Abstract
West Nile virus (WNV) is a mosquito-borne member of flaviviruses that causes significant morbidity and mortality especially among children. There is currently no approved vaccine or antiviral therapeutic for human use. In a previous study, we described compounds containing the 8-hydroxyquinoline (8-HQ) scaffold as inhibitors of West Nile virus serine protease (NS2B/NS3pro) in a high throughput screen (HTS) using the purified WNV NS2B/NS3pro as the target. In this study, we analyzed potencies of some commercially available as well as chemically synthesized derivatives of 8-HQ by biochemical assays. An insight into the contribution of various substitutions of 8-HQ moiety for inhibition of the protease activity was revealed. Most importantly, the substitution of the N1 of the 8-HQ ring by –CH– in compound 26 significantly reduced the inhibition of the viral protease by this naphthalen-1-ol derivative. The kinetic constant (Ki) for the most potent 8-HQ inhibitor (compound 14) with an IC50 value of 2.01 ± 0.08 μM using the tetra-peptide substrate was determined to be 5.8 μM. This compound inhibits the WNV NS2B/NS3pro by a competitive mode of inhibition which is supported by molecular modeling.
1. Introduction
West Nile virus, a member of mosquito-borne Flaviviridae family, belongs to the Japanese encephalitis serocomplex which includes Japanese encephalitis virus, West Nile virus (WNV), Kunjin virus, and Murray Valley encephalitis virus. WNV causes significant morbidity and mortality (for reviews, see Brinton (2002); Gould and Solomon (2008), Lindenbach and Rice (2003) and Weaver and Barrett (2004). WNV was previously unknown in the U.S. In 1999, the first epidemic of WNV occurred in New York City. Since then reported cases of WNV spread to most of the U.S. by infection cycle between certain species of mosquitoes and bird species serving as reservoirs. Because human→mosquito→human transmission cycle does not occur, humans are considered as dead-end hosts. Most WNV infections are either asymptomatic or associated with mild flu-like symptoms, which could, in a small number of cases, advance to encephalitis (Gould and Solomon, 2008). There are currently no vaccines or antiviral therapeutics available for treatment of WNV-infected patients.
WNV encodes a positive-strand RNA of about 11 kb in length that is translated to a single polyprotein precursor which produces 10 mature proteins by co- and post-translational processing. The three proteins, the capsid (C), precursor/mature membrane protein (prM/M), and the envelope (E) form the virion; the seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) are expressed in the infected cells and are required for viral replication (for reviews, see (Beasley, 2005; Lindenbach and Rice, 2003)).
The two component viral serine protease, NS2B-NS3, plays a crucial role in viral replication as it is required for processing of the polyprotein precursor prior to the assembly of the viral replicase complex. This requirement makes the viral protease an excellent target for development of antiviral therapeutics. The viral protease cleavage sites have in common a pair of basic amino acids, R and K, (or Q at the P2 position of DENV2 NS2B-NS3 cleavage site) followed by G, S, or A at the P1′ position (Lindenbach and Rice, 2003) (according to the nomenclature of (Schechter and Berger, 1967)). The serine protease catalytic triad is located within the N-terminal 185 amino acids (Bazan and Fletterick, 1989; Chambers et al., 1990; Preugschat et al., 1990). NS2B is the required cofactor for NS3 protease activity (Chambers et al., 1991; Falgout et al., 1991; Wengler et al., 1991; Zhang et al., 1992). The NS2B is an integral membrane protein in the endoplasmic reticulum (Clum et al., 1997) containing hydrophobic regions flanking a hydrophilic region of ~45 amino acid residues which forms a complex with the NS3 protease domain (Arias et al., 1993; Chambers et al., 1993; Clum et al., 1997; Falgout et al., 1993). Using an E. coli-expressed and purified DENV2 NS2B/NS3pro, an in vitro protease assay using a fluorogenic tri-peptide substrate was established. This assay demonstrated that NS2B stimulates the protease activity of the NS3pro domain ~3300-fold with the Boc-G-R-R-7-amino-4-methyl coumarin (AMC) substrate (Yusof et al., 2000). Subsequent studies optimized the expression levels, solubility and catalytic properties of the DENV2 and WNV NS2B-NS3 proteases (Leung et al., 2001; Li et al., 2005; Nall et al., 2004; Shiryaev et al., 2007). The crystal structures of the WNV NS2B-NS3 protease complex containing a covalently bound tetra-peptide substrate-based inhibitor (Erbel et al., 2006) or a non-covalent complex with the trypsin inhibitor, aprotinin (Aleshin et al., 2007) have been solved. Similarly, the crystal structures of DENV2 (Erbel et al., 2006), DENV1 (Chandramouli et al., 2010) and WNV NS2B/NS3pro (Aleshin et al., 2007) in the absence of a substrate-based inhibitor have also been solved. These studies provide information regarding the substrate specificity as well as the role of cofactor NS2B peptide in activation of the NS3pro domain.
In a previous study, we reported the adaptation of the in vitro WNV protease assay with the tri-peptide substrate, Boc-G-K-R-AMC to a high throughput screening (HTS) format. We screened ~32,000 compounds and identified 98 compounds with several distinct chemical scaffolds. One group of these compounds contained an 8-HQ moiety as a common scaffold. In this study, we analyzed some commercially available as well as chemically synthesized derivatives of 8-HQ including one compound in which 8-HQ was replaced with a naphthalen-1-ol ring.
2. Materials and Methods
2.1 Materials
The WNV NS2B-NS3pro expression plasmid encoding the protease precursor used in this study contains the hydrophilic domain of NS2B cofactor (45 amino acids) and the NS3pro domain (186 residues + 2 residues LN from HindIII site at the C-terminus of NS3pro) separated by five C-terminal amino acids (QYTKR↓) of NS2B (P1 to P5). The expression and purification of this protease has been described previously (Mueller et al., 2007, 2008). The WNV NS2B-NS3 protease precursor underwent autoproteolysis at KR↓ to yield a non-covalently associated NS2B/NS3pro complex which was purified by TalonR affinity matrix (Clontech) (Mueller et al., 2007). The fluorogenic peptide substrates, t-butyl-oxycarbonyl (Boc)-Gly-Lys-Arg-7-amino-4-methyl coumarin (AMC) and Benzoyl (Bz)-norleucine (Nle)-Lys-Arg-Arg-AMC were purchased from Bachem (Torrance, CA). For later experiments, Bz-Nle-Lys-Arg-Arg-AMC was also custom-synthesized by NeoBioScience (Cambridge, MA). AMC was purchased from Anaspec, Inc (Fremont, CA). The 8-HQ compounds with codes (molecular weights and ID numbers within parentheses) are: 10 (M.W. 357.41; T0511-7984), 13 (M.W. 383.47; LT00013768), compound B (M.W. 349.41; T0510-2459; ref. (Mueller et al., 2008)) were purchased from Ryan Scientific Inc., Mt. Pleasant, S.C. The following compounds with IDs, II-11-, series were synthesized by Dr. Kyungae Lee: 14 (M.W. 439.53; II-11-1), 15 (M.W. 347.43; II-11-2), 16 (447.53; II-11-3), 17 (M.W. 489.59; II-11-5), 18 (M.W. 438.54; II-11-6), 19 (M.W. 453.56; II-11-7), 20 (M.W. 349.41; II-11-10), 21 (M.W. 363.43; II-11-13), 22 (M.W. 398.48; II-11-11), 23 (M.W. 333.41; 7211190047), 24 (M.W. 323.37; 7211800112), 25 (M.W. 334.39; II-11-4), 26 (M.W. 396.48; II-26-3). Compounds 20 (T0508-1930) and 23 (F0870-0049) were re-purchased from Ryan Scientific Inc. Compound 14 was re-synthesized by Dr. Huiguo Lai and it was found to be at least 98% pure as analyzed by NMR.
2.2. In vitro protease assays
All assays were done in triplicate in black 96-well plates. Protease activity experiments were performed in vitro using purified viral protease NS2B-NS3-pro from WNV. The reaction mixture of 100 μL/well contained 200 mM Tris pH 9.5, 30% glycerol, 27 nM WNV, 2% DMSO and fluorogenic WNV substrate Boc-GKR-AMC at 100 μM for percent inhibition experiments as indicated. To validate the assay conditions, aprotinin (bovine pancreatic trypsin inhibitor, BPTI) was used as a positive control (Ki for WNV NS2B/NS3pro ≈ 162 nM at a final concentration of 2–10 μM. Assay mixtures containing 2% dimethyl sulfoxide (DMSO) were used for the no-inhibitor control. Furthermore, a control containing only 2% DMSO and the substrate was used to achieve the minimum value of relative fluorescence units (RFU) for background subtraction. In all assays, the inhibitor-enzyme complex was allowed to form by pre-incubation for 15 min at room temperature (RT) before addition of the substrate. After 5 min incubation, reactions were monitored by the release of free AMC, every 1.5 min for 30 min. The fluorescence emission was recorded at 465 nm following excitation at 385 nm using the SpectraMax Gemini EM (Molecular Devices). The relative fluorescence units (RFU) were converted to absolute AMC product concentrations using Excel, where the data were transformed using the slope from the linear regression of the AMC calibration curve generated from the fluorescence values versus micromolar (μM) concentrations of free AMC. RFUs, obtained in a standard assay in the absence of inhibitors, were taken as 100% protease activity (or 0% inhibition), the background value (no protease) as 0% activity (or 100% inhibition).
2.3. Determination of IC50
To determine the 50% inhibitory concentration (IC50) of a compound, protease assays were performed as described above except in the presence of various concentrations of an inhibitor (0.3 – 40 μM), serially diluted in 100% DMSO. The final DMSO concentration was 2% in each well. RFU values were converted into percent inhibition. The data were fitted to a sigmoid dose-response function using non-linear regression analysis of GraphPad Prism 5 and IC50 values were obtained. The IC50 values were also determined using the tetra-peptide substrate in the range of 5 nM to 25 μM as described in the text.
2.4. Kinetic analysis
To determine the Km and Vmax values of compound 14, four different concentrations of the inhibitor (0 to 5 μM) were assayed at twelve tetra-peptide substrate concentrations ranging from 0–50 μM. Ki value was calculated from these data using SigmaPlot.
2.5. Molecular docking of compounds 14, 17, 18, 22 and 26 into WNV protease
Compounds 14, 17, 18, 22 and 26 were sketched in ZINC (UCSF) and converted to SMILES strings (http://zinc.docking.org/choose.shtml). The NCI/CADD Group SMILES Translator and Structure File Generator Molecular (http://cactus.nci.nih.gov/translate/) was used to generate coordinates in the .mol2 format. AutoDockTools was used to add hydrogen atoms and electric charges. Molecular docking was performed using AutoDock Vina (http://mgltools.scripps.edu) implementing grid-based scoring. Each compound was docked in 1,000 orientations and the best scoring orientations were energy-minimized.
3. Results and Discussion
3.1. Inhibition of WNV NS2B/NS3pro by 8-HQ derivatives
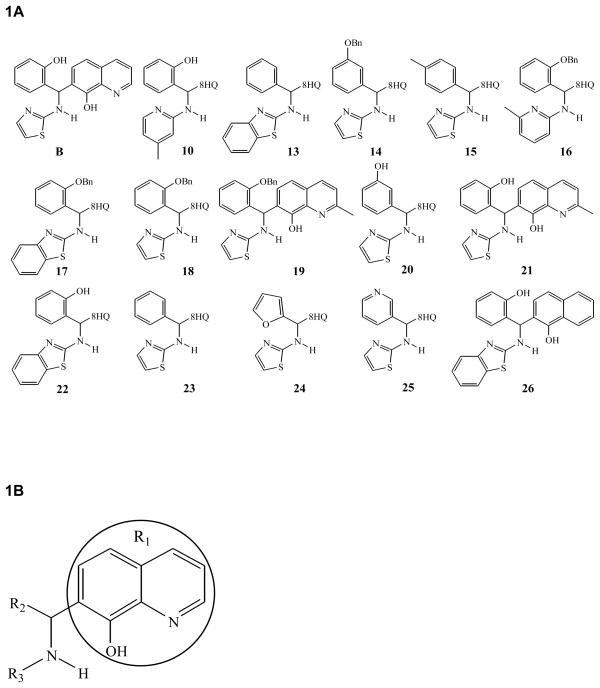
In our previous study, we reported identification of small molecule inhibitors of WNV NS2B/NS3pro using a fluorogenic tri-peptide substrate, Boc-G-K-R-AMC, in a high throughput screening initiative. The 98 compounds selected as primary “hits” with drug-like physicochemical and pharmacological properties were divided into five groups based on structural similarity. Two groups of compounds are substituted at position 7 of the 8-HQ ring (R1) and are linked to either an –NH–C=O or an –NH– group (Mueller et al., 2008). The lead compound B, a 8-HQ derivative belonging to the –NH– group, was reported to have a Ki value of 3.4 ± 0.6 μM (Mueller et al., 2008). In this study, fifteen 8-HQ compounds including the compound B having the 8-HQ scaffold (Fig. 1) and one additional compound (compound 26) containing a naphthalen-1-ol instead of the 8-HQ scaffold were characterized for their percent inhibition of WNV NS2B/NS3pro in order to develop a structure-activity relationship and mode of inhibition.
Figure 1. Structures of 8-HQ derivatives.
A. The structures of sixteen compounds that were analyzed in this study are shown. B. The general structure of the 8-HQ scaffold (R1) is shown containing various aryl substituents at R2 and R3.
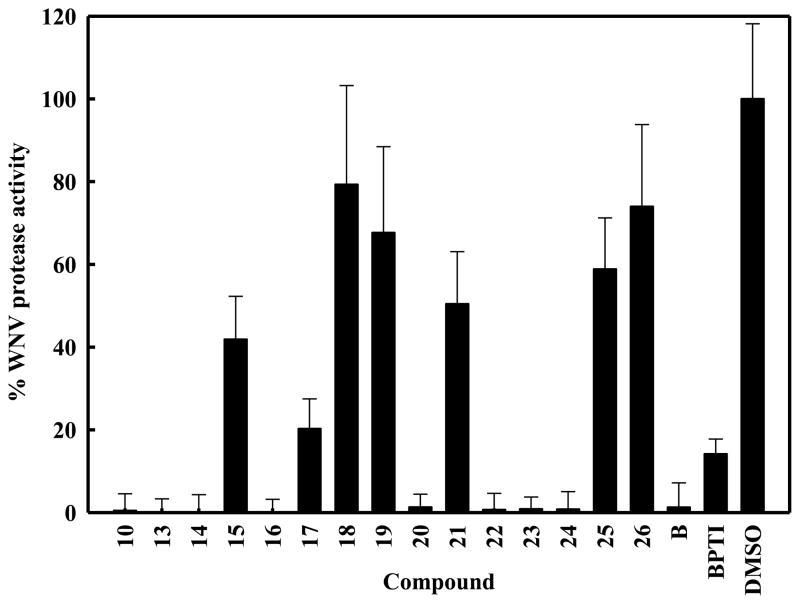
The in vitro protease assays were performed using the WNV protease NS2B/NS3-pro and the tri-peptide substrate in the absence or the presence of an inhibitor at a fixed concentration of 40 μM. Eight compounds and the compound B, characterized previously as a lead inhibitor of WNV NS2B/NS3pro (Mueller et al., 2008) showed ≥ 70% inhibition (Fig. 2). It is noteworthy that compound 26 containing the naphthalen-1-ol scaffold with the substitution of N1 of 8-HQ with –CH–, reduced the inhibitory activity of compound 22 from ~99% to 26% (discussed further below). These results indicated that the 8-HQ moiety is a major determinant for inhibition of WNV NS2B/NS3pro by this class of compounds. In compounds 15 and 18, addition of a methyl group, even in the para position of the –CH–, or a benzyloxy (–OBn) group in the ortho position in R2 (Fig. 1) inhibited the protease activity by ~60% and 20%, respectively. Comparison of the activities of compounds 18, 20, 21 and 23 indicate that either no substitution or the position of the benzyl ring substitution in R2 is an important determinant for inhibition of WNV protease. Substitution in the ortho position to the –CH– in the R2 tends to interfere with the inhibitory activity of the compounds, perhaps due to steric hindrance in which –OH seems to be more favorable than -OBn, and moving this –OH substitution to the meta position (compounds 20 vs 21) relieves this interference. This steric interference in the inhibitory activity is again revealed when the activities of compounds 17 and 22 pair or 18 and 14 pair is compared. Comparison of activities of compounds 17 and 18 (Fig. 2) reveal that benzothiazole ring contributes better inhibition of the protease than thiazole ring at the R3 position. Moreover, compounds 17 and 22 have the same R3 group, the benzothiazole ring. However, in the R2 benzyl ring, the –OH is preferred over –OBn in the ortho position in the compounds 17 vs 22 or without any substitution as in compound 13.
Figure 2. Inhibition of WNV NS2B-NS3pro by 8-HQ derivatives.
Aprotinin was used as a positive control (100%) and DMSO alone (2%) as a negative control (no inhibitor). Compounds were used at a final concentration of 40 μM. (A) Eight compounds decreased the protease activity by more than 70%. Error bars indicate standard deviation of the means.
In compounds 18 and 14, R3 group is the same, a thiazole ring, whereas in the R2 group, the meta position is preferred for the benzyloxy substitution than the ortho position or even –OH substitution at the ortho position (Compound B). The benzyloxy group is flexible and can be freely rotated along the oxygen-carbon single bond. The butterfly-shaped molecule can interact with the protease residues by π-π interactions. In addition, the benzyloxy group provides an extra aromatic ring to interact with the protease residues.
Substitution of the R2 benzyl ring with a pyridyl ring contributed to a significant reduction of inhibitory activity (Compounds 23 vs 25), whereas with a furan ring did not affect the activity (Compounds 23 and 24). Comparison of activities of compounds 18 and 19 reveals that both have –OBn substitution in the ortho position of the R2 benzyl ring except that the compound 19 has a –CH3 group substitution in the 8-HQ (R1) ring. This 8-HQ (R1) substitution has a marginal contribution towards inhibition of the WNV protease activity.
3.2. Determination of IC50 values
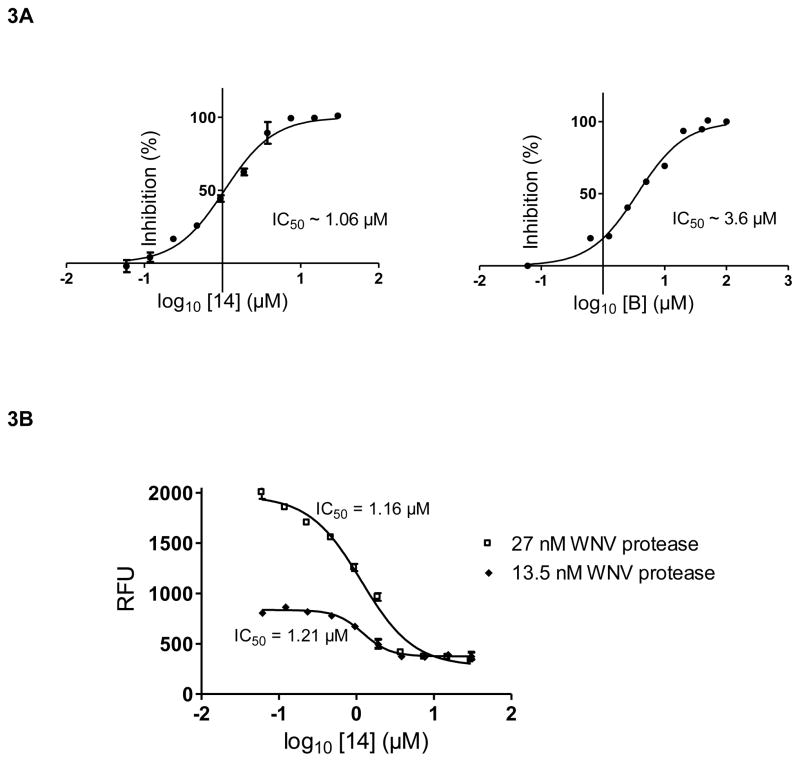
Next, the IC50 values were determined for six compounds using the tri-peptide substrate and concentrations of the inhibitor between zero and 40 μM (Table 1). The IC50 determinations for the compounds 14 and B are shown in Fig. 3. The compound B (Ki = 3.4 ± 0.6 μM) described in a previous study (Mueller et al., 2008) was used for validation of the IC50 values of unknown compounds analyzed in this study. The RFU values were converted into % inhibition and plotted versus the Log10 of compound concentrations resulting in sigmoid concentration-response curves. Under the conditions of our assay when the enzyme concentration is less than the inhibitor concentration and the Ki value, the apparent IC50 value should not change with different enzyme concentrations, especially when the mode of inhibition is competitive. Therefore, we determined the apparent IC50 values at two different concentrations, 27 nM and 13.5 nM, of WNV NS2B/NS3pro to validate this expectation. For tight binding inhibitors, the apparent value will simply be close to half of the concentration of the enzyme which is not the case with compound 14. The IC50 values determined at two different enzyme concentrations, 27 and 13.5 nM, were 1.16 and 1.21μM, respectively. The experiment was repeated 4 times and the values ranged from 0.78 to 1.7 μM. The results of one representative experiment are shown in Fig. 3B. Moreover, at high inhibitor concentrations, both curves reached a minimum RFU level, the same value (~300 RFU) as for the background control with no protease. The IC50 values were also determined using the tetra-peptide substrate, Bz-Nle-Lys-Arg-Arg-AMC, for compounds 13 and 23 (Table 1). From the IC50 values of these 8-HQ inhibitors, compound 14 was identified to be the most potent inhibitor. Therefore, mode of inhibition study and molecular modeling were performed with compound 14 (see below).
Table 1.
Kinetics parameters for the tetra-peptide substrate and compound 14 against WNV NS2BH-NS3pro at 37 °C
| Inhibitors | IC50 (μM)a | Inhibitors | IC50 (μM)a | IC50 (μM)b |
|---|---|---|---|---|
| B | 3.6 ± 2.0 | 13 | 2.60 ± 0.09 | 6.18 ± 0.27 |
| 10 | 13.4 ± 3.9 | 14 | 1.06 ± 0.07 | 2.01 ± 0.08 |
| 16 | 3.4 ± 0.8 | 23 | 9.85 ± 0.34 | 12.86 ± 0.54 |
Tri-peptide substrate Boc-GKR-AMC;
Tetra-peptide substrate Bz-Nle-KRR-AMC.
Figure 3. A. Determination of IC50 for compounds 14 and B.
The IC50 values were determined using the tri-peptide substrate as described under Materials and Methods. The x-values are displayed as the log of compound concentration (0–40 μM). The error bars represent standard deviation. The experiments in triplicate were repeated 4 times, and the IC50 values were determined to be 1.06 ± 0.07 μM for compound 14 and 3.6 +/− 2.0 for compound B. B. The IC50 of compound 14 was determined at two different WNV NS2B/NS3pro concentrations and tri-peptide as the substrate. The experiments in triplicate were repeated 4 times. The values ranged from 0.78 to 1.7 μM. The results from one representative experiment are shown.
3.3. Kinetics of inhibition of WNV NS2B/NS3pro by compound 14
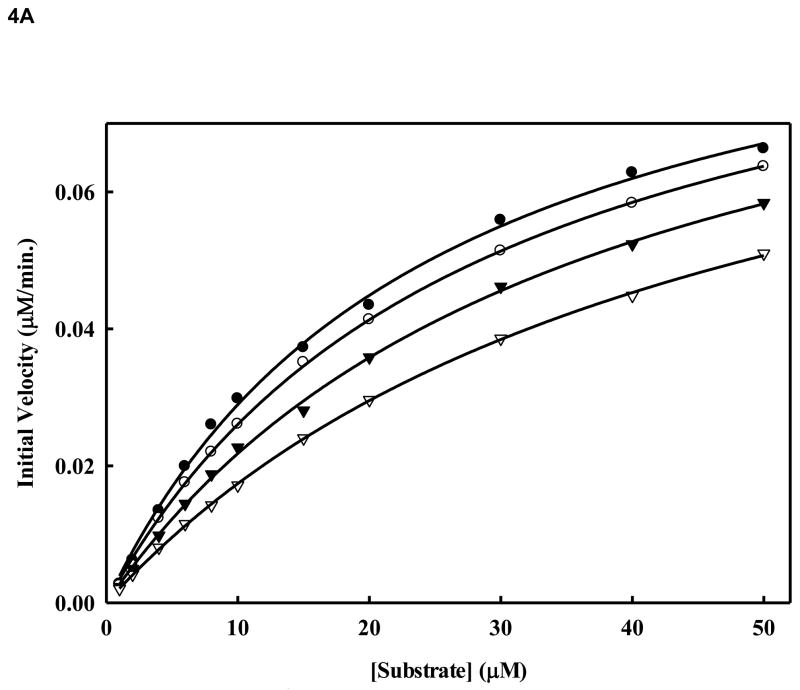
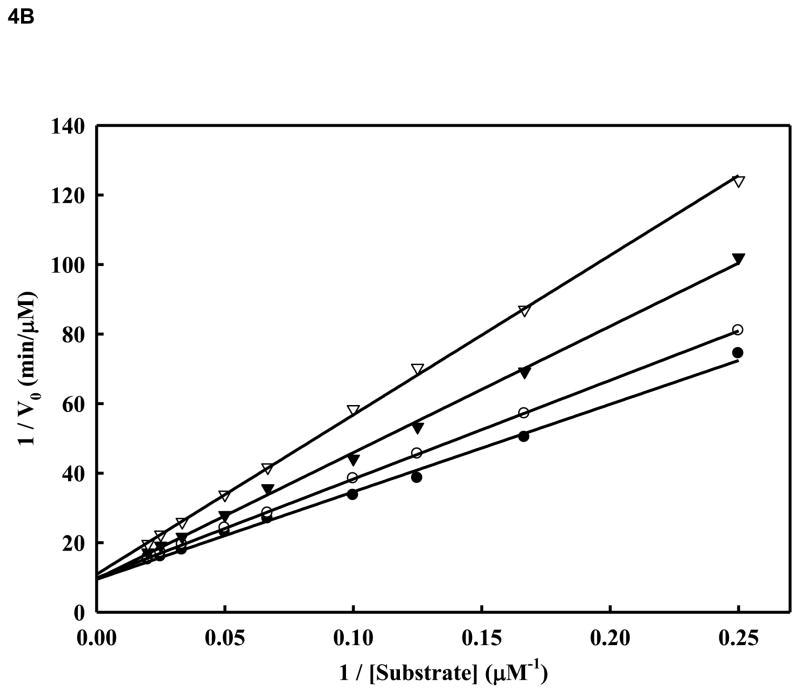
We sought to analyze the mode of inhibition of the WNV NS2B/NS3pro-catalyzed cleavage of the tetra-peptide substrate by compound 14. We performed kinetic analysis to determine the Km, kcat and Vmax values in the presence and absence of the inhibitor (compound 14) at four different concentrations (Fig. 4A and Fig. 4B and Table 2). The highest inhibitor concentration used was 5 μM to avoid potential nonspecific inhibition due to aggregation which could interfere with the kinetics of true inhibition (Jadhav et al., 2010). In Fig. 4C, for each of the four inhibitor concentrations (0, 1, 2, and 5 μM), we followed the kinetics at 12 substrate concentrations (0,1, 2, 4, 6, 8, 10, 15, 20, 30, 40 and 50 μM). As shown in Table 2 and Fig. 4A and 4B, the apparent Michaelis-Menten constants (Kmapp) increased and the kcat/Km decreased proportionally to the increase in inhibitor concentrations in the range of 0, 1.0, 2.0 and 5.0 μM. The corresponding kcat values were within ~5% variance (Table 2). The results indicate that compound 14 at increasing concentrations reduced the affinity of the enzyme for the substrate as is evident from the increase of Km values. This type of inhibition is a characteristic feature of competitive mode of inhibition.
Figure 4. A. Inhibition of WNV NS2BH-NS3pro protease activity by compound 14.
Initial reaction rates of cleavage of the tetra-peptide-AMC substrate catalyzed by WNV NS2BH-NS3 protease (28 nM) in 200 mM Tris HCl (pH 9.5), 6.0 mM NaCl, 30 % glycerol and 0.1 % CHAPS at 37 C were determined by varying the tetra-peptide substrate concentrations in the range of 1, 2, 4, 6, 8, 10, 15, 20, 30, 40 and 50 μM at each concentration of inhibitor fixed at 0 (solid circle), 1.0 μM (open circle), 2.0 μM (solid triangle) and 5.0 μM (open triangle). The reactions were initiated by the addition of WNV NS2BH/NS3pro protease and the fluorescence intensity at 460 nm was monitored with an excitation at 380 nm. Reactions were less than 5% completion in all cases to maintain valid steady-state measurements. The solid lines are fitted lines using the Michaelis-Menten equation. B. Lineweaver-Burk plot of compound 14. The same experiments as described in Fig. 4A were used to plot this graph. The concentrations of the inhibitors are 0 (solid circle), 1.0 μM (open circle), 2.0 μM (solid triangle) and 5.0 μM (open triangle), respectively. The concentrations of tetra-peptide substrate are (4, 6, 8, 10, 15, 20, 30, 40 and 50 μM range).
Table 2.
Kinetics parameters for the tetra-peptide substrate and compound 14 against WNV NS2BH-NS3pro at 37 °C
| [Inhibitors] | Km | kcat | kcat/Km |
|---|---|---|---|
| μM | μM | s−1 | M−1·s−1 |
| 0 | 24.56 ± 1.43 | 0.061 ± 0.001 | 2486 ± 185 |
| 1 | 28.36 ± 0.85 | 0.059 ± 0.002 | 2080 ± 137 |
| 2 | 35.96 ± 1.70 | 0.059 ± 0.002 | 1641 ± 139 |
| 5 | 45.93 ± 1.08 | 0.058 ± 0.002 | 1263 ± 75 |
3.4. Molecular Docking
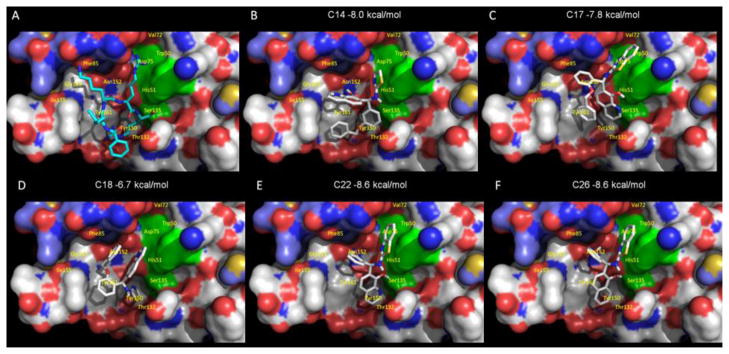
Crystal structure of the tetra-peptide substrate-based inhibitor bound to the WNV NS2B/NS3pro was reported previously (Aleshin et al., 2007; Erbel et al., 2006). To understand how the N1 of 8-HQ in compounds 14 and 22 may be important for inhibitory activity of WNV protease we predicted the free energy of binding of compound 14 and 22 to the active site of WNV NS2B/NS3pro by molecular docking. We compared the orientation posed by molecular docking and predicted free energy of binding values using compounds that exhibit similar (e.g., compound 22) or distinct inhibitor activities (e.g., compounds 18 and 26 in Fig 2). AutoDock Vina was used to dock compounds 14, 17, 18, 22 and 26 in the active site bound by Bz-Nle-Lys-Arg-Arg-H in the crystal structure (PDB code 2FP7 (Erbel et al., 2006) (Fig. 5A). All compounds were predicted to interact with the active site by hydrophobic and polar contacts to the residues shown in Fig. 5. Although each compound was predicted to bind to the active site with free energy of binding values of −6.7 kcal per mol or lower, compounds that exhibited greater than 75 % inhibition of protease activity, including compounds 14 and 22, were predicted to bind better (−8.0 kcal and −8.6 kcal per mol for compound 14 and 22 shown in Fig. 5B and 5E, respectively). The benzothiazole moiety of more active compounds 17 and 22 is predicted to form contacts with Asp75 at the active site. This interaction is not present in the less active compound 18 which has a thiazole moiety at this position (Fig. 5C and 5E, vs 5D).
Figure 5. Molecular docking of compounds 14 and 26 onto WNV protease structure.
The WNV protease crystal structure coordinates (PDB ID code 2FP7) were used for molecular docking. AutoDock Vina was used to dock each compound in the active site bound by Bz-Nle-Lys-Arg-Arg-H substrate-based inhibitor (Erbel et al., 2006). The solvent-accessible surface of the protein is shown in color-coded atoms (white, carbon; red, oxygen; blue, nitrogen). The catalytic triad (H51, D75, and S135) is shown as a green surface. A. The structure of the Bz-Nle-Lys-Arg-Arg-H substrate-based inhibitor bound WNV NS2B-NS3pro (Erbel et al., 2006). B. Compound 14 is shown in the orientation posed by molecular docking. Compounds 17, 18, 22 and 26 are shown in panels C, D, E and F, respectively.
Comparison of the orientation posed by molecular docking permits analysis of potential interactions in the context of functional protease activity. Compound 22, a potent protease inhibitor, and compound 26, a significantly less active inhibitor, are structurally nearly identical and the predicted docking scores suggest that the overall interactions between the compounds and the protease are likely to be similar. The difference in inhibition between the compounds 22 and 26 may result from potential interactions with Ser135. Although they are predicted to bind similarly to the active site, the N1 of 8-HQ in compound 22, Fig.5E, is positioned in close proximity to the active site Ser135 (3.2 Å) with a potential to form a H bond with OH of Ser135, in contrast to the less active compound 26 with a carbon at this position (4.5 Å; see Fig 5F) which is close to the acceptable limit for a distance of van der Waals contacts. Since a H bond at this position would not be possible as a contact point between compound 26 and the active site, the naphthen-1-ol ring of compound 26 may assume a different orientation in binding the WNV NS2B/NS3pro active site compared to the more active compound 22. Solving the crystal structure of the WNV NS2B/NS3pro in the presence of compounds 14 and 22 could reveal the mode of inhibition of WNV NS2B/NS3pro by this class of compounds containing the 8-HQ scaffold.
In conclusion, our results reveal, (1) some potent inhibitors of WNV NS2B/NS3pro by compounds belonging to the 8-HQ class, (2) the N1 of 8-HQ ring is important for inhibition of protease activity, and (3) the mode of inhibition of this class of compounds is by competing with the substrate binding to the active site. Further work to solve the structure of the protease in complex with the inhibitor is necessary to gain insight into the precise orientation of the compound in the active site.
Acknowledgments
We thank Dr. Prasanth Viswanathan for the early work with the assays of the compounds. This work was supported in whole or in part, by National Institutes of Health grants, AI070791, AI070791-03S1 and U01-AI082068 (to R. P.) and the NIH grant awarded to the New England Regional Center of Excellence (NSRB) (U54 AI057159).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aleshin AE, Shiryaev SA, Strongin AY, Liddington RC. Structural evidence for regulation and specificity of flaviviral proteases and evolution of the Flaviviridae fold. Protein Sci. 2007;16:795–806. doi: 10.1110/ps.072753207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias CF, Preugschat F, Strauss JH. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993;193:888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- Bazan JF, Fletterick RJ. Detection of a trypsin-like serine protease domain in flaviviruses and pestiviruses. Virology. 1989;171:637–639. doi: 10.1016/0042-6822(89)90639-9. [DOI] [PubMed] [Google Scholar]
- Beasley DW. Recent advances in the molecular biology of west nile virus. Curr Mol Med. 2005;5:835–850. doi: 10.2174/156652405774962272. [DOI] [PubMed] [Google Scholar]
- Brinton MA. THE MOLECULAR BIOLOGY OF WEST NILE VIRUS: A New Invader of the Western Hemisphere. Annu Rev Microbiol. 2002;56:371–402. doi: 10.1146/annurev.micro.56.012302.160654. [DOI] [PubMed] [Google Scholar]
- Chambers TJ, Weir RC, Grakoui A, McCourt DW, Bazan JF, Fletterick RJ, Rice CM. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci U S A. 1990;87:8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ, Grakoui A, Rice CM. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991;65:6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers TJ, Nestorowicz A, Amberg SM, Rice CM. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J Virol. 1993;67:6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli S, Joseph JS, Daudenarde S, Gatchalian J, Cornillez-Ty C, Kuhn P. Serotype-specific structural differences in the protease-cofactor complexes of the dengue virus family. J Virol. 2010;84:3059–3067. doi: 10.1128/JVI.02044-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clum S, Ebner KE, Padmanabhan R. Cotranslational membrane insertion of the serine proteinase precursor NS2B-NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J Biol Chem. 1997;272:30715–30723. doi: 10.1074/jbc.272.49.30715. [DOI] [PubMed] [Google Scholar]
- Erbel P, Schiering N, D’Arcy A, Renatus M, Kroemer M, Lim SP, Yin Z, Keller TH, Vasudevan SG, Hommel U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- Falgout B, Pethel M, Zhang YM, Lai CJ. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falgout B, Miller RH, Lai CJ. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: Identification of a domain required for NS2B-NS3 protease activity. Journal of Virology. 1993;67:2034–2042. doi: 10.1128/jvi.67.4.2034-2042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould EA, Solomon T. Pathogenic flaviviruses. Lancet. 2008;371:500–509. doi: 10.1016/S0140-6736(08)60238-X. [DOI] [PubMed] [Google Scholar]
- Jadhav A, Ferreira RS, Klumpp C, Mott BT, Austin CP, Inglese J, Thomas CJ, Maloney DJ, Shoichet BK, Simeonov A. Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of a thiol protease. J Med Chem. 2010;53:37–51. doi: 10.1021/jm901070c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D, Schroder K, White H, Fang NX, Stoermer MJ, Abbenante G, Martin JL, Young PR, Fairlie DP. Activity of recombinant dengue 2 virus NS3 protease in the presence of a truncated NS2B co-factor, small peptide substrates, and inhibitors. J Biol Chem. 2001;276:45762–45771. doi: 10.1074/jbc.M107360200. [DOI] [PubMed] [Google Scholar]
- Li J, Lim SP, Beer D, Patel V, Wen D, Tumanut C, Tully DC, Williams JA, Jiricek J, Priestle JP, Harris JL, Vasudevan SG. Functional profiling of recombinant NS3 proteases from all four serotypes of dengue virus using tetrapeptide and octapeptide substrate libraries. J Biol Chem. 2005;280:28766–28774. doi: 10.1074/jbc.M500588200. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- Mueller NH, Yon C, Ganesh VK, Padmanabhan R. Characterization of the West Nile virus protease substrate specificity and inhibitors. Int J Biochem Cell Biol. 2007;39:606–614. doi: 10.1016/j.biocel.2006.10.025. [DOI] [PubMed] [Google Scholar]
- Mueller NH, Pattabiraman N, Ansarah-Sobrinho C, Viswanathan P, Pierson TC, Padmanabhan R. Identification and biochemical characterization of small-molecule inhibitors of west nile virus serine protease by a high-throughput screen. Antimicrob Agents Chemother. 2008;52:3385–3393. doi: 10.1128/AAC.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nall TA, Chappell KJ, Stoermer MJ, Fang NX, Tyndall JD, Young PR, Fairlie DP. Enzymatic characterization and homology model of a catalytically active recombinant West Nile virus NS3 protease. J Biol Chem. 2004;279:48535–48542. doi: 10.1074/jbc.M406810200. [DOI] [PubMed] [Google Scholar]
- Preugschat F, Yao CW, Strauss JH. In vitro processing of dengue virus type 2 nonstructural proteins NS2A, NS2B, and NS3. J Virol. 1990;64:4364–4374. doi: 10.1128/jvi.64.9.4364-4374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27:157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- Shiryaev SA, Kozlov IA, Ratnikov BI, Smith JW, Lebl M, Strongin AY. Cleavage preference distinguishes the two-component NS2B-NS3 serine proteinases of Dengue and West Nile viruses. Biochem J. 2007;401:743–752. doi: 10.1042/BJ20061136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Barrett AD. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol. 2004;2:789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wengler G, Czaya G, Farber PM, Hegemann JH. In vitro synthesis of West Nile virus proteins indicates that the amino-terminal segment of the NS3 protein contains the active centre of the protease which cleaves the viral polyprotein after multiple basic amino acids. J Gen Virol. 1991;72:851–858. doi: 10.1099/0022-1317-72-4-851. [DOI] [PubMed] [Google Scholar]
- Yusof R, Clum S, Wetzel M, Murthy HM, Padmanabhan R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J Biol Chem. 2000;275:9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]
- Zhang L, Mohan PM, Padmanabhan R. Processing and localization of Dengue virus type 2 polyprotein precursor NS3-NS4A-NS4B-NS5. J Virol. 1992;66:7549–7554. doi: 10.1128/jvi.66.12.7549-7554.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]