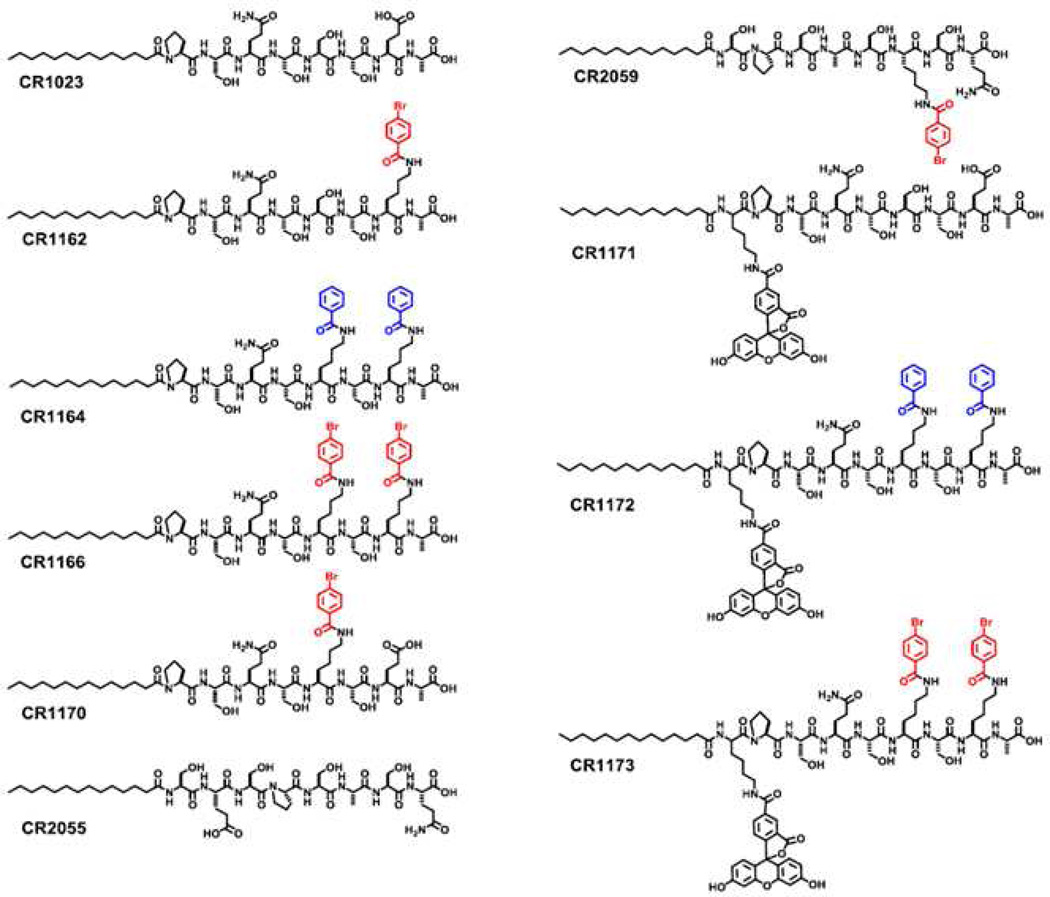
Abstract
GIPC (GAIP-interacting protein, C terminus) represents a new target class for the discovery of chemotherapeutics. While many of the current generation of anticancer agents function by directly binding to intracellular kinases or cell surface receptors, the disruption of cytosolic protein-protein interactions mediated by non-enzymatic domains is an underdeveloped avenue for inhibiting cancer growth. One such example is the PDZ domain of GIPC. Previously we developed a molecular probe, the cell permeable octapeptide CR1023 (N-myristoyl-PSQSSSEA), which diminished proliferation of pancreatic cancer cells. We have expanded upon that discovery using a chemical modification approach, and here report a series of cell permeable, side chain-modified lipopeptides that target the GIPC PDZ domain in vitro and in vivo. These peptides exhibit significant activity against pancreatic and breast cancers, both in vitro and in animal models. CR1166 (N-myristoyl-PSQSK(εN-4-bromobenzoyl)SK(εN-4-bromobenzoyl)A), bearing two halogenated aromatic units on alternate side chains, was found to be the most active compound, with pronounced down-regulation of EGFR/1GF-1R expression. We hypothesize that these organic acid-modified residues extend the productive reach of the peptide beyond the canonical binding pocket, which defines the limit of accessibility for the native proteinogenic sequences that the PDZ domain has evolved to recognize. Cell permeability is achieved with N-terminal lipidation using myristate, rather than a larger CPP (cell-penetrating peptide) sequence. This, in conjunction with optimization of targeting through side chain modification, has yielded an approach that will allow the discovery and development of next-generation cellular probes for GIPC PDZ as well as other PDZ domains.
Keywords: GIPC, PDZ domain, peptides, lipopeptides, structure-based design, sequence alignment, breast cancer, pancreatic cancer, myristoylation
Pancreatic and breast cancers are diseases with high levels of mortality. Statistics for pancreatic cancer are especially grim, with significant death rates within one to two years after diagnosis, and this ultimately accounts for 6% of all cancer-related deaths.1,2–3 Breast cancer, too, exacts a large toll, and is responsible for 3% of all deaths in American women.4–5 A drug development strategy that could disrupt signaling pathways required by both pancreatic and breast cancers could therefore represent an attractive and high-impact approach for therapeutic treatment.
The protein GIPC (GAIP-interacting protein, C terminus) appears to be a viable candidate for such an intervention. GIPC is overexpressed both in pancreatic and breast cancers,6–9 and abnormal activation of GIPC-mediated interactions has been shown to cause or contribute to pancreatic and breast cancers. Several studies including ours have implicated the PDZ domain of GIPC (“GIPC PDZ”) as a critical player in the biology of normal and malignant cells. 7,8,10,11 GIPC PDZ binds to several different cellular proteins, one of which is GAIP (Gα interacting protein), an RGS (regulators of G protein signaling) protein. Association with RGS-GAIP6–8 is responsible for the stability of the tyrosine kinase receptor IGF-1R.
Like the majority of PDZ domains, the binding mode of GIPC PDZ involves direct association with the C-terminal tail of its endogenous partner proteins.6 Based on the accumulated biological data, coupled with the typical mode of PDZ domain binding, what follows is that disruption of this GIPC PDZ-specific interaction by selective inhibitors should block protein-protein interactions within cellular signaling pathways that are required for cancer growth. This was the basis for our initial report in which we disclosed the design, preparation and evaluation of the cell-permeable, linear lipopeptide CR1023 (N-myristoyl-PSQSSSEA, Figure 1).8,12 This chemical probe bears the last eight residues of the C-terminal sequence of GAIP, and was found to inhibit proliferation of pancreatic cancer cells (AsPC-1) in vitro and tumor growth in vivo.8
Figure 1.
Chemical structures of modified and unmodified peptides used in this study.
Beginning with CR1023 as our lead, our objective was to generate potent, cell-permeable GIPC PDZ domain-targeting peptides that would exhibit enhanced efficacy in our biological assays. We had several options, based on our prior experience in devising ligand-design strategies for the PDZ domain. These involved designs based not only on linear peptides composed of standard13 or nonproteinogenic residues,14 but also cyclic,15,16 multivalent,17 and chemically-modified peptides.18 After reviewing all these approaches, we elected to use the binding sequence represented by CR1023, and implement the organic acid-modification strategy that had successfully generated high-affinity inhibitors for the PDZ3 domain of PSD-95.18
This methodology as first reported, in which the side chains of selected residues within a lead sequence were acylated with organic acid substructures, originally involved a chemical library prepared in parallel format. This represents a systematic way in which to introduce ‘non-peptide’ chemical diversity into a conventional peptide, potentially allowing access to non-canonical binding regions of a PDZ domain. This in turn may enhance affinity, selectivity, bioavailability, or metabolic stability, but these benefits have yet to be experimentally validated, and were not specifically investigated in this report.
For the current study, we decided to pursue a more directed approach, and generated a smaller series of peptides as a pilot study, before advancing to a larger screening effort (Figure 1). Here we report on the design and preparation of these novel chemically-modified peptide probes, their biological efficacy as evaluated by both cell-based and animal experiments, and attempts to rationalize the molecular mechanism through homology modeling of the GIPC PDZ protein itself.
RESULTS AND DISCUSSION
Critical early events in tumor progression involve the proliferation, survival, and apoptosis of cancer cells. GIPC appears to be one particular modulator of such progression through the agency of its PDZ domain. The selective inhibition of protein-protein interactions mediated by this particular domain, with subsequent disruption of the signaling pathways associated with GIPC activity, appears to be a promising target for developing novel anticancer peptide ligands. Here we present the molecular design approach pursued towards this end, followed by a series of in vitro and in vivo biological studies performed in cultured cancer cells and an animal tumor model.
Molecular Design—Organic Acid Modification of Peptide Side Chains
For this study, a total of ten peptides were designed and prepared (Figure 1 & SI-Fig.1). Using CR1023 as a starting point, we replaced the existing residues at either the “−1” or “−3” position with lysine, so that that the amine-bearing side chain could be used for chemical modification through acylation with an organic acid. The motivation was to have a simple organic acid–such as benzoate–serve as a form of scaffold or ‘placeholder’ organic moiety, one that could easily be diversified through chemical substitution in the context of a chemical library if later warranted. Thus the design rationale is twofold: first, by using organic acids which display chemical character distinct from that of the standard amino acids, new modes of molecular contact may be implemented; and second, by using a longer ‘amine donor’ residue like lysine (in contrast with those bearing shorter chains, such as ornithine), it is possible to position the appended organic component in a region outside the canonical binding pocket. In this way, it may be possible to incur favorable binding interactions not accessible to the proteinogenic amino acids.
As a first step towards maximizing activity, several brominated sequences were prepared, since halogen substituents are often disproportionately found in biologically and medicinally active compounds. Peptides were constructed based on the ordered sequence found in CR1023, in a limited set that represented monosubstituted and disubstituted analogs (CR1162, CR1164, CR1166 and CR1170).
As support compounds, we generated fluorescently-labeled counterparts to the primary peptides for use in imaging studies (CR1171, CR1172 and CR1173). In addition, we prepared two negative control peptides in which the native residues have been scrambled so as to maintain gross molecular properties (e.g., mass, electrostatics), but no longer represent ordered sequences that should be recognized by GIPC PDZ (CR2055 and CR2059).
One important design consideration was the manner in which to promote membrane permeability, which is a frequently cited problem for many peptides. The peptides were designed so that a myristoyl fatty acid, a saturated 14-carbon alkyl acid, would function as the cargo delivery system. Although not as commonly employed as cell-penetrating peptides (CPPs), there are various examples in the literature for lipidated peptides and lipophilic drug conjugates being used as drug delivery systems.19–21 In particular, myristate has shown promising results for membrane transport, yet only a limited number of myristoylated cargo delivery studies have been reported.22–28 Myristoylated peptides have shown minimal adverse effects on cell viability studies, being non-toxic even up to 100 µM concentrations.29 From a preparative standpoint, they also represent a greatly reduced synthetic burden, as only a single coupling step is required per fatty acid equivalent, in contrast to the multiple coupling steps and residues needed to assemble a functional CPP sequence.
Biological Activity Overview
A series of cell- and animal-based experiments were performed to evaluate the biological efficacy of the peptides in Figure 1. To summarize in advance, what we consistently observe is the enhanced efficacy of the organic acid-modified CR1166 (N-myristoyl-PSQSK(εN-4-bromobenzoyl)SK(εN-4-bromobenzoyl)A), bearing the two bromobenzoate moieties, over that of the original parent sequence peptide CR1023. These experiments were conducted alongside the control compounds CR2055 (scrambled unmodified sequence) and CR2059 (scrambled sequence with single site of modification), as well as other analog compounds. Cell-based assays involved pancreatic (AsPC-1) and breast cancer cell lines (MDA-MB-231-WT).
CR1166 Inhibits Pancreatic And Breast Cancer Cell Proliferation
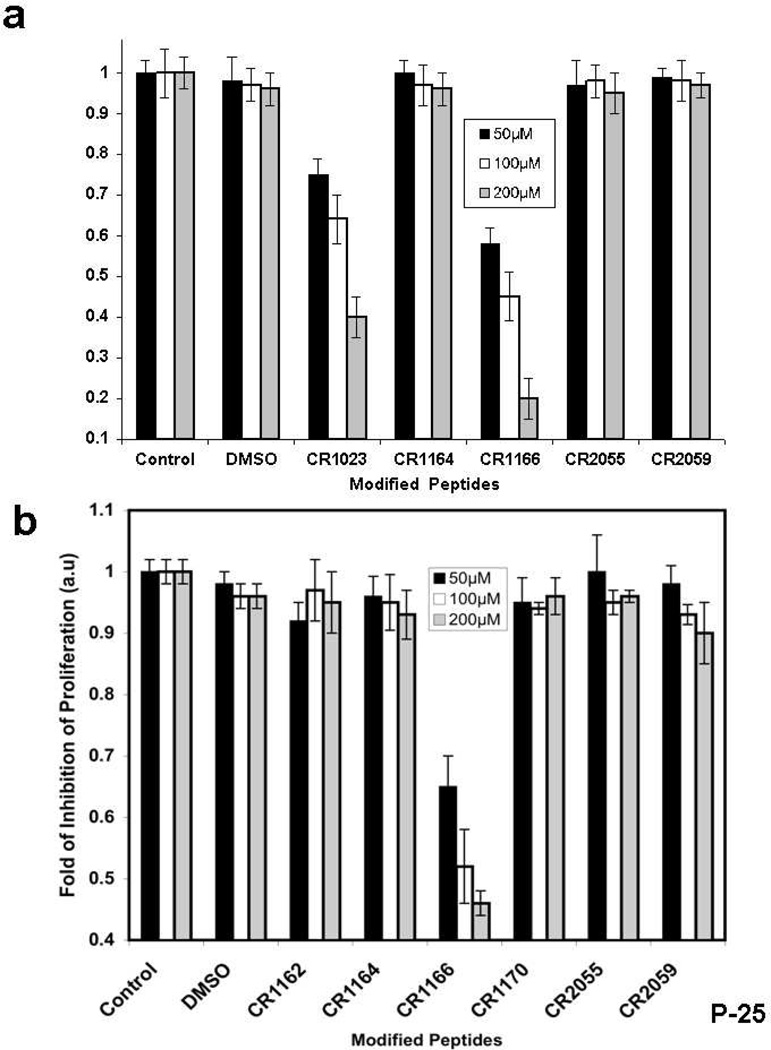
Dose-dependent (50–200 µM) proliferation assays were performed against AsPC-1 and MDA-MB-231-WT cell lines, using radioactive [3H]-thymidine (Figure 2a-b). CR1166 shows significant inhibition of breast cancer cell proliferation in a dose-dependent manner, whereas the control peptides were devoid of activity (Figure 2b). Increasing the concentration of compound CR1166 from 50–100 µM increases the inhibition of proliferation from 42% to 80% in AsPC-1 (Figure 2a), while with CR1023 it increases only from 25 to 60% under similar conditions. Likewise, increasing the concentration of compound CR1166 from 50–100 µM increases the inhibition of proliferation from 32% to 56% in MDA-MB-231-WT (Figure 2b), but for CR1023 it increases only from 19 to 33% (data not shown for compound CR1023).
Figure 2. (a-d): Inhibition of proliferation and induction of apoptosis in pancreatic cancer and breast cancer: a-b.
Inhibition of cell proliferation using different modified GIPC peptides in (a) AsPC1 cells and (b) MDA-MB-231-WT cells for 48 h. With a [3H]-thymidine incorporation assay, a dose-dependent significant inhibition of cell proliferation was observed in both cell lines when cells were treated with compound CR1166 compared to parent active peptide CR1023. No inhibition was observed with control peptides (CR2055, CR2059), or peptides (CR1162, CR1170) or DMSO. Inhibition of proliferation of AsPC1 cells was significantly increased with increasing concentration of CR1023 and CR1166 peptides. CR1166 is the best peptide for inhibition of cell proliferation in both cell lines compared to CR1023 and the control peptides. AsPC1 cells were incubated with different peptides for 48 hours at different three concentrations (50, 100 and 200 µM). DMSO indicates cells treated with DMSO (required for 200 µM peptides dissolved in DMSO). (c-d). Induction of apoptosis in AsPC-1 and MDA-MB-231-WT cells. Apoptosis assay of (c) AsPC-1 and (d) MDA-MB-231-WT cells treated with DMSO, and compounds CR1023, CR1164, CR1170, CR2055, and CR2059 in a dose-dependent manner (50–200µM) for 48 hours. Significant apoptosis was observed after treatment with CR1023 and CR1166 in both cell lines in a dose-dependent manner. CR1166 is most effective among all peptides. No inhibition was observed with either the control peptide CR2055 or DMSO. Apoptosis assay of AsPC-1 and MDA-MB-231-WT cells treated with DMSO and modified GIPC peptides in a dose-dependent manner (50–200µM) for 48 h of treatment using Annexin V-FITC Apoptosis Kit. Significant induction of apoptosis was observed by compound CR1166.
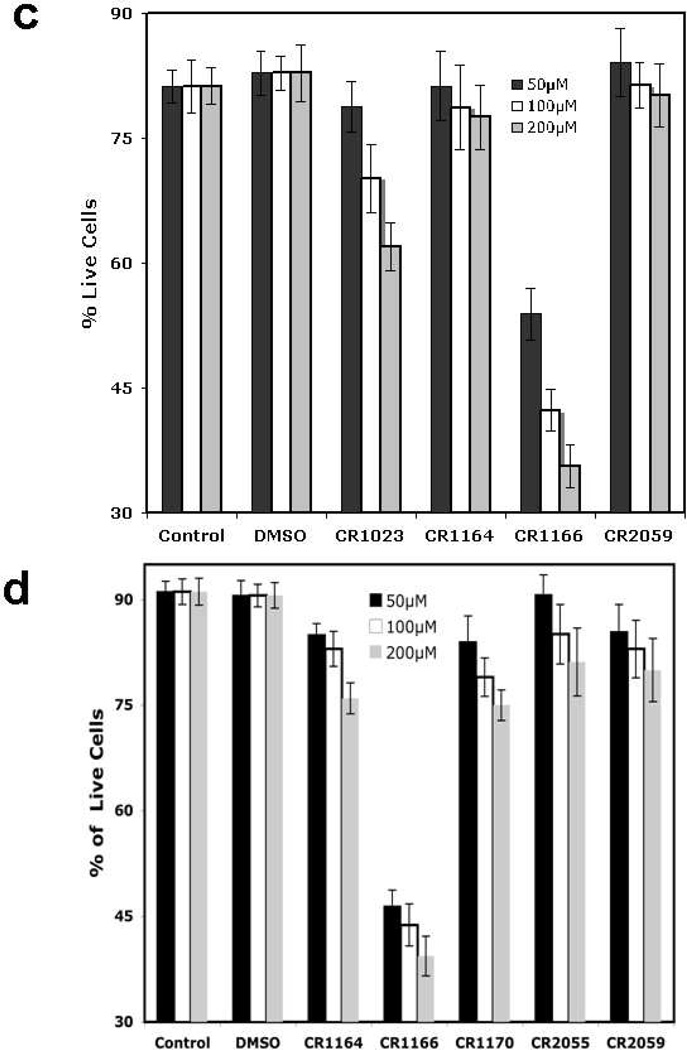
CR1166 is Cytotoxic to Pancreatic and Breast Cancer Cells
To assess the cytotoxic and cell viability effect of the peptides, an MTS assay was conducted in AsPC-1 and MDA-MB-231-WT cells using variable concentration (50–200 µM) (see SI-Fig. 2a-b). Both CR1023 and CR1166 treatment resulted in significant reduction of cell viability, in a dose-dependent manner, compared to CR1162, CR1164, CR2055 and CR2059. However, peptide CR1166 shows 50%, 45%, and 35% further reduction of cell viability compared to CR1023 at 50, 100 and 200 µM concentrations, respectively, compared to the parent active compound CR1023 in AsPC-1 cells. A similar effect is observed in MDA-MB-231-WT cells (SI-Fig. 2b).
CR1166 Induces Apoptosis in Pancreatic and Breast Cancer Cells
Dose-dependent (50–200 µM) apoptotic assays (using the annexin/PI method) were performed against AsPC1 and MDA-MB-231-WT cells (Figure 2c and Figure 2d). A significant induction of apoptosis was observed for CR1023 and CR1166, whereas the control and other peptides showed little effect. Increasing concentrations of CR1166 from 50 to 200 µM induces apoptosis (from 47% to 65%) with 53% to 35% reduction in live AsPC-1 cells within 48 h of treatment. CR1023, though, shows 78% to 62% reduction in live AsPC-1 cells at the same time point. On the other hand, increasing concentrations of CR1166 from 50 to 200 µM induces apoptosis from 54% to 61% in MDA-MB-231-WT cells within 48 h of treatment, at the same time point, the value range for CR1023 is 30% to 35% (data not shown).
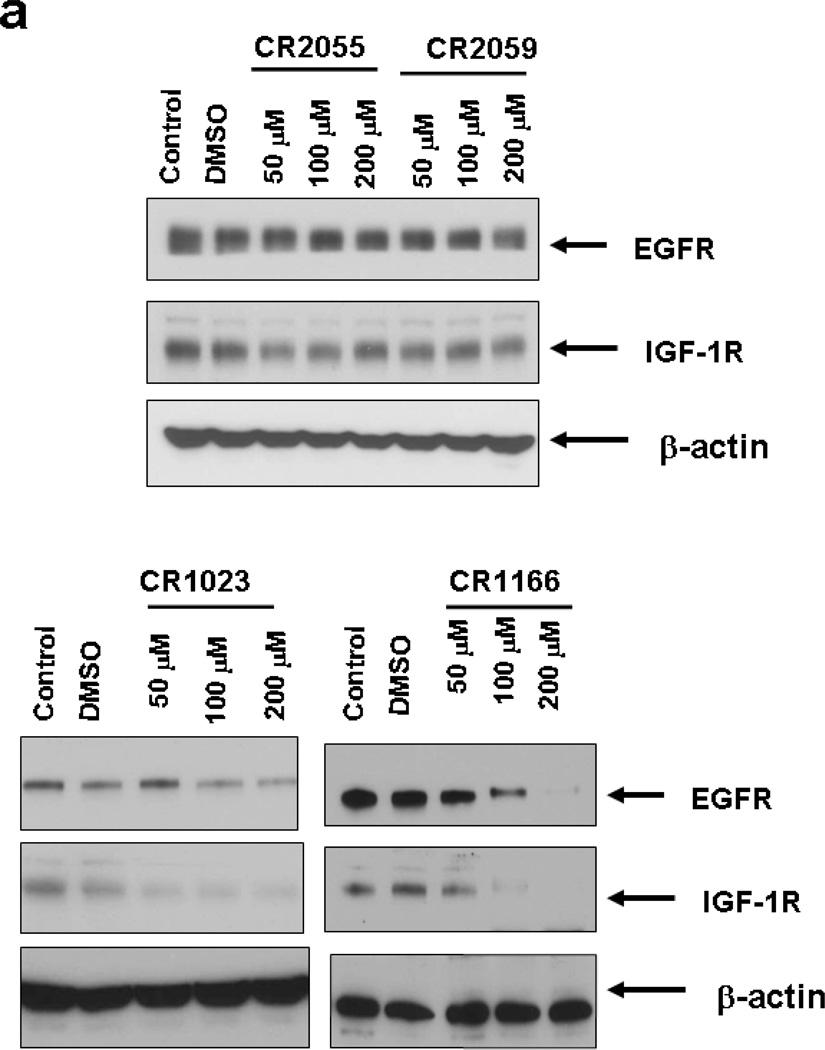
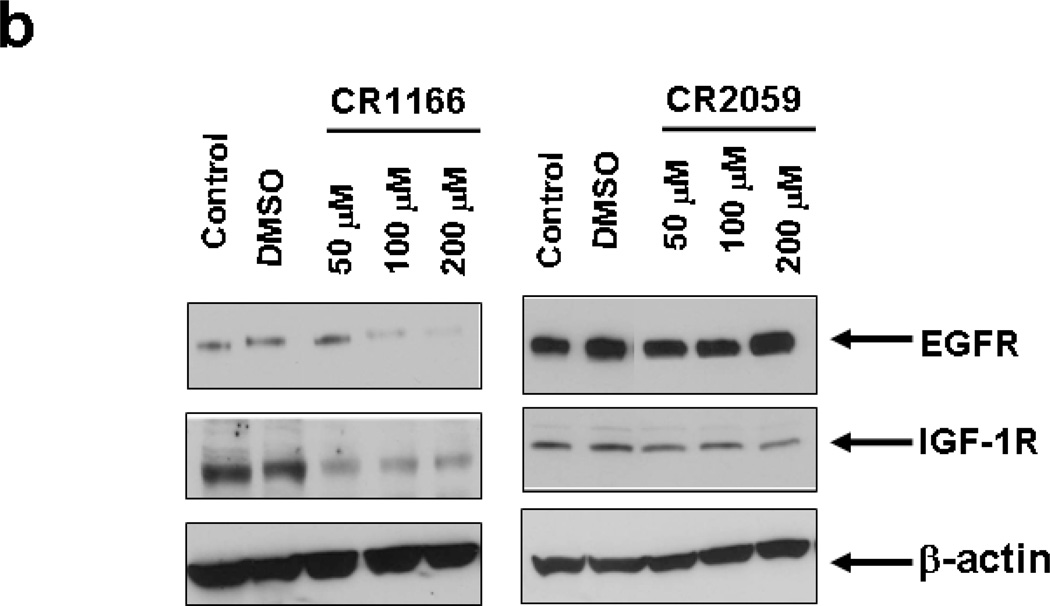
CR1166 Down-Regulates EGFR and IGF-1R Expression
EGFR and IGF-1R are overexpressed in pancreatic and breast carcinomas, and their cellular levels are regulated by GIPC.7, 8 Given this linkage, we investigated whether any of the peptides would affect IGF-1R and EGFR protein expression in both AsPC-1 and MDA-MB-231A-WT cells. The levels of EGFR and IGF-1R in both the pancreatic (Figure 3a) and breast cancer cells (Figure 3b) were analyzed after treatment with the peptides (48 hours at 50, 100 and 200 µM) using Western blot analysis. Immunoblots depict dose-dependent regulation of EGFR and IGF-1R expression in AsPC-1 (Figure 3a) and MDA-MB-231A-WT cell lines (Figure 3b) when treated with CR1023 and CR1166, compared to the cells that were untreated (DMSO) or treated with the control and other peptides. In both cell lines CR1166 is the most effective peptide in down-regulating selectively EGFR and IGF-1R expression.
Figure 3. (a-b): Treatment of compound CR1166 down-regulates EGFR and IGF-1R expression in AsPC-1 and MDA-MB-231-WT cells.
a-b. Western blot analysis of the proteins obtained from control (a) AsPC-1 cells or (b) MDA-MB-231-WT cells and cells treated with different modified Myr-GIPC peptides at different doses (50 µM – 200µM) for 48 hours. As determined by Western blotting, myristoylated GIPC peptides, most notably CR1166, regulates EGFR and IGF-1R expression in (a) AsPC-1 cells and (b) MDA-MB-231-WT cells. DMSO indicates cells treated with DMSO (required for 200 µM peptides dissolved in DMSO).
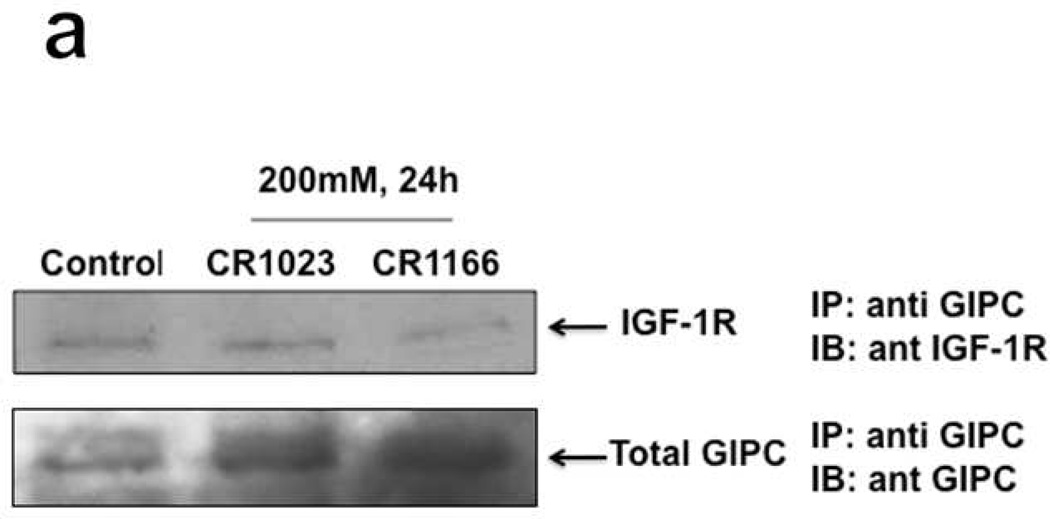
CR1166 Inhibits Association Between GIPC and IGF-1R
GIPC association with IGF-1R is important for IGF-1R stability. An immunoblot against IGF-IR after immunoprecipitation for GIPC (IP:GIPC IB:IGF-1R) indicates that indirect evidence of selective targeting to IGF-1R is enhanced by CR1166, as it shows the down regulation of IGF-1R when the cells were treated with that compound (Figure 4a). Inhibition of the interaction between IGF1R and GIPC is more pronounced with CR1166 than with CR1023. Our previous results demonstrated that GIPC regulates the IGF-1R level in pancreatic cancer, and it is PDZ domain–dependant.7,8 In confirmation of these results, we have treated the AsPC1 cells with CR1166 and collected the cell lysate in RIPA buffer. After I.P. with a GIPC (N-19) antibody, Western blot analysis indicated significant down-regulation of IGF-1R expression while the AsPC1 cells were treated with CR1166, compared to untreated cells or those treated with compound CR1023. These results unambiguously show that blocking GIPC with CR1166 reduced protein levels of IGF-IR significantly in the in vitro model.
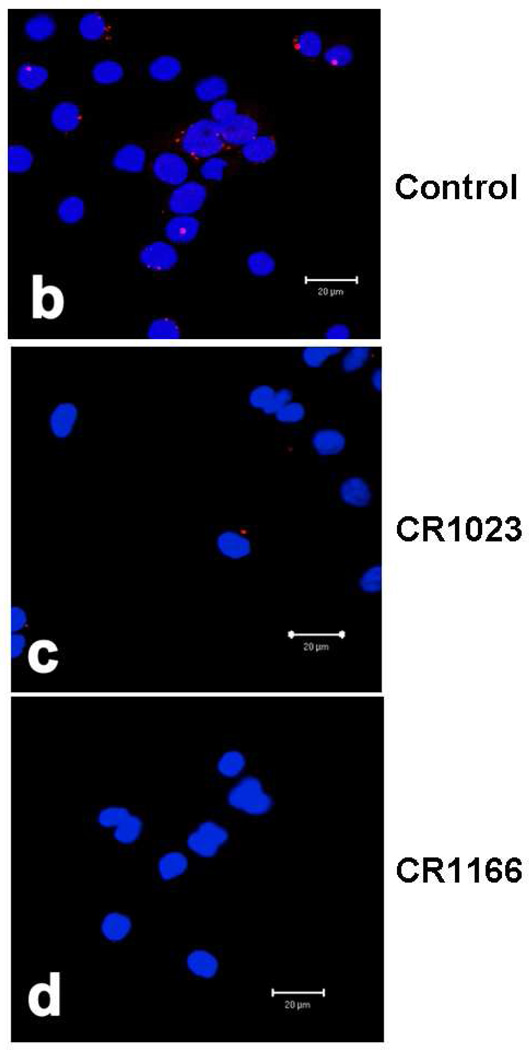
Figure 4. (a-d). a. Co-immunoprecipitation of IGF-IR and GIPC in AsPC1 cells.
(a), immunoblot against IGF-IR after immunoprecipitation for GIPC (IP:GIPC/IB:IGF-1R). AsPC1 cells were treated with 200 µM peptide (CR1023 and CR1166) for 24 hours. This immunoblot showed that CR1166 treatment reduces the association between GIPC and IGF-IR more than CR1023 peptide. (b-d). Co-localization of GIPC and IGF-1R using CR1023 and CR1166. (b) Control, (c) treatment with CR1023, and (d) treatment with CR1166, which inhibits the association between GIPC and IGF-1R in AsPC1 cells.
CR1166 Inhibits Co-localization of GIPC and IGF-1R
The co-localization of GIPC and IGF-1R was visualized and located in control untreated AsPC1 cells using a Duolink II Fluorescence kit (Figure 4b). Red fluorescence indicates the co-localization of GIPC and IGF-1R in AsPC1 cells. Minimal red color fluorescence can be seen when the cells were treated with CR1023 (Figure 4c). However, there is no red color when the AsPC1 cells were incubated with CR1166 (Figure 4d). These results indirectly support the contention that there is inhibition of the binding between GIPC and IGF-1R in the presence of CR1166, and that this peptide selectively binds to the PDZ domain of GIPC.
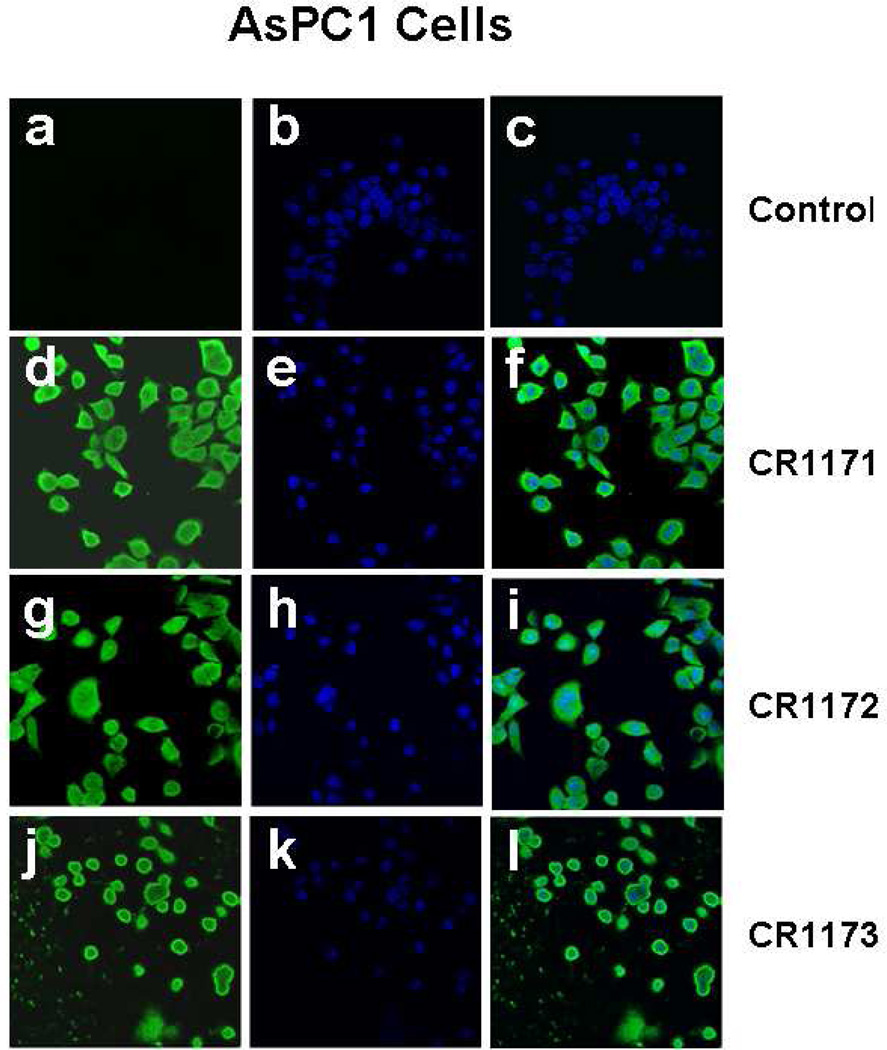
CR1166 Shows Prominent Membrane Localization in Pancreatic and Breast Cancer Cells
To detect whether these myristoylated peptides were able to cross the cell membrane and compete for intracellular GIPC-binding interactions, we synthesized 5-carboxyfluorescein analogs of the compounds (CR1171, CR1172, and CR1173, corresponding to the sequences of CR1023, CR1164, and CR1166, respectively). The green (for fluorescein) and blue (for DAPI) fluorescent microscopy images were collected for AsPC-1(Figures 5 a-l) and MDA-MB 231-WT cells (SI-Figures 3 a-l) treated with CR1171, CR1172, and CR1173 using a LSM 510 confocal laser scan microscope. The presence of green fluorescence in Figures 5d, -g, -j clearly depicts the internalization of compounds CR1171 and CR1172 in AsPC-1 cells. In a similar vein, the presence of green fluorescence in SI-Figures 3d, -g, and -j provide visual confirmation of the internalization of CR1171 and CR1172 in MDA-MB-231-WT cells. Untreated AsPC-1 and MDA-MB-231-WT cells were devoid of the green fluorescent signal (Figure 5a and SI-Figures 3a) Interestingly, compound CR1173 is localized only on the membrane of AsPC-1(Figures 5j-l) and MDA-MB-231-WT cells (SI-Figures 3j-l), whereas CR1171 and CR1172 stained the entire cells under the same conditions. We speculate the reason why compound CR1173 is localized on the surface of the membrane is based upon improved binding contacts (Figures 6a-b) and/or increased membrane permeability. The better defined membrane localization of CR1166 compared to the more disperse localization of CR1023 demonstrates the impact of the halogenated aryl groups, and how modification of organic moieties appended to a peptide ligand can impart enhanced properties, such as cellular distribution.
Figure 5. (a-l). (a-l) Localization Compound CR1023, CR1164 and CR1166, in AsPC-1 cells.
Confocal images of AsPC1 cells treated with FAM-conjugated peptides (CR1171, CR1172 and CR1173). (a-c) Control experiment, cells treated with DMSO, (d-f) cells treated with compound CR1171, (g-I) cells treated with compound CR1172, (j-l) cells treated with compound CR1173. Green fluorescence emissions due to presence of fluorescein in peptide were collected through a 505–550 nm band pass filter in conjunction with an argon ion laser excitation of 488 nm. The blue fluorescence emissions were collected through a 420–480 nm band pass filter in conjunction with a diode laser excitation of 405 nm for DAPI. No green fluorescence was observed in control cells treated with DMSO due to absence of fluorescein.
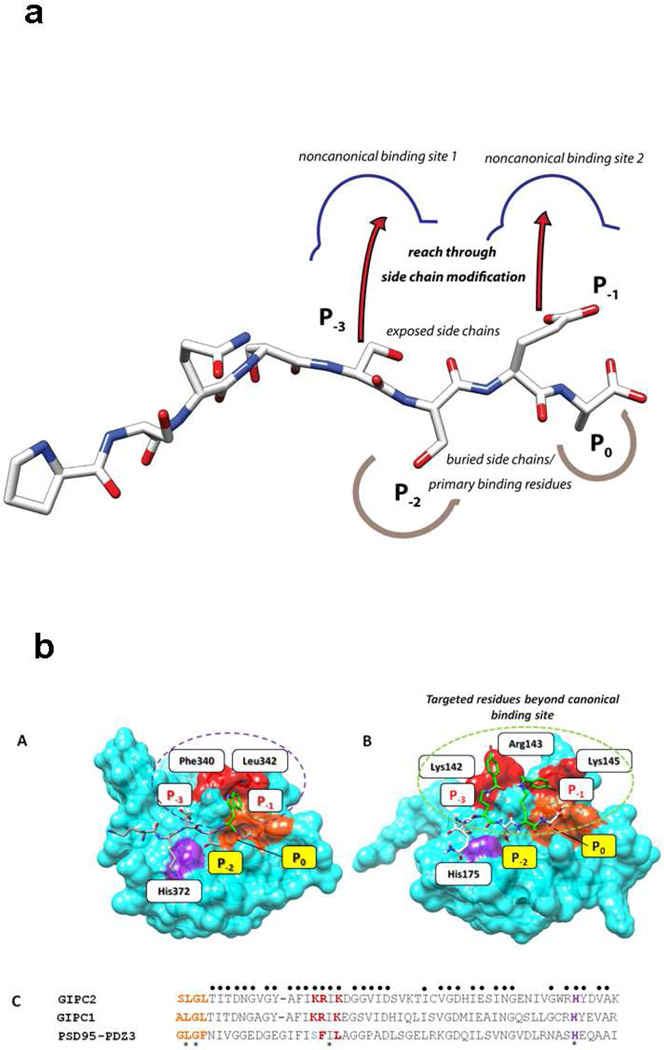
Figure 6. Design of the modified compounds based on the sequence of parent active compound, PSQSSSEA (CR1023,).
a. The P−1 and/or P−3 positions are replaced by Lys acylated with an organic acid (halogenated or non-halogenated). If only P−3 is modified, P−1 is Glu, if only P−1 position is modified, P−3 is Ser. b. Structure-based design rationale for halogenated benzoyl-modified ligands for GIPC PDZ domains, based on the complex between PDZ3 and KKETWV. Positions P0 and P−2 denote conserved primary binding determinants. (A) Key residues of PDZ3 domain binding interaction with the KKETWV are shown with conserved residues for canonical binding sites and the P−1 interaction site. (B) Expected binding mode of the pharmacophore region of CR1166 [QSK(4-bromobenzoyl)SK(4-bromobenzoyl)A] in the GIPC PDZ domain. The residues (Gly-Leu-Gly-Phe) of the carboxylate binding GLGF loop (of PDZ3) and its counterpart SLGL (in GIPC2) are colored in orange, and the His crucial for P−2 binding is shown in purple. Additional residues contacting ligands in both PDZ3 and GIPC PDZ complexes are red. The red surface represents the targeted region (P−3 and/or P−1) in PDZ proteins. In PDZ3 hydrophobic residues (Phe and Leu) line the cavity, in GIPC these are positively charged resides (Arg and Lys). In the design, either P−1 or P−3, or both, positions will be replaced by Lys acylated with halogenated benzoic acid. (C) Multiple sequence alignment of selected region of PDZ domains of GIPC2, RGS-GAIP interacting protein (GIPC1) and PSD-95-PDZ3. A sequence alignment of GIPC2, RGS-GAIP interacting protein (GIPC1) and PSD95-PDZ3 was performed with T-Coffee (www.ebi.ac.uk/t-coffee). The sequences were arranged according to structure-based alignment. The key residues identical in all three regions are marked with an asterisk, those identical to either GIPC PDZ domain regions are marked with a dot. The color scheme is as in A. The alignment scores for all three PDZ regions and for the two GIPC PDZ proteins are 81% and 98%, respectively. The amino acid sequences are as follows: GIPC2 chain A (PDB ID: 3GGE), RGS-GAIP (GIPC1) (accession AAC67550.1), and PSD95-PDZ3 (PDB ID: 1TP5). Either P−1 (Glu) or P−3 (Ser) or both positions are replaced by Lys acylated with organic acid (halogenated or non-halogenated). Structures were generated from Chem3D Pro program v11.0.1, AutoDock Tools 1.5.4 and rendered using UCSF Chimera v1.5.
Enhanced Activity of Diarylbrominated CR1166
Overall, CR1166 has the highest activity compared to no activity in CR1164. This is most likely due to the new potential contacts that the halogen makes with the conserved P−3 and P−1 interaction regions of the GIPC PDZ domain, while retaining the P−2 and P0 ‘anchoring residues’ that are seen with many PDZ domain-peptide interactions (Figure 6a). Similar effects have been observed in other protein-ligand complexes, e.g., flavopiridol, a kinase inhibitor currently undergoing clinical trials, possesses a halogenated-phenyl group instead of unsubstituted phenyl which increases its inhibitory effect by six-fold.33,34,35 Organic halogen atoms contribute to a variety of noncovalent protein-ligand interactions such as halogen bonds, hydrogen bonds, and electrostatic-type interactions that enhance the stability of ligand-target complexes.36,37,38 Furthermore, the attachment of bulky halogen atoms can increase binding contacts39,40,41 and also enhance membrane permeability.42 Ohmori et al.43 and Hidaka et al.44 showed that halogen modifications in the compound Nemonapride can be used to overcome low selectivity over D4 receptors and that removal of chlorine was deleterious for dopaminergic receptors selectivity. All these data are supportive of the proposition that the aromatic halogenation of CR1166 results in a stronger binding interaction with the GIPC PDZ domain.
Homology Modeling and Rationalizeing the Effect of Organic Acid Modification
An empirically determined structure of a protein target can aid in generating explanations for a ligand’s efficacy, through the exercise of careful molecular modeling. The absence of such structural information for GIPC PDZ, though, alone or in complex with a bound ligand, motivated us to construct a simple homology model based on characterized PDZ domain protein. With this, we can make qualified rationalizations about the degree to which our molecular modifications affect ligand binding/activity.
Using the X-ray crystallographic structure of PSD-95-PDZ3 complex with KKETWV (PDB entry 1TP5) as a starting point, followed by sequence alignment with the GIPC PDZ domain to identify binding pockets (as well as possible targeting regions), and finally, manually docking the key binding residues of CR1023 and CR1166, we could propose regions accessible by these ligands on the PDZ surface (Figure 6b). Docking the residues of CR1166 into the model in a manner consistent with PDZ domain binding (Figure 6a) suggested that the dual 4-bromobenzoyl modifications can interact with two non-canonical binding sites created by regions which are positively charged, but highly conserved among GIPC PDZ domains (Figure 6b). Observation of these binding pockets suggests that the dual 4-bromobenzoyl modifications occupy regions outside the canonical binding site, regions formed by the Lys-Arg-Ile-Lys residues of the GIPC PDZ domain. With optimal positioning, the modified side chain moieties might form favorable hydrogen bonds, halogen bonds, and electrostatic interactions.
Overall, these modeled results correlate with our design hypothesis where increased activity could be linked to targeting two non-canonical binding regions. These are highly conserved among human, rat, and mouse GIPC proteins but absent in a representative “Class I” PDZ domain, the third PDZ domain (PDZ3) of PSD-95.6 At this point, such model explanations are speculative, and more detailed bioenergetic analysis will require structural biological characterization, coupled with calorimetric binding assays to quantitate the individual free energy contributions of the side chains.
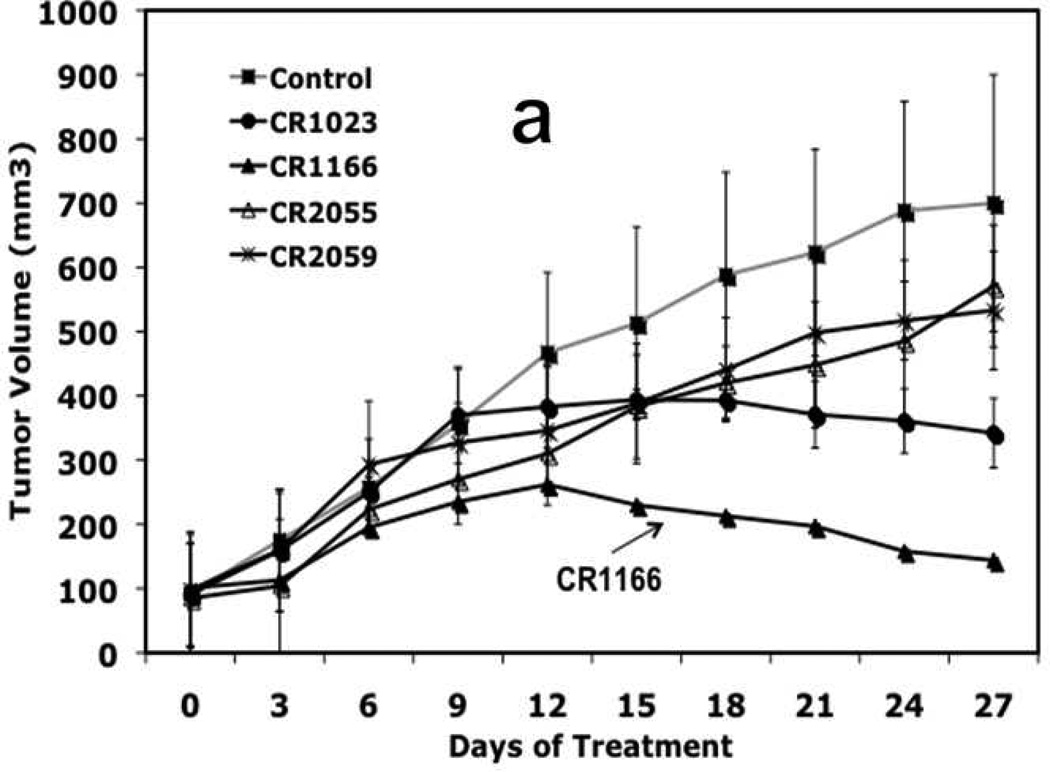
CR1166 Reduces Pancreatic and Breast Tumor Growth
In vivo tumor model experiments using female SCID mice were performed. The pancreatic tumor model was established by taking 5×106 cells from AsPC-1 cells expressing GFP/luciferase, resuspending in 50 µL of sterile PBS, and injecting into the mouse right flank.30,31 The breast tumor model was established by taking 2×106 cells of MDA-MB-231-WT, resuspending in 50µL of sterile PBS, and injecting into the mammary fat pad of the mice. When tumor volume reached 50–100 mm3, the compounds (CR1023, CR1164, CR1166, CR1170, CR2055, and CR2059) were intratumorally injected on alternate days over a period of one month.
To monitor pancreatic tumor burden, the mice were non-invasively imaged using an IVIS 200 Bioluminescence Imaging system (Xenon Corp.) before being sacrificed30. SI-Fig. 4 depicts comparative bioluminescence images of the pancreatic tumors expressing GFP/luciferase from the mice treated with active compounds CR1023 and CR1166, along with CR1164, CR2055, and CR2059. From SI-Fig. 4 it is evident that there is a significant reduction of pancreatic tumor growth when the mice were treated with CR1166 compared to CR1023 or DMSO (control group). However, there is little or no effect in tumor reduction when mice were treated with negative control peptides CR2055 or CR2059.
CR1166 Suppresses EGFR and IGF-1R Expression in Pancreatic and Breast Tumors
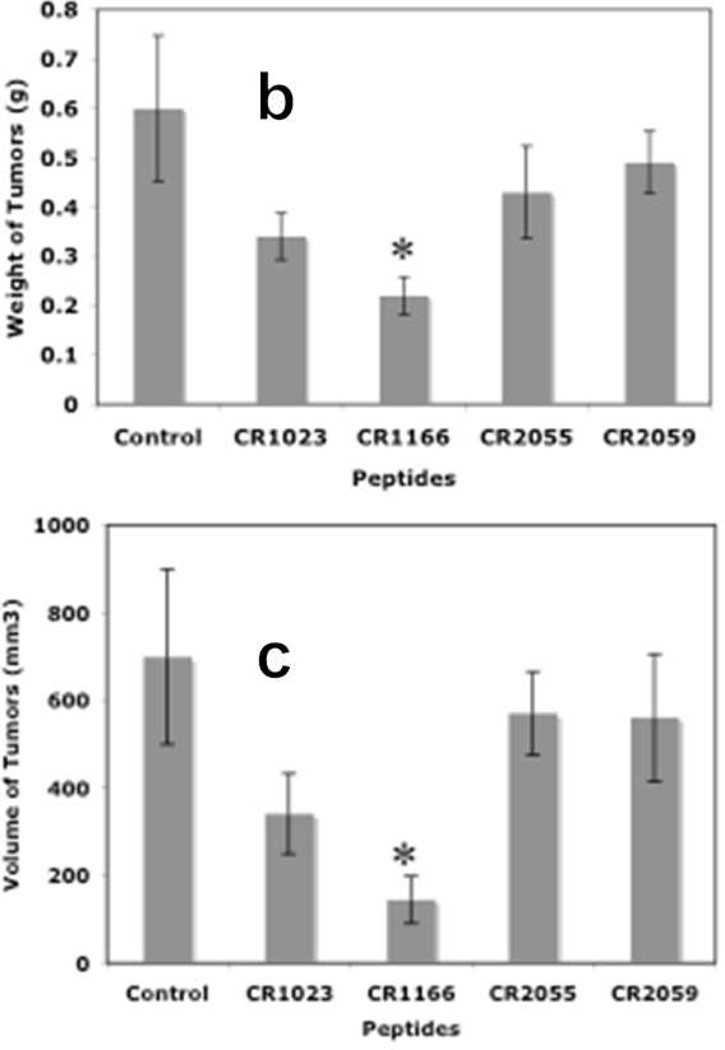
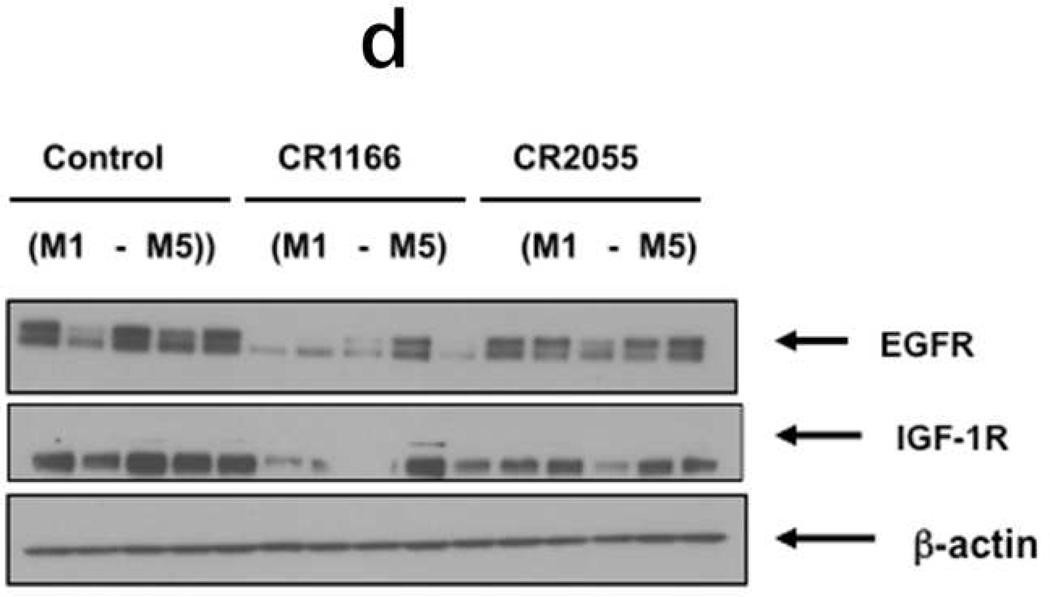
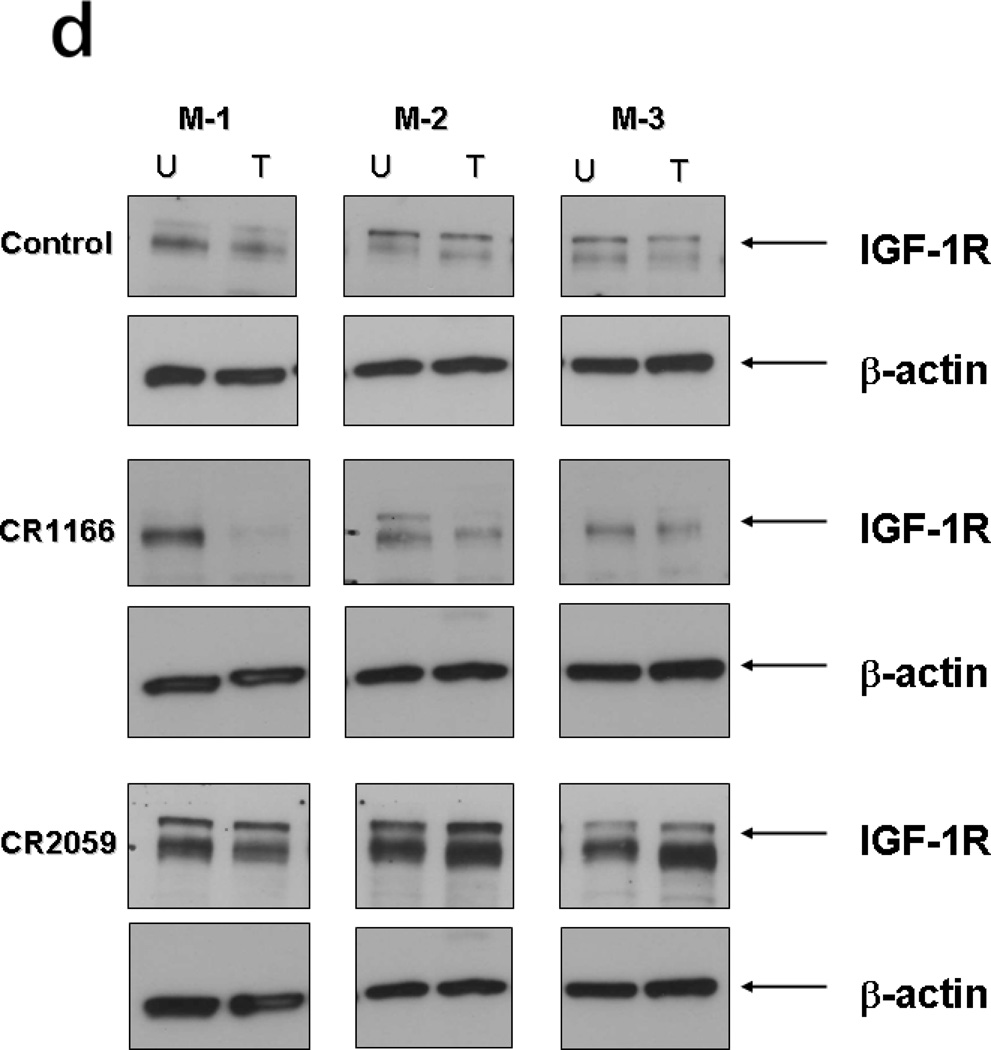
After bioluminescence imaging, we examined the effect of the peptides on tumor growth (weight and volume) and EGFR and IGF-1R expression from tumors collected from different groups. CR1166 suppressed the pancreatic tumor growth over time compared to CR1023 and control peptides CR2055 and CR2059 (Figure 7 a-c). CR1166-treated mice showed significant tumor regression (i.e., 4.8, 2.3, 3.8, and 3.9 times) as compared to control DMSO-PBS-treated mice and mice treated with CR1023, CR2055 and CR2059, respectively. A portion of each tumor was evaluated for IGF-1R and EGFR expression, and a significant reduction of both IGF-1R and EGFR expression was observed in mice treated with CR1166 (Figure 7d). These results correlate with our in vitro data.
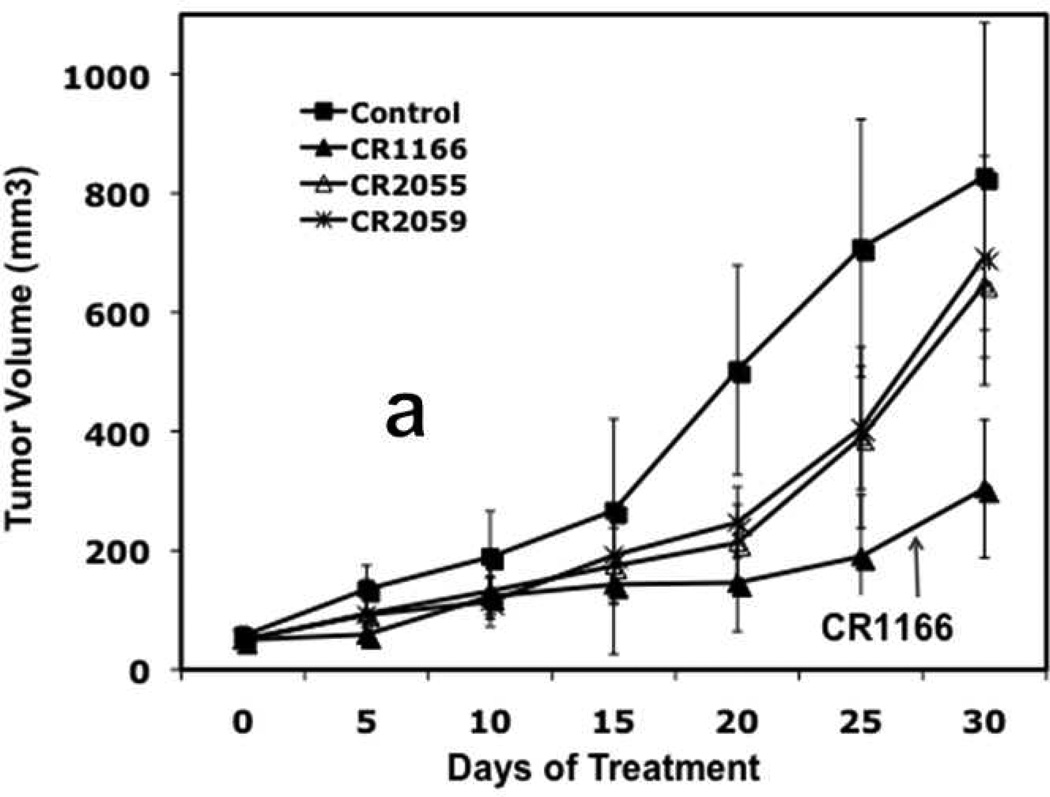
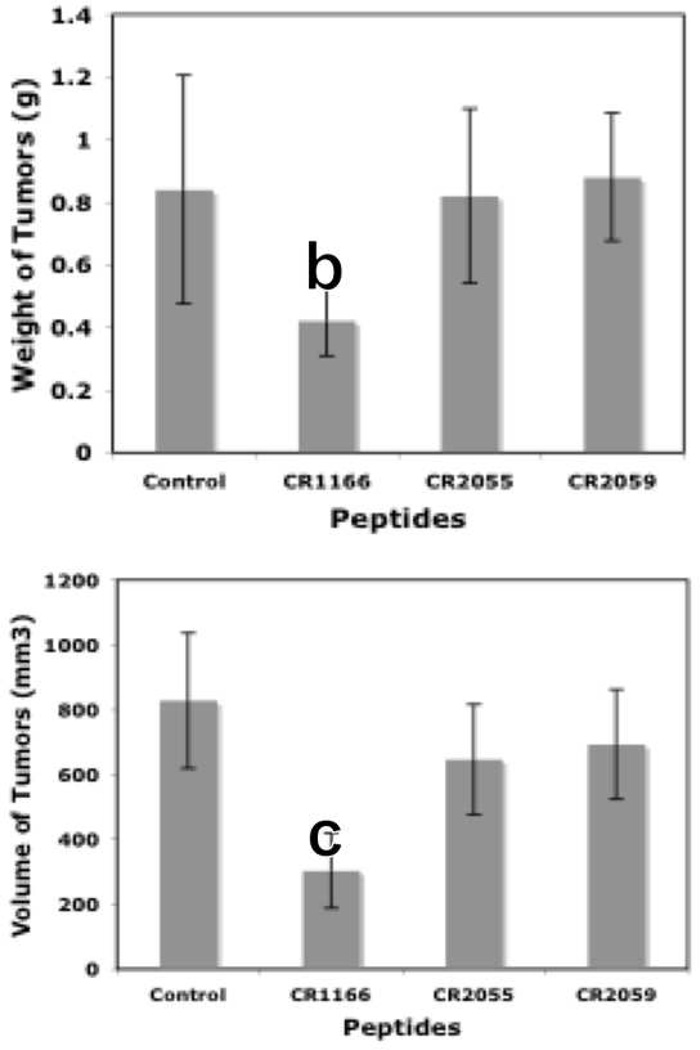
Figure 7. (a-d), In vivo effect of intratumoral injection of peptides on tumor weight, volume, EGFR and IGF-1R expression in pancreatic tumor model.
a. Effect of administration of peptides on the tumor growth over time. b-c. Tumor weight and tumor volumes decreased after peptide treatment, especially CR1166, compared to CR1023. Tumors collected after sacrificing the SCID mice and measuring with digital slide calipers using the (1/2ab2) formula. d. EGFR and IGF-1R expression is decreased in the harvest tumors after CR1166 treatment compared to tumors treated with DMSO alone.
Similarly, CR1166 suppressed breast tumor growth over time compared to CR1023 (data not shown) and control peptides CR2055 and CR2059 (Figure 8 a-c). Tumors were then harvested, and their average mass and volume measured (Figure 8b and Figure 8c). It is evident from the data that CR1166-treated mice clearly demonstrate significant breast tumor regression (i.e., 2.7, 1.4, 2.1, and 2.2 times) as compared to the control DMSO-PBS-treated mice and mice treated with CR1023 (data not shown), CR2055, and CR2059, respectively. A portion of each tumor was evaluated for IGF-1R expression, and a reduction of IGF-1R expression was observed in mice treated with CR1166 (Figure 8d). These results also correlate with our in vitro data in breast cancer cells.
Figure 8. (a-d), In vivo effect of intratumoral injection of peptides on tumor weight, volume, EGFR and IGF-1R expression in breast tumor model.
a. Effect of administration of peptides on the tumor growth over time. b-c. Tumor weight and tumor volumes decreased after peptide treatment, especially CR1166, compared to control peptides. Tumors collected after sacrifice of the SCID mice and measured by digital slide calipers using the (1/2ab2) formula. d. EGFR and IGF-1R expression is decreased in the harvest tumors after CR1166 treatment (T) compared to tumors treated with DMSO alone (U) and CR2055 and CR2059.
CR1166 Lowers Proliferation Activity of Breast Tumors
Proliferation, being a key feature in tumor progression, can be estimated by the nuclear antigen Ki-67. 32 The proliferation marker Ki67 showed a significantly lower proliferation activity (~5 fold) in the CR1166 treatment group of mice compared with the control group in the breast cancer tumors (SI-Fig. 5).
Summary
We have successfully developed a cell-permeable lipopeptide that inhibits the PDZ domain of GIPC both in vitro and in vivo, and exhibits significant inhibitory effect against pancreatic and breast cancer cells and tumors. These results add weight to the assertion that GIPC is a chemotherapeutic drug target, and enhances the prospect of peptide-based drug leads serving as next-generation therapeutic agents. In addition, our results add validation to a chemically-modified design strategy for developing cell-permeable peptide probes that target specific PDZ domain-containing proteins in vivo. Finally, at the level molecular design, CR1166 is an example of a peptide's bioactivity being improved through halogen substitution, which has commonly been seen with organic small molecules, but much less so for peptides.
METHODS
Peptide Synthesis, Purification and Characterization
Compounds were prepared using Fmoc-based solid-phase peptide synthesis (SPPS) protocols, with modifications as required to generate the organic acid-modified analogs. These SPPS methods employed microwave heating (CEM Discover S-class microwave synthesizer). Purification was carried out using reverse-phase HPLC, and characterization by mass spectrometry. Detailed procedures for all compounds are provided in the Supporting Information.
Cell Culture and Functional Activity Tests
Cell proliferation assay, MTS assay (SI-Fig. 2). Apoptosis assay, western blot analysis, along with confocal fluorescence microscopy(SI-Fig. 3), Ki67 staining (SI-Fig. 5) are described in detail in the
In vivo Experiments in Mice
Experiments on six-week-old female SCID (severe combined immuno deficient) mice were performed along with bioluminescence image as described in the Supporting Information (SI-Fig. 4).
Statistical Analysis
The data in the bar graphs represent the mean ± standard error of the mean (S.E.M) of at least three independent experiments, each performed with triplicate samples. Statistical analyses were performed using a Student’s t test, with a two-tailed value of P<0.05 considered significant.
Homology Modeling
Peptide ligands were modeled into the GIPC PDZ binding pocket using VMD molecular graphics software (v. 1.8.6). The manual docking of proposed binding residues of compounds involved an iterative process of manual predocking of the conserved P0 (Ala) and P−2 (Ser) residues to the canonical GIPC PDZ domain binding pockets. Further detail is provided in the caption to Figure 6 and in the Supporting Information.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by National Institutes of Health (NIH) grant CA150190 to DM and MS. Additional funding was provided as a generous gift from Bruce and Martha Atwater to DM. CRP is also grateful to DST, Govt. of India, New Delhi, India for the award of Ramanujan Fellowship (GAP 0305\DST\CP).
Footnotes
SUPPORTING INFORMATION
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Anderson KE, Mack T, Silverman D. Cancer of the pancreas. 3rd ed. New York: Oxford University Press; 2006. [Google Scholar]
- 2.Saif MW. Urgent Calls for Effective Earlier Disease Detection and Multi-Modal Therapeutic Strategies in Pancreatic Cancer. J. Pancreas. 2009;10:464–465. [Google Scholar]
- 3.ACS Breast Cancer Facts & Figures. [accessed January14];2009–2010 http://www.cancer.org/docroot/STT/STT_0.asp.
- 4.WHO, World Health Organization. Vol. 2008. World Health Organization; 2008. Agency for Research on Cancer: Fact Sheet No. 297,” July 2008. [Google Scholar]
- 5.ACS, American Cancer Society, “What Are the Key Statistics fo Breast Cancer?,”. American Cancer Society. 2008 http://www.cancer.org/2008CAFFfinalsecured.pdf.
- 6.De Vries L, Lou XJ, Zhao G, Zheng B, Farquhar MG. GIPC, a PDZ domain containing protein, interacts specifically with the C terminus of RGS-GAIP. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12340–12345. doi: 10.1073/pnas.95.21.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muders MH, Dutta SK, Wang L, Lau JS, Bhattacharya R, Smyrk TC, Chari ST, Datta K, Mukhopadhyay D. Expression and regulatory role of GAIP-interacting protein GIPC in pancreatic adenocarcinoma. Cancer Res. 2006;66:10264–10268. doi: 10.1158/0008-5472.CAN-06-2321. [DOI] [PubMed] [Google Scholar]
- 8.Muders MH, Vohra PK, Dutta SK, Wang E, Ikeda Y, Wang L, Udugamasooriya DG, Memic A, Rupashinghe CN, Baretton GB, Aust DE, Langer S, Dam K, Simons M, Spaller MR, Mukhopadhyay D. Targeting GIPC/Synectin in Pancreatic Cancer Inhibits Tumor Growth. Clin. Cancer Res. 2009;15:4095–4103. doi: 10.1158/1078-0432.CCR-08-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudchenko S, Scanlan M, Kalantarov G, Yavelsky V, Levy C, Estabrook A, Old L, Chan GL, Lobel L, Trakht I. A human monoclonal autoantibody to breast cancer identifies the PDZ domain containing protein GIPCI as a novel breast cancer-associated antigen. Bmc Cancer. 2008;8:248. doi: 10.1186/1471-2407-8-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kedlaya RH, Bhat KMR, Mitchell J, Darnell SJ, Setaluri V. TRP1 interacting PDZ-domain protein GIPC forms oligomers and is localized to intracellular vesicles in human melanocytes. Arch. Biochem. Biophys. 2006;454:160–169. doi: 10.1016/j.abb.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirikoshi H, Katoh M. Expression of human GIPC1 in normal tissues, cancer cell lines, and primary tumors. Int. J Mol. Med. 2002;9:509–513. [PubMed] [Google Scholar]
- 12.Wang L, Lau JS, Patra CR, Cao Y, Bhattacharya S, Dutta S, Nandy D, Wang E, Rupasinghe CN, Vohra P, Spaller MR, Mukhopadhyay D. RGS-GAIP–Interacting Protein Controls Breast Cancer Progression. Mol. Cancer Res. 2010;8:1591. doi: 10.1158/1541-7786.MCR-10-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saro D, Li T, Rupasinghe C, Paredes A, Caspers N, Spaller MR. A thermodynamic ligand binding study of the third PDZ domain (PDZ3) from the mammalian neuronal protein PSD-95. Biochemistry. 2007;46:6340–6352. doi: 10.1021/bi062088k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saro D, Klosi E, Paredes A, Spaller MR. Thermodynamic analysis of a hydrophobic binding site: probing the PDZ domain with nonproteinogenic peptide ligands. Org. Lett. 2004;30:3429–3432. doi: 10.1021/ol049181q. [DOI] [PubMed] [Google Scholar]
- 15.Piserchio A, Salinas GD, Li T, Marshall J, Spaller MR, Mierke DF. Targeting specific PDZ domains of PSD-95, structural basis for enhanced affinity and enzymatic stability of a cyclic peptide. Chem. Biol. 2004;11:469–473. doi: 10.1016/j.chembiol.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 16.Li T, Saro D, Spaller MR. Thermodynamic profiling of conformationally constrained cyclic ligands for the PDZ domain. Bioorg. Med. Chem. Lett. 2004;14:1385–1388. doi: 10.1016/j.bmcl.2003.09.103. [DOI] [PubMed] [Google Scholar]
- 17.Klosi E, Saro D, Spaller MR. Bivalent peptides as PDZ domain ligands. Bioorg. Med. Chem. Lett. 2007;17:6147–6150. doi: 10.1016/j.bmcl.2007.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Udugamasooriya DG, Sharma SC, Spaller MR. A chemical library approach to organic-modified peptide ligands for PDZ domain proteins: a synthetic, thermodynamic and structural investigation. Chembiochem. 2008;9:1587–1589. doi: 10.1002/cbic.200800126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toth I, Griffith IP, Del Olmo Fernandez E, Hafeez RA, Holley JL, Ward P, Gibbons WA. Lipidic peptides. VIII. Cellular uptake studies of lipidic amino acid, its oligomers and highly lipophilic drug conjugates. Int. J. Pharm. 1992;79:39–45. [Google Scholar]
- 20.Khalil IA, Futaki S, Niwa M, Baba Y, Kaji N, Kamiya H, Harashima H. Mechanism of improved gene transfer by the N-terminal stearylation of octaarginine: enhanced cellular association by hydrophobic core formation. Gene Ther. 2004;11:636–644. doi: 10.1038/sj.gt.3302128. [DOI] [PubMed] [Google Scholar]
- 21.Gras-Masse H. Lipid vector for the delivery of peptides towards intracellular pharmacological targets. J. Mol. Recognit. 2003;16:234–239. doi: 10.1002/jmr.631. [DOI] [PubMed] [Google Scholar]
- 22.Reig F, Haro I, Polo D, Egea MA, Alsina MA. Interfacial interactions of hydrophobic peptides with lipid bilayers. J Colloid Interface Sci. 2002;246:60–69. doi: 10.1006/jcis.2001.8056. [DOI] [PubMed] [Google Scholar]
- 23.Kubelt J, Menon AK, Muller P, Herrmann A. Transbilayer movement of fluorescent phospholipid analogues in the cytoplasmic membrane of Escherichia coli. Biochemistry. 2002;41:5605–5612. doi: 10.1021/bi0118714. [DOI] [PubMed] [Google Scholar]
- 24.Losonczi JA, Tian F, Prestegard JH. Nuclear magnetic resonance studies of the N-terminal fragment of adenosine diphosphate ribosylation factor 1 in micelles and bicelles: influence of N-myristoylation. Biochemistry. 2000;39:3804–3816. doi: 10.1021/bi9923050. [DOI] [PubMed] [Google Scholar]
- 25.Victor K, Cafiso DS. Structure and position of the N-terminal membrane-binding domain of pp60src at the membrane interface. Biochemistry. 1998;37:3402–3410. doi: 10.1021/bi9721501. [DOI] [PubMed] [Google Scholar]
- 26.Ben-Tal N, Honig B, Peitzsch RM, Denisov G, McLaughlin S. Binding of small basic peptides to membranes containing acidic lipids: theoretical models and experimental results. Biophys J. 1996;71:561–575. doi: 10.1016/S0006-3495(96)79280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sankaram MB. Membrane interaction of small N-myristoylated peptides: implications for membrane anchoring and protein-protein association. Biophys J. 1994;67:105–112. doi: 10.1016/S0006-3495(94)80459-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peitzsch RM, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: Pertinence to myristoylated proteins. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- 29.Nelson AR, Borland L, Allbritton NL, Sims CE. Myristoyl-Based Transport of Peptides into Living Cells. Biochemistry. 2007;46:14771–14781. doi: 10.1021/bi701295k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patra CR, Bhattacharya R, Wang EF, Katarya A, Lau JS, Dutta S, Muders M, Wang SF, Buhrow SA, Safgren SL, Yaszemski MJ, Reid JM, Ames MM, Mukherjee P, Mukbopadhyay D. Targeted delivery of gemcitabine to pancreatic adenocarcinoma using cetuximab as a targeting agent. Cancer Res. 2008;68:1970–1978. doi: 10.1158/0008-5472.CAN-07-6102. [DOI] [PubMed] [Google Scholar]
- 31.Stephan S, Datta K, Wang EF, Li JP, Brekken RA, Parangi S, Thorpe PE, Mukhopadbyay D. Effect of rapamycin alone and in combination with antiangiogenesis therapy in an orthotopic model of human pancreatic cancer. Clin. Cancer Res. 2004;10:6993–7000. doi: 10.1158/1078-0432.CCR-04-0808. [DOI] [PubMed] [Google Scholar]
- 32.(a) Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449 doi: 10.1038/nature06188. 557-U4, [DOI] [PubMed] [Google Scholar]; (b) Urruticoechea A, Smith IE, Dowsett M. Proliferation marker Ki-67 in early breast cancer. J. Clin. Oncol. 2005;23:7212–7220. doi: 10.1200/JCO.2005.07.501. [DOI] [PubMed] [Google Scholar]
- 33.Manhani KK, Arcuri HA, da Silveira NJF, Uchoa HB, de Azevedo WF, Canduri F. Molecular models of protein kinase 6 from Plasmodium falciparum. J. Mol. Modeling. 2005;12:42–48. doi: 10.1007/s00894-005-0002-1. [DOI] [PubMed] [Google Scholar]
- 34.Krystof V, Cankar P, Frysova I, Slouka J, Kontopidis G, Dzubak P, Hajduch M, Srovnal J, de Azevedo WF, Orsag M, Paprskarova M, Rolcik J, Latr A, Fischer PM, Strnad M. 4-arylazo-3,5-diamino-1H-pyrazole CDK inhibitors: SAR study, crystal structure in complex with CDK2, selectivity, and cellular effects. J. Mol. Med. 2006;49:6500–6509. doi: 10.1021/jm0605740. [DOI] [PubMed] [Google Scholar]
- 35.Perez PC, Caceres RA, Canduri F, de Azevedo WF. modeling and dynamics simulation of human cyclin-dependent kinase 3 complexed with inhibitors. Comput. Biol. Med. 2009;39:130–140. doi: 10.1016/j.compbiomed.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Lu YX, Wang Y, Zhu WL. Nonbonding interactions of organic halogens in biological systems: implications for drug discovery and biomolecular design. Phys. Chem. Chem. Phys. 2010;12:4543–4551. doi: 10.1039/b926326h. [DOI] [PubMed] [Google Scholar]
- 37.Metrangolo P, Resnati G. Halogen Bonding: Fundamentals and Applications. Berlin: 2008. [Google Scholar]
- 38.Lima LMA, Barreiro EJ. Bioisosterism: A useful strategy for molecular modification and drug design. Curr. Med. Chem. 2005;12:23–49. doi: 10.2174/0929867053363540. [DOI] [PubMed] [Google Scholar]
- 39.Monforte AM, Logoteta P, Ferro S, De Luca L, Iraci N, Maga G, De Clercq E, Pannecouque C, Chimirri A. Design, synthesis, and structure-activity relationships of 1,3-dihydrobenzimidazol-2-one analogues as anti-HIV agents. Bioorg. Med. Chem. 2009;17:5962–5967. doi: 10.1016/j.bmc.2009.06.068. [DOI] [PubMed] [Google Scholar]
- 40.Greenbaum DC, Mackey Z, Hansell E, Doyle P, Gut J, Caffrey CR, Lehrman J, Rosenthal PJ, McKerrow JH, Chibale K. Synthesis and structure-activity relationships of parasiticidal thiosemicarbazone cysteine protease inhibitors against Plasmodium falciparum, Trypanosoma brucei, and Trypanosoma cruzi. J. Med. Chem. 2004;47:3212–3219. doi: 10.1021/jm030549j. [DOI] [PubMed] [Google Scholar]
- 41.Himmel DM, Das K, Clark AD, Hughes SH, Benjahad A, Oumouch S, Guillemont J, Coupa S, Poncelet A, Csoka I, Meyer C, Andries K, Nguyen CH, Grierson DS, Arnold E. Crystal structures for HIV-1 reverse transcriptase in complexes with three pyridinone derivatives: A new class of non-nucleoside inhibitors effective against a broad range of drug-resistant strains. J. Med. Chem. 2005;48:7582–7591. doi: 10.1021/jm0500323. [DOI] [PubMed] [Google Scholar]
- 42.Gerebtzoff G, Li-Blatter X, Fischer H, Frentzel A, Seelig A. Halogenation of drugs enhances membrane binding and permeation. Chembiochem. 2004;5:676–684. doi: 10.1002/cbic.200400017. [DOI] [PubMed] [Google Scholar]
- 43.Ohmori J, Maeno K, Hidaka K, Nakato K, Matsumoto M, Tada S, Hattori H, Sakamoto S, sukamoto S, Usuda S, Mase T. Dopamine D3 and D4 receptor antagonists: synthesis and structure-activity relationships of (S)-(+)-N-(1-benzyl-3-pyrrolidinyl)-5-chloro-4-[(cyclopropylcarbonyl)amino]-2-methoxybenzamide (YM-43611) J. Med. Chem. 1996;29:2764–2772. doi: 10.1021/jm9601720. [DOI] [PubMed] [Google Scholar]
- 44.Hidaka K, Tada S, Matsumoto M, Ohmori J, Maeno K, Yamaguchi T. YM-50001: A novel, potent and selective dopamine D4 receptor antagonist. Neuroreport. 1996;7:2543–2546. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.