwatershed from the python package Mahotas (20), which is part of the larger PythonVision project for computer vision. This watershed function is an implementation of the standard ordered-queue, heap-based watershed algorithm (17). To facilitate GUI interactions, we developed a Python program using the packages WxPython and Matplotlib. We also made heavy use of numpy, scipy, and the Python Imaging Library. Ultra high performance functions were implemented in Cython. The complete program, known as SeedWater Segmenter (SWS), is available for download under a BSD license at Google Code (http://code.google.com/p/seedwater/).
Completely manual vector segmentation was performed using Inkscape (www.inkscape.org) to generate an SVG file and a custom Python program to extract the SVG/XML data and convert it to polygons for accurate geometric comparisons using the python package “shapely.”
RESULTS
Embryonic epithelial tissues in Drosophila are characterized by connected sheets of cells wrapped over a curved surface with yolk beneath. Two main cell types make up the tissues: epidermis cells and amnioserosa cells, each with very different average sizes (Figure 1A). These tissues undergo a number of morphological changes during embryonic development including extreme cell shape changes, cell rearrangements, bulk tissue motion and cell death. In studying these changes and the forces underlying them, researchers often use laser microsurgery to ablate one or more cells, creating artificial (and sometimes very large) perturbations to the remaining cells (16,21,22). Each of the above creates problems when segmenting and tracking cells in time-lapse images of living embryos. We have designed our segmentation and tracking software so that it provides the user with tools that can handle these difficulties. Below, we start with an image that has an initial set of automatically generated seeds and show that our manual correction tools are sufficient to correct common segmentation errors easily and effectively.
In the first round of manual intervention, we add and remove seeds to correctly identify the location of all the cells in the image, assisting the Gaussian technique manually. By right-clicking to delete seeds and left-clicking to add seeds, this process proceeds very quickly (Figure 2). Also, because of our centroid-based tracking system, adding and deleting entire cells becomes largely unnecessary after the first frame. Note that the user must typically make some decisions regarding which partially-visible cells are worth segmenting. The tools for adding and deleting cells allow the user to make such decisions on the first frame and yet revisit these decisions later as cells enter or leave the viewing area.
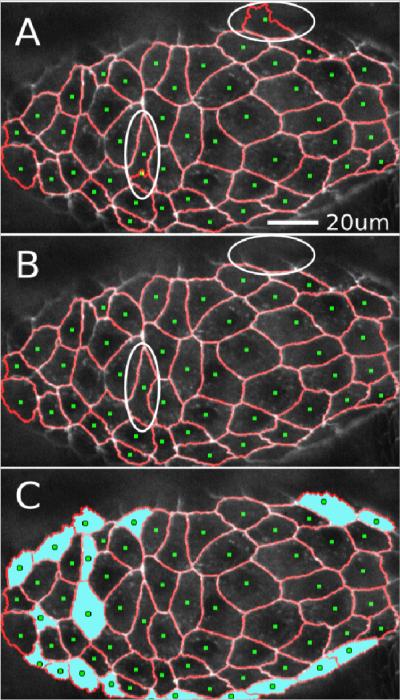
Figure 2.
Adding and deleting seeds manually.
(A) Initial automatic segmentation of an image (σ = 2.5 μm). Over-segmented regions with unwanted seeds are circled. The upper circled region highlights a cell at the edge of the imaging plane with a poorly defined boundary. The lower-left circled region has two seeds dividing a single cell.
(B) Segmentation after manual removal of unwanted seeds.
(C) Segmentation after manual addition of seeds to correct under-segmented regions (cyan fill). Seeds were added for sixteen cells around the margins of the tissue. These cells had been considered part of the background by the automatic algorithm. Seeds were also added for three internal cells that had not automatically received their own seeds.
Although add and delete are important tools in identifying which regions are cells, the user also needs the ability to decide which sub-regions belong to which cells (see Figure 1, arrow #6). Advantageously, the watershed algorithm allows for more than one seed of each value to be present in a single cell. This “extra seed” feature gives us the ability to use manual seed intervention not only to identify each cell, but also to define which sub-regions belong in which cell. By placing an extra seed in a subregion, it is very easy to identify that sub-region as part of a particular cell (Figure 3A–E).
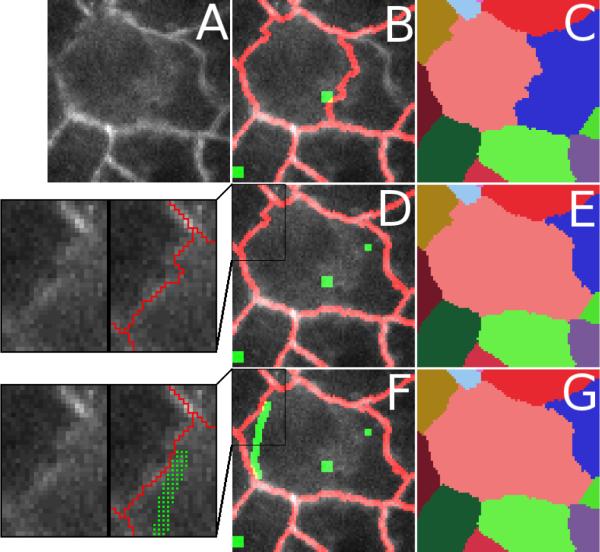
Figure 3.
Adding multiple extra seeds to correct mis-segmentation of cellular subregions. (A) Original image to be segmented. (B, C) Initial segmentation shown as an overlay with green seeds and red segment boundaries in (B) and as a false-colored cell ID map in (C). The subregion just to the right of the central seed is misassigned to an adjacent cell (blue). The upper left boundary of the central cell (pink) is also not satisfactory. (D, E) By adding a single extra seed, the originally mis-segmented subregion is reassigned to the appropriate cell (pink instead of blue). For the user, this is a two-click process: a left-click on the region that needs to be expanded followed by a right click to place the extra seed. The inset to the left of (D) shows a close-up of a remaining problematic boundary (with and without overlay). (F, G) This boundary is improved by adding a polyline of extra seeds (green line in (F) that appears as distinct seeds in the inset). For the user, creating a line of seeds works as above except multiple right clicks are used. A line segment of seeds is added between each successive right-click location. As shown in the inset to the left of (F), the result is a slight improvement in the overlap of the watershed boundary and the imaged cell-cell boundary. The best location for this boundary was determined by visually tracking its motion in previous and subsequent frames of the image stack (not shown).
This model works very well for most problem cases, but after segmenting a number of data sets, we realize that some boundaries simply “misbehave” unless extra seeds are placed essentially along the whole boundary. This can happen for boundaries that are particularly discontinuous, have a low signal-to-noise ratio or have unusual image topology (such as a gradient that makes one watershed region likely to invade another). To quickly handle such problem cases, we implemented a tool that inserts freehand lines of seeds. With this tool, the user can essentially draw the boundary in directly (Figure 3F–G).
This seed model is simple, intuitive and easy to teach to new users. It benefits from the consistency and speed of the watershed algorithm, yet still allows users to correct the segmentation as much as is needed.
We also tried several preprocessing filters including denoise, Gaussian blur, unsharp mask, and CLAHE (23) to see if we could reduce the subsequent need for manual intervention. Filtered and unfiltered image sets generally yield a similar number of errors in cell identification, but filtered images tend to yield artifacts during boundary generation. The one exception was CLAHE filtering, which enhances poorly lit regions for easier viewing and watershed segmentation without compromising the shapes of boundaries that already segmented well. We thus determined that it was preferable to segment unfiltered or CLAHE-filtered images – giving the truest possible representation of cell morphology.
Tracking
Once all cells have been identified and the segmentation has been adjusted to associate all subregions with the correct cell, the next step is to generate seeds for the next frame. Rather than finding these seeds using the same Gaussian minima approach as the first frame, we generate subsequent seeds from the centroids of each cell in the previous frame (Figure 4). This has the added advantage of automatic tracking because the seed value will be the same in the second frame as in the first.
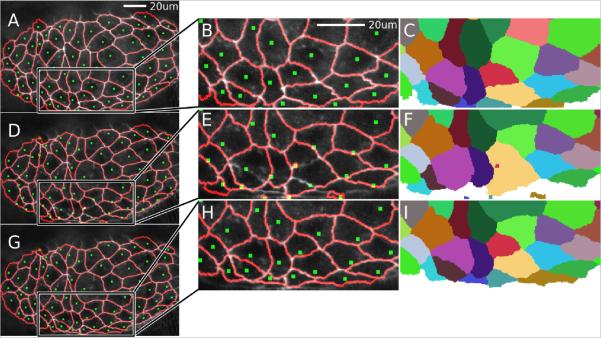
Figure 4.
Cell tracking when the frame-to-frame movements of cells are large.
(A–C) Complete manually assisted segmentation of a cluster of amnioserosa cells. The segmentation overlay shows seeds (green squares) and segment outlines (red lines). (B) is a close-up of the boxed region and (C) is the corresponding false-colored cell ID map.
(D–F) Automatic tracking and segmentation of the next frame after laser ablation of a central cell and with a large time interval between frames (70 seconds). The large time interval exaggerates cell motion between frames and causes the centroid-based algorithm to track cells improperly in some regions, especially near the bottom middle of the image (zoomed region in E, F). The errors are clearly discernable in (F) compared to (C). Note that even in this relatively extreme case, the automatic tracking performs very well for most cells in the image, particularly outside the boxed region. Tracking generally works well unless the cell moves over half its diameter.
(G–I) Corrected tracking and segmentation after using the “Lasso” and “Move Seeds” tools. The “Lasso” tool works by clicking to form a polygon that encircles multiple seeds. These seeds are then moved using the arrow keys to position them properly. This seed adjustment process is quite fast (a few seconds) by starting with bulk motion and then adjusting individual seeds as needed.
In rare cases, a cell may move so much from one frame to the next that the previous centroid no longer falls within the cell's next boundary. This is easy to correct by simply selecting the seed point and moving it to a more appropriate location. In fact, if there is bulk motion, seeds can be “lassoed” and moved in bulk (Figure 4). In addition, if two seeds switched cells, their identifier values can be swapped with “Swap Values”; and if a seed was assigned an incorrect ID (which can happen if it is deleted and re-inserted) then “Change Value” can directly change the assigned ID. In practice, these tracking problems occur infrequently and are relatively simple to fix.
Robustness to Noise
To analyze the resilience of our watershed algorithm to additive and multiplicative noise, we segmented an artificial image of two “cells” separated by a single, central bright line (100 pixels long, 1 pixel wide, and with a brightness S above the background). Automatic watershed segmentation is largely unaffected by noise up to S, begins to fail more often than it succeeds when the noise is 2S (signal-to-noise ratio or SNR = 0.5), and almost always fails for SNR = 0.25. At this latter level, a Gaussian filter can rescue about 40% of the segmentations. These results hold for both additive and multiplicative noise. Despite the automatic segmentation failures, users can distinguish the appropriate boundary down to SNR ~ 0.25. Thus, the SNR-range of 0.25–1 is the real “sweet spot” for a semi-automatic routine like SWS, especially if the noise is spatially varied within this range. We find similar results when the central dividing line has a Gaussian profile (with σ = 1 pixel).
Speed
We chose a watershed algorithm because it is robust and extremely fast, lending itself to highly responsive interactivity. Upon a change in the seeds, our segmentation program must process a GUI event, update the seed data, re-run the watershed algorithm, and redraw the frame. Nevertheless, all of this is completed in approximately half a second on a typical desktop PC. This short lag means that the user can interactively perform hundreds of operations in a matter of a few of minutes.
To measure the speed and impact of manual intervention on a real image set, we timed how long it took to segment a typical 190-frame xyt stack with approximately 64 cells per frame (62 cells appeared in all frames; 3 cells appeared in just a subset) (Figure 5A–B). First, to illustrate how quickly SWS converges on a solution, we performed minimal tracking and generally let the algorithm do the work automatically. During this “minimal tracking” phase, we inspected the intermediate segmentations, but only to ensure that each seed stayed within the proper region to maintain ID consistency through time; we ignored any problems with borders and sub-regions. This process took 50 minutes (about 16 s per frame). The initial segmentation was then improved by three more rounds of manual intervention, completing segmentation to user-desired accuracy in approximately 6 hours (under 2 minutes per frame). Figure 5A–B shows how the overall user-defined segmentation accuracy improved with time, albeit with diminishing returns. After the first pass segmentation, manual intervention changed the cellular assignment of only 1.4% of the pixels in the entire image set, but in so doing, it changed the borders of 12% of the cells. As shown in Figure 5C, the errors were distributed very non-uniformly over the segmented cell population, with just a few cells having very large errors. We found that the most time consuming part of manual intervention was simply inspecting the current segmentation to see if it was satisfactory. When errors were identified, the corrections were implemented very quickly.
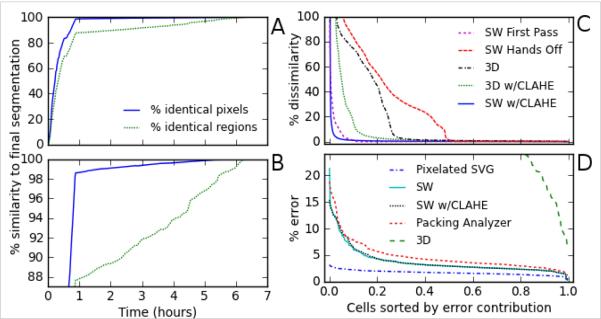
Figure 5.
Comparison of segmentation speed and accuracy for a typical data set: 190 frames with an average of 64 cells per frame.
(A, B) Improved accuracy versus time spent on manual intervention using SWS. Both graphs represent the same data; (B) simply has a tighter zoom in the y-axis to more clearly show the data after 50 minutes. Intermediate segmentations were saved after each change in watershed borders or at least every 60 s. The accuracy at each intermediate time point was assessed based on either the percentage of pixels whose assignment matched the final segmentation or the percentage of cells whose boundaries matched the final segmentation. The “first pass” segmentation was performed with minimal tracking, generally letting the algorithms do the work automatically (50 min), achieving a pixel-based accuracy of 98.6% and a cell-based accuracy of 88%. We then performed three more rounds of manual intervention and adjustment that improved the segmentation and tracking to user-desired accuracy in approximately six hours (~ 2 min per frame). The efficacy of manual intervention will vary with user experience and imaging quality, but this set is representative. The diminishing returns of continued manual intervention are most evident in the pixel-based comparison, but even this is somewhat linear over large time periods because we made successive passes through the entire stack.
(C) Distribution of errors over all segmented cells for selected intermediate segmentations and other techniques. Errors are defined as deviations from the final SWS segmentation. The x-axis is a normalized list of cell indices sorted from largest to smallest relative error. The y-axis is the number of erroneous pixels for each cell divided by the average area of all cells. We compared a “Hands Off” SWS segmentation with no user intervention and a “First Pass” SWS segmentation with minimal manual tracking assistance. For comparison, we include a 3D watershed segmentation, and SWS on an CLAHE-filtered image set.
(D) Distribution of errors over all segmented cells in comparison to a “gold standard” manual segmentation using a vector editor. These comparisons are limited to five evenly spaced frames (every 47th of the full data set). Absolute errors are thus compared for SWS segmentation, Packing Analyzer, and 3D watershed segmentation. As a baseline, we include errors induced by pixelating the vector segmentation.
Segmentation Quality
To place the performance of SWS in context, we compare its final segmentation to several automatic segmentation and tracking methods (Figure 5C). First, we compare to a completely automatic watershed segmentation and tracking (SWS in a “hands off” mode). This process took less than four minutes and correctly segmented 45% of the cells, but the rest were fairly bad. In fact, over 20% were half wrong or worse and about 6% were essentially all wrong, i.e., completely outside their proper boundary. This highlights the improvement achieved simply by supervising the tracking in our “first pass” segmentation above. Second, we compare to a 3D watershed algorithm. This process used only the initial seeds in the first frame and segmented the entire image set in only 90 s. It was much more effective than “hands off” SWS, but not nearly as accurate as semi-automatic tracking. With a 3D watershed, almost one fourth of cells had an error >15%. This poorly segmenting subset could be reduced to one tenth of the cells using CLAHE pre-filtering. This is just shy of the performance of semi-automatic tracking. As a final comparison, we pre-filtered with CLAHE and used the exact same seeds as in the final SWS set. Although we expected this pre-filter to slightly improve edge delineation, we instead found that the small changes introduced by CLAHE pre-filtering did not always work well with our manually corrected seeds. This caused a small number of regions to fill improperly, leading to large errors in a few cells (Figure 5C).
To get an absolute measure of SWS performance, we created a “gold standard” based on an expert's manual segmentation using a vector editor. This manual segmentation took over an hour per frame. We thus restrict the gold standard comparison to just five image frames (every 47th from the 190-frame set).
As a baseline, simply pixelating the vector segmentation leads to a fairly uniform error distribution with a per-cell average of 1.7%; any pixel-based segmentation of this image set will have at least this minimum error. In comparison, the final SWS segmentation had an average error of 3.7%. A handful of cells had 10–20% errors, but the overwhelming majority had errors just below the average (Figure 5D). Close inspection revealed that these errors were largely just ambiguity in the exact boundary location. We also tried pre-filtering with CLAHE and readjusting the seeds to correct some of the CLAHE-induced errors, but this eventually yielded results virtually identical to the unfiltered SWS version. We also compared the performance of another semi-automatic routine, Packing Analyzer (3), but did not attempt to use its tracking system. This process took about five min per frame and yielded results similar to the SWS segmentation (4.7% error). Finally, we performed 3D watershed segmentation on this five-frame subset. Although 3D watershed performed surprisingly well on the full data set, it performed very poorly in this subset: 7 of every 10 cells had errors > 30%. This breakdown is expected because the cell boundaries now move substantially between frames (much more than the typical boundary width). A summary of our results on the accuracy and speed of various segmentation methods is presented in Table 1.
TABLE 1.
| Comparison to SWS (190 frames) | Approx. Time Per Frame | Pixel-based % Dissimilarity | % Cells with Dissimilarity >5% | % Identical Cells |
|---|---|---|---|---|
| Final SWS (reference) | 110 s | - | - | - |
| First Pass SWS | 16 s | 1.4% | 5.6% | 88% |
| “Hands Off” SWS | 1.25 s | 28.1% | 48.5% | 45% |
| 3D Watershed | 0.5 s | 19.9% | 27.1% | 1.4% |
| 3D w/CLAHE | 0.5 s | 9.7% | 14.7% | 1.0% |
| SWS w/CLAHE uncorrected | 110 s | 1.3% | 2.1% | 6.8% |
| Comparison to manual vector segmentation (5 frames) | Approx. Time Per Frame | Pixel-based % Error | % Cells With Error >5% |
|---|---|---|---|
| Vector Segmentation | 2 hours | - | - |
| Pixelation of Vector Segment. | - | 1.7% | 0% |
| Final SWS | 110 s | 3.7% | 14.3% |
| SWS w/CLAHE corrected | 110 s | 3.8% | 16.3% |
| Packing Analyzer | 300 s | 4.7% | 26.6% |
| 3D Watershed | 0.5 s | 65.0% | 100% |
Measuring Cell Shape Oscillations
As a demonstration of SWS performance, we use it here to investigate cell shape oscillations in the amnioserosa (24–27). Prior work has shown that these cells undergo cyclical changes in apical area with a period of 230 ± 76 s (24). Adjacent cells have a tendency to oscillate out of phase, but they can also quickly switch between in-phase and out-of-phase oscillations. When the cells are visualized with a cell-boundary marker like GFP-armadillo (a β-catenin like protein) and viewed as oscillations in apical cell area, there is no evidence for long-range, wave-like propagation (24). On the other hand, when these same cells are visualized with a different fluorescent marker – GFP-moesin, which highlights concentrations of filamentous actin (21) – one can clearly discern wave-like propagation of f-actin accumulations that are correlated with sub-cellular contractions (16). To address this discrepancy, we performed SWS segmentation of a time-lapse image set of GFP-cadherin-labeled amnioserosa cells (66 cells over 63 image frames spanning 1300 s). We then calculated the autocorrelation functions of both cell area and triple-junction velocity (triple junctions were defined as points that touch three or more cells). Both functions showed clear oscillatory behavior; first minima at 144 s for cell area and 123 s for triple-junction velocity; subsequent maxima at 267 s in both cases. We then used the triple-junction velocities to calculate a time-and-space pair correlation function (Figure 6). The Δ=0 line of this function is the velocity autocorrelation and clearly shows the peaks described above. In addition, a density plot of the pair correlation function shows that the extrema move to longer time delays τ at larger spatial separations Δ, i.e., the correlations and anti-correlations propagate. Their propagation speed of 0.14 μm/s is close to that observed previously for apical accumulations of f-actin, 0.2 μm/s (16). The propagation is however limited and decays within 30 μm (about one and a half cells). Thus, one can observe contraction waves in the amnioserosa using just cell boundary labeling, if one uses a sub-cellular analysis based on triple-junction velocities and one has a complete and accurate segmentation that allows averaging over a large number of triple-junction pairs. The apparent discrepancy in previous observations results from whether one chooses to monitor the oscillations with cellular or sub-cellular resolution.
Figure 6.
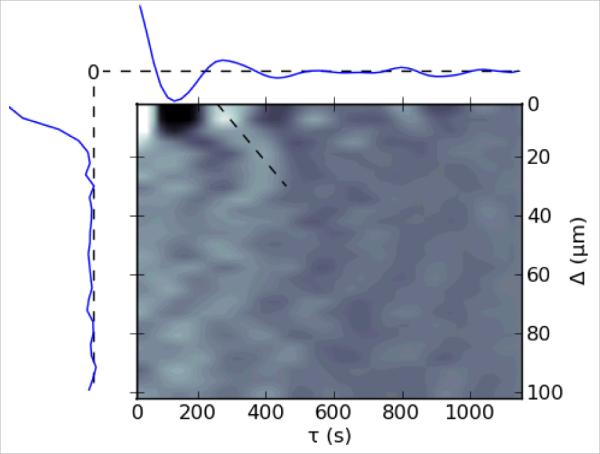
Time-and-space pair correlation function of triple-junction velocities for a data set with 66 segmented cells. The x-axis is the time separation between points (τ) and the y-axis is the distance between points (Δ). Correlations are normalized so that the peak at (0,0) has a value of 1. The Δ=0 axis (autocorrelation function) is plotted above the density plot, and the τ=0 axis (distance correlation) is plotted to the left. Dashed lines in these two plots represent zero correlation. The minima and maxima of the autocorrelation appear as dark and bright spots on the Δ=0 axis of the density plot with the first of each occurring at 123 and 267 s, respectively. In the full pair correlation function (density plot), the extrema move to longer time delays as the distance between the pair increases. This wave-like propagation is demarcated by the angled dashed line, which has a slope and thus velocity of 0.14 μm/s.
DISCUSSION
Image segmentation and the quantitative measurement of cell morphology are invaluable in the attempt to link physical and biological processes (28,29). Segmentation is a very wide field and a large number of approaches have been previously implemented. These range from direct drawing of segmentation boundaries to automatic registration of an entire image stack (13). Before designing our own segmentation tool, we examined several similar tools including ITK-SNAP (12), FIJI's TrakEM2 (9), CellProfiler (4), and Packing Analyzer (3); none of these was able to segment our typical (and thus imperfect) time-lapse image sequences with the speed and accuracy we desired. ITK-SNAP, FIJI's TrakEM2, and the more recent V3D (30) have tools for direct 3D segmentation of structures. These work very well for the smooth and continuous boundaries of 3D xyz structures in MRI images but have difficulty with some of our cells as 3D xyt structures. These 3D tools either lack the ability to reliably generate non-overlapping regions (because they are focused on single anatomical structures) or provide insufficient manual adjustments. The other tools perform 2D segmentation that can then be repeatedly applied to a sequence of images. Some focus on high segmentation accuracy for a small number of cells (e.g., fast marching tool and pen tool of FIJI's TrackEM2 (9) or the interactive tools of Intelligent Scissors (31–33)); others focus on speed and sacrifice some accuracy for high-throughput analysis of large numbers of cells (CellProfiler (4)). The tool designed to handle image segmentation tasks most similar to ours is Packing Analyzer. It is also a hybrid manual/automatic system (3), but is parameter-based and implements post-process correction and tracking. Packing Analyzer works well with many of our best data sets, but is not as efficient nor as flexible as needed for our more difficult data sets.
SWS fills a niche as a user tool for extremely accurate tracking of cells in living tissues when image quality suffers sporadic, but typical problems. By using a simple seeded watershed approach, it gives users a readily understandable way to manipulate segmentation results, save their progress, and even redo and undo changes. Furthermore, because it is a parameter-less system, users with no prior experience in image segmentation can get up to speed in minutes with very little instruction. SWS's approach is based on a simple and useful paradigm – manual segmentation correction using direct seed manipulation. It is easy to use and understand, but also powerful and efficient.
SWS is presently a stand-alone tool, but it also has potential as a post-processing engine. For instance, one could use an alternative segmentation to generate seed points, load these seeds to initialize SWS, and then use its manual correction tools to improve the quality of the final segmentation. In this way, SWS can also be a powerful tool for “rescuing” imperfect segmentations of existing image sets. The key concept could also be integrated into existing tools like ImageJ, ITK, or V3D, combining efficient manual correction with powerful visualization and analysis tools.
We have considered creating a fully 3D version of SWS using a 3D watershed algorithm, but interactivity would suffer. Compared to 2D, a 3D watershed segmentation is slow, purely based on a linear increase in computation time with the number of frames to be segmented. This could potentially be alleviated on a massively parallel architecture. There are also artifacts created in 3D watersheds of time-lapse images: some cells fall into two different xyt watersheds, one giving way to the other in time; other xyt watersheds split into parallel time branches, creating separated regions with a single ID.
Other possible future improvements include tools to aid in visualization and identification of segmentation errors. Decision-making is often the most time-consuming part of segmentation, so it could be beneficial to visually flag cells that have large shape changes, little overlap with the previous frame, or discontinuities in the paths of centroids or triple-junctions. One can imagine providing a library of user-selectable (and user-expandable) routines for flagging suspicious segmentations. We see this as the direction in which segmentation efficiency can be most rapidly improved.
Supplementary Material
Acknowledgments
Research support acknowledgement: This work supported by the Human Frontier Science Program (RGP0021/2007C); National Science Foundation (IOB-0545679); and National Institutes of Health (1R01GM099107).
REFERENCES
- 1.Pawley JB. Handbook of biological confocal microscopy. 3rd ed. Springer; New York: 2006. [Google Scholar]
- 2.Held M, Schmitz MH, Fischer B, Walter T, Neumann B, Olma MH, Peter M, Ellenberg J, Gerlich DW. CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nature Methods. 2010;7:747–754. doi: 10.1038/nmeth.1486. [DOI] [PubMed] [Google Scholar]
- 3.Aigouy B, Farhadifar R, Staple DB, Sagner A, Röper J-C, Jülicher F, Eaton S. Cell Flow Reorients the Axis of Planar Polarity in the Wing Epithelium of Drosophila. Cell. 2010;142:773–786. doi: 10.1016/j.cell.2010.07.042. [DOI] [PubMed] [Google Scholar]
- 4.Lamprecht MR, Sabatini DM, Carpenter AE. CellProfiler™: free, versatile software for automated biological image analysis. Biotechniques. 2007;42:71. doi: 10.2144/000112257. [DOI] [PubMed] [Google Scholar]
- 5.Li K, Miller ED, Chen M, Kanade T, Weiss LE, Campbell PG. Cell population tracking and lineage construction with spatiotemporal context. Medical Image Analysis. 2008;12:546–566. doi: 10.1016/j.media.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sbalzarini IF, Koumoutsakos P. Feature point tracking and trajectory analysis for video imaging in cell biology. Journal of Structural Biology. 2005;151:182–195. doi: 10.1016/j.jsb.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Pinidiyaarachchi A, Wählby C. Seeded watersheds for combined segmentation and tracking of cells. Image Analysis and Processing-ICIAP 2005. 2005:336–343. [Google Scholar]
- 8.McCullough DP, Gudla PR, Harris BS, Collins JA, Meaburn KJ, Nakaya M-A, Yamaguchi TP, Misteli T, Lockett SJ. Segmentation of Whole Cells and Cell Nuclei From 3-D Optical Microscope Images Using Dynamic Programming. IEEE Transactions on Medical Imaging. 2008;27:723–734. doi: 10.1109/TMI.2007.913135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardona A, Saalfeld S, Preibisch S, Schmid B, Cheng A, Pulokas J, Tomancak P, Hartenstein V. An integrated micro-and macroarchitectural analysis of the Drosophila brain by computer-assisted serial section electron microscopy. PLoS Biology. 2010;8:e1000502. doi: 10.1371/journal.pbio.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hahn HK, Peitgen HO. Proc. SPIE Medical Imaging. San Diego, CA: 2003. IWT-interactive watershed transform: a hierarchical method for efficient interactive and automated segmentation of multidimensional gray-scale images; pp. 643–653. [Google Scholar]
- 11.Lin G, Chawla MK, Olson K, Barnes CA, Guzowski JF, Bjornsson C, Shain W, Roysam B. A multi-model approach to simultaneous segmentation and classification of heterogeneous populations of cell nuclei in 3D confocal microscope images. Cytometry A. 2007;71A:724–736. doi: 10.1002/cyto.a.20430. [DOI] [PubMed] [Google Scholar]
- 12.Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, Gee JC, Gerig G. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 13.Khairy K, Keller PJ. Reconstructing embryonic development. Genesis. 2011;49:1–26. doi: 10.1002/dvg.20698. [DOI] [PubMed] [Google Scholar]
- 14.Miura K. Microscopy Techniques. Vol 95. Springer; Berlin / Heidelberg: 2005. Tracking movement in cell biology; pp. 1304–1307. [DOI] [PubMed] [Google Scholar]
- 15.Oda H, Tsukita S. Real-time imaging of cell-cell adherens junctions reveals that Drosophila mesoderm invagination begins with two phases of apical constriction of cells. Journal of Cell Science. 2001;114:493. doi: 10.1242/jcs.114.3.493. [DOI] [PubMed] [Google Scholar]
- 16.Ma X, Lynch HE, Scully PC, Hutson MS. Probing embryonic tissue mechanics with laser hole drilling. Physical Biology. 2009;6:1–12. doi: 10.1088/1478-3975/6/3/036004. [DOI] [PubMed] [Google Scholar]
- 17.Beucher S, Meyer F. The Morphological Approach to Segmentation: The Watershed Transformation. In: Dougherty E, editor. Mathematical Morphology in Image Processing. CRC Press; New York: 1993. pp. 433–481. [Google Scholar]
- 18.Vincent L, Soille P. Watersheds in digital spaces: an efficient algorithm based on immersion simulations. IEEE Transactions on Pattern Analysis and Machine Intelligence. 1991;13:583–598. [Google Scholar]
- 19.Lotufo R, Falcao A. The ordered queue and the optimality of the watershed approaches. Mathematical Morphology and its Applications to Image and Signal Processing. 2002:341–350. [Google Scholar]
- 20.Coelho LP. [Accessed May 26, 2011];Mahotas: Image Processing in Python. Available at: http://luispedro.org/software/mahotas.
- 21.Kiehart DP, Galbraith CG, Edwards KA, Rickoll WL, Montague RA. Multiple forces contribute to cell sheet morphogenesis for dorsal closure in Drosophila. Journal of Cell Biology. 2000;149:471–490. doi: 10.1083/jcb.149.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutson MS, Tokutake Y, Chang M-S, Bloor JW, Venakides S, Kiehart DP, Edwards GS. Forces for Morphogenesis Investigated with Laser Microsurgery and Quantitative Modeling. Science. 2003;300:145–149. doi: 10.1126/science.1079552. [DOI] [PubMed] [Google Scholar]
- 23.Zuiderveld K. Contrast limited adaptive histogram equalization. In: Heckbert PS, editor. Graphics gems IV. Boston, MA: 1994. pp. 474–485. [Google Scholar]
- 24.Solon J, Kaya-Çopur A, Colombelli J, Brunner D. Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell. 2009;137:1331–1342. doi: 10.1016/j.cell.2009.03.050. [DOI] [PubMed] [Google Scholar]
- 25.Blanchard GB, Murugesu S, Adams RJ, Martinez-Arias A, Gorfinkiel N. Cytoskeletal dynamics and supracellular organisation of cell shape fluctuations during dorsal closure. Development. 2010;137:2743–2752. doi: 10.1242/dev.045872. [DOI] [PubMed] [Google Scholar]
- 26.David DJV, Tishkina A, Harris TJC. The PAR complex regulates pulsed actomyosin contractions during amnioserosa apical constriction in Drosophila. Development. 2010;137:1645–1655. doi: 10.1242/dev.044107. [DOI] [PubMed] [Google Scholar]
- 27.Azevedo D, Antunes M, Prag S, Ma X, Hacker U, Brodland GW, Hutson MS, Solon J, Jacinto A. DRhoGEF2 Regulates Cellular Tension and Cell Pulsations in the Amnioserosa during Drosophila Dorsal Closure. In: Gottardi C, editor. PLoS ONE. Vol. 6. 2011. p. e23964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernández-González R, Muñoz-Barrutia A, Barcellos-Hoff MH, Ortiz-de-Solorzano C. Quantitative in vivo microscopy: the return from the “omics.”. Current Opinion in Biotechnology. 2006;17:501–510. doi: 10.1016/j.copbio.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 29.Hutson MS, Ma X. Mechanical aspects of developmental biology: perspectives on growth and form in the (post)-genomic age. Phys Biol. 2008;5:015001. doi: 10.1088/1478-3975/5/1/015001. [DOI] [PubMed] [Google Scholar]
- 30.Peng H, Ruan Z, Long F, Simpson JH, Myers EW. V3D enables real-time 3D visualization and quantitative analysis of large-scale biological image data sets. Nature Biotechnology. 2010;28:348–353. doi: 10.1038/nbt.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortensen EN, Barrett WA. Interactive segmentation with intelligent scissors. Graphical Models and Image Processing. 1998;60:349–384. [Google Scholar]
- 32.Ben-Zadok N, Riklin-Raviv T, Kiryati N. IEEE International Symposium on Biomedical Imaging: From Nano to Macro. IEEE; Boston, MA: 2009. Interactive level set segmentation for image-guided therapy; pp. 1079–1082. [Google Scholar]
- 33.Kang Y, Engelke K, Kalender WA. Interactive 3D editing tools for image segmentation. Medical Image Analysis. 2004;8:35–46. doi: 10.1016/j.media.2003.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.