Abstract
The need for novel antibiotics is greater now than perhaps any time since the pre-antibiotic era. Indeed, the recent collapse of most pharmaceutical antibacterial groups, combined with the emergence of hypervirulent and pan-antibiotic-resistant bacteria have, in effect, created a “perfect storm” that has severely compromised infection treatment options and led to dramatic increases in the incidence and severity of bacterial infections. Simply put, it is imperative that we develop new classes of antibiotics for the therapeutic intervention of bacterial infections. In that regard, RNA degradation is an essential biological process that has not been exploited for antibiotic development. Herein we discuss the factors that govern bacterial RNA degradation, highlight members of this machinery that represent attractive antimicrobial drug development targets and describe the use of high-throughput screening as a means of developing antimicrobials that target these enzymes. Such agents would represent first-in-class antibiotics that would be less apt to inactivation by currently encountered enzymatic antibiotic-resistance determinants.
Introduction
Infectious diseases are the second-leading cause of death worldwide1. Despite this, there has been a mass exodus of pharmaceutical antimicrobial discovery programs, leaving a void in the drug pipeline that, without intervention, will inevitably result in a healthcare crisis. Indeed, the Infectious Diseases Society of America recently warned of antibiotic-resistant ESKAPE bacterial pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) and the desperate need for new agents to treat these insidious organisms2. Most current antibiotics are derivatives of molecules discovered over 50 years ago and are losing their foothold as effective means of treating infections due to the emergence of antibiotic resistance3. Simply put, bacterial antibiotic resistance is outpacing new drug development making it imperative to expand antibiotic drug development to other essential cellular processes in order to create novel agents for the therapeutic intervention of current and emerging antibiotic-resistant bacteria4. RNA turnover is one such essential biological process that is rich in antimicrobial targets but has not been exploited for antibiotic drug discovery. Accordingly, this review is intended to bring to light the fundamental differences in RNA turnover between host and bacterial pathogen, distinguish those ribonucleases (RNases) that are attractive antibacterial targets, and provide methods to take advantage of these targets for drug development with the ultimate goal of expanding our antibiotic arsenal.
mRNA Turnover: Pathogen and Host
Many currently available antibiotics target essential pathways involved in cell wall synthesis, folate metabolism, protein translation, RNA transcription, or DNA replication3. These antibiotics are engineered to exert broad antimicrobial activity against an array of bacterial pathogens by targeting essential prokaryotic enzymes within the aforementioned pathways without causing off-target toxic effects toward human counterparts. In that regard, a simple comparison of the physiological characteristics of messenger RNA illustrates wholesale differences between the host and pathogen. For instance, bacteria couple transcription and translation and their mRNA is degraded rapidly (average half-life of ≤ 2.0 min), does not bear a 5’ 7-methylguanosine (m7G) cap, and is rarely 3’ polyadenylated. Mammalian cells diverge from their prokaryotic ancestors in that they compartmentalize their RNA metabolic steps and their mRNA has a longer half life (minutes to days), is 5’ m7G capped, and is polyadenylated at the 3’ terminus5. Therefore, it is not surprising that the molecular machinery that governs bacterial and eukaryotic mRNA degradation differs, and consequently these differences could be exploited for antibiotic drug discovery. As a prerequisite to this approach, one must first appreciate the basic similarities and differences in transcript turnover between the host and pathogen, the RNases involved, and their properties, which are briefly described below. For a more comprehensive report of RNA degradation in these two kingdoms, please refer to several recent excellent reviews6–11.
The major initiator of bacterial mRNA decay is considered to be a multi-protein complex termed the degradosome. This complex is best characterized in the Gram-negative model organism, Escherichia coli, and consists of at least four subunits: RNA helicase B (RhlB), enolase, polynucleotide phosphorylase (PNPase), and RNase E (Figure 1A)12. RNase E is the central component of the E. coli degradosome, establishing a scaffold for the assembly of other degradosome subunits and performing the initial endoribonucleolytic event during substrate mRNA decay12. RNase E preferentially cleaves 5’ monophosphorylated transcripts, thus the rate of mRNA decay is accelerated by the enzyme RppH, which converts the 5’ triphosphate group to 5’ monophosphate13. Resulting cleavage products are subsequently digested in a 3’→5’ fashion by the concerted activities of the degradosome-associated exoribonuclease PNPase and RhlB RNA helicase or by degradosome-independent 3’→5’ exoribonucleases, such as RNase II and RNase R12, 14–16. Other endoribonucleases also contribute to mRNA degradation, including RNase G, RNase I, and RNase III17–19. Most of these RNases cannot degrade to the single nucleotide, resulting in short RNA fragments that are further broken down by the enzyme Oligoribonuclease (Orn; 3’→5’ exoribonuclease)20. Additionally, the endoribonuclease RNase P is known to cleave mRNA transcripts that contain riboswitches and can cleave near stem-loop structures within E. coli mRNAs21–23. Resulting cleavage products contain a 5’ loop structure that acts to stabilize select transcripts23. RNase E, Orn, and RNase P are essential enzymes in the Gram-negative model organism E. coli, thus they may be good antibiotic drug discovery targets24–26.
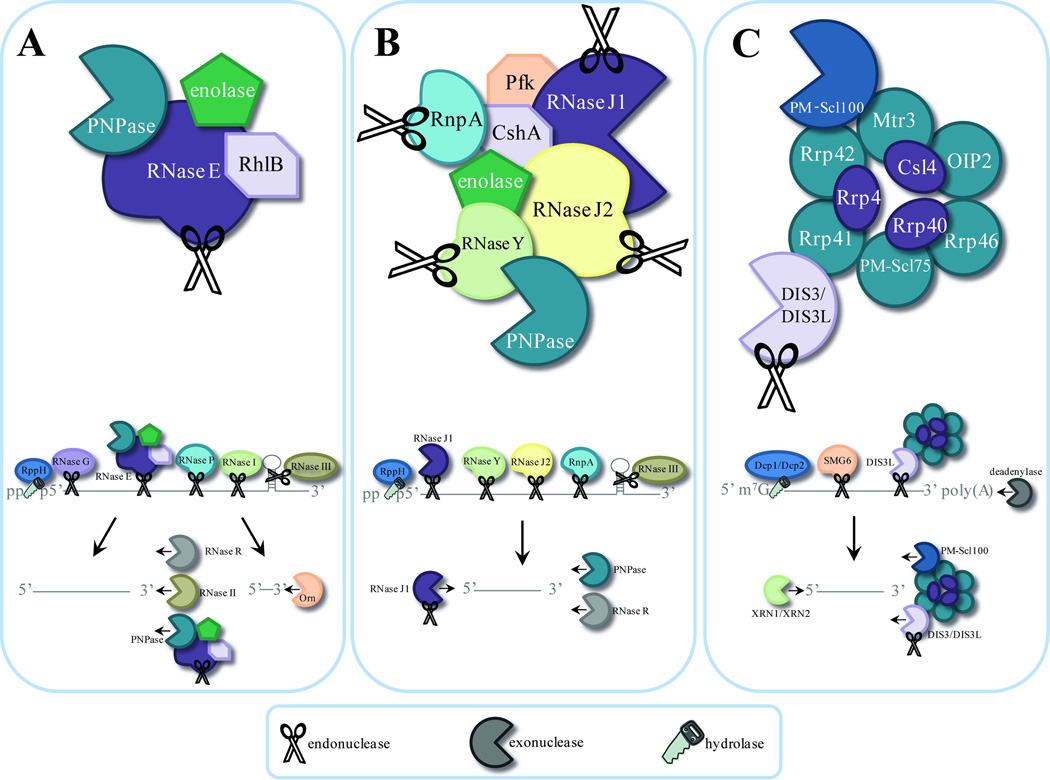
Figure 1. Mechanisms of mRNA decay.
A. Model of mRNA degradosome and degradation pathways in Escherichia coli. The E. coli degradosome includes RNA helicase B (RhlB), enolase, polynucleotide phosphorylase (PNPase), and RNase E. Initiation of mRNA decay occurs with the internal cleavage by RNase E. This cleavage favors 5’ monophosphorylated transcripts, which is achieved through the action of RppH. Degradosome-independent endoribonucleases RNase G, RNase P, and RNase I cleave single-stranded RNA (ssRNA), while RNase III recognizes and cleaves double-stranded RNA secondary structures (dsRNA). Resulting cleavage products are further digested by the degradosome PNPase or by the action of RNase R and RNase II in a 3’→5’ manner into fragments that are degraded into single nucleotides by the 3’→5’ endoribonuclease Orn. B. Proposed model of mRNA degradosome-like complex in Staphylococcus aureus. The Gram-positive degradosome-like complex includes RNase J1, RNase J2, RNase Y (also known as CvfA and YmdA), enolase, RNA helicase (CshA), PNPase, phosphofructokinase (Pfk), and RnpA. In Gram-positive bacteria, internal cleavage by RNase J1 initiates mRNA degradation. RNase J1 preferentially cleaves 5’ monophosphorylated mRNA molecules that have been stripped of pyrophosphate by RppH. Other members of the degradosome, including RNase Y, RNase J2, and RnpA, also cleave transcripts endonucleolytically. Secondary dsRNA structures are recognized and cleaved by the endoribonuclease RNase III. Resulting RNA pieces are then degraded in a 3’→5’ fashion by the degradosome member PNPase and the degradosome-independent RNase R. Fragments are then broken down in the 5’→3’ direction by RNase J1. C. The human exosome contains two structures: a ring structure composed of Rrp41, Rrp42, Mtr3, OIP2, Rrp46, and PM-Scl75, and a cap structure containing Rrp4, Rrp40, and Csl4. These exosome core components associate with the 3’→5’ exoribonucleases DIS3 and PM-Scl100 in the nucleus, or the dual endo- and 3’→5’ exo-ribonuclease DIS3L in the cytoplasm. Degradation of mRNA is initiated by removal of the poly(A) tail by deadenylase activity, decapping of the 5’ end by the Dcp1/Dcp2 complex, or by the endoribonucleolytic activity of the exosome-associated cytoplasmic DIS3L. The resulting mRNA is then vulnerable to 3’→5’ degradation by the exosome-associated PM-Scl100, DIS3, or DIS3L, and 5’→3’ decay by the exoribonucleases XRN2 (nucleus) or XRN1 (cytoplasm).
Researchers have long speculated that mRNA degradation within Gram-positive bacteria is also mediated by an RNA degradosome. However, the absence of an RNase E ortholog has thwarted efforts to identify this complex. Only recently, studies have revealed that Bacillus subtilis and Staphylococcus aureus are indeed capable of forming degradosome-like complexes consisting of at least eight subunits, including RNase J1, RNase J2, RNase Y (also known as CvfA and YmdA), enolase, RNA helicase (CshA), PNPase, phosphofructokinase (Pfk), and the protein component of the ribonucleoprotein complex RNase P, RnpA (Figure 1B)27, 28. Current studies have begun to unravel the mechanism(s) by which components of the Gram-positive degradosome subunits contribute to mRNA decay. Those studies have predominantly focused on understanding RNA degradation in the Gram-positive model organism, B. subtilis, where RNase J1, which exhibits endo- and 5’→3’ exoribonuclease activities, is likely to initiate degradation. Internal cleavage is thought to be initiated by RNase J1 and resulting fragments are subsequently exonucleolytically digested by RNase J1 in the 5’→3’ direction in concert with PNPase and CshA in the 3’→5’ direction27–29. Like RNase E, RNase J1 preferentially cleaves 5’ monophosphorylated mRNA, and an RppH equivalent (bsRpph or YtkD) has been identified in B. subtilis30. Another essential component of the B. subtilis degradosome-like complex is RNase Y, an endoribonuclease that contributes to bulk RNA degradation and has also been hypothesized to be the functional equivalent to the E. coli RNase E31, 32. B. subtilis transcripts are also degraded by a combination of other endoribonucleases, such as RNase J2, and the degradosome-independent RNase III33. Additionally, RNase P which is a ribonucleoprotein complex composed of an RNA subunit (RnpB) and a protein subunit (RnpA) affects the mRNA turnover properties of specific B. subtilis transcripts21, 34, 35. Studies in S. aureus have revealed that the protein component of RNase P, RnpA, affects bulk cellular mRNA turnover, as RnpA-depleted cells show increased mRNA stability, suggesting that RnpA acts to globally destabilize transcripts, albeit through an unknown mechanism36. RnpA is an essential member of the degradosome-like complex in both B. subtilis and S. aureus28.
When comparing the Gram-positive B. subtilis and S. aureus mRNA degradosomes, the components are conserved, however the interactions between their subunits vary, and they have different physiological characteristics. For instance, B. subtilis RNase Y is an essential enzyme, but it is not required for S. aureus viability; conversely, RNase J2 is an essential S. aureus gene but allelic deletions in B. subtilis are not lethal37–39. Thus, while the overarching mechanisms by which these two Gram-positive bacteria degrade RNA molecules may be conserved, the subunits’ behaviors and properties are likely to differ. In that regard, given that the latter is a life-threatening human pathogen, the essential components of the S. aureus degradosome may serve as the more practical and effective targets for antibiotic development. S. aureus RNase J1, RNase J2, and RnpA, are essential members of the organism’s RNA degradation apparatus and may represent antibiotic targets.
Eukaryotic cells are highly specialized and compartmentalized, and as such, individual steps in the mRNA degradation pathway occur in distinct locations within the cell and are carried out by correspondingly unique combinations of RNases9, 10. Human mRNA is synthesized, 5’ m7G capped, and 3’ polyadenylated within the nucleus. Degradation of the mRNA molecule can occur within the nucleus or at any point during or after transport to the cytoplasm by endonucleolytic cleavage, 5’ decapping (decapping enzymes Dcp1/Dcp2), and/or removal of the 3’ poly(A) tail by a variety of unique deadenylases (PAN2–PAN3, CCR4–NOT and poly(A)-specific ribonuclease (PARN)), each of which have varying roles in mRNA deadenylation among eukaryotes10. These events in-turn create substrates that are susceptible to 5’→3’ exoribonucleases, such as XRN2 (nucleus) and XRN1 (cytoplasm), or decay in the 3’→5’ direction via the exosome (Figure 1C)10. The core of the exosome contains two distinct structures formed by nine subunits and is thought to be essential within humans40, 41. The RNA-binding cap structure (Rrp4, Rrp40, and Csl4) recognizes the mRNA substrate and passes it through the hexameric ring composed of PM-Scl75 (Rrp45), Rrp41, Rrp42, Mtr3, OIP2 (Rrp43), and Rrp4640, 42. Although the ring structure possesses conserved exoribonuclease domains (similar to bacterial PNPase and RNase PH), it has lost the ability to directly cleave RNA9. Instead, the exosome coordinates the assembly of 3’→5’ exoribonucleases PM-Scl100 (Rrp6) and DIS3 in the nucleus, or the 3’→5’ exo- and endo-ribonuclease DIS3L in the cytoplasm, and as a unit with these RNases, degrades mRNA41, 43–46. Although the exosome accounts for the majority of mRNA turnover, internal cleavage of the transcript can also occur via endoribonucleases (SMG6), ribozymes, and the RNA interference pathway, each of which produces substrates that are susceptible to 5’→3’ and 3’→5’ exoribonucleases6.
Exploiting Essential mRNA Turnover Machinery in Bacteria
As previously mentioned, ideal antibiotics exhibit broad-spectrum antimicrobial activity against an expansive repertoire of bacterial pathogens and obviously must not be toxic to the host. In that regard, even a superficial understanding of the bacterial and host mRNA turnover pathways illustrates that many of the essential RNases involved in bacterial mRNA decay act via endonucleolytic cleavage, while eukaryotic decay occurs predominantly in an exonucleolytic fashion. Thus, development of agents that inhibit essential bacterial endonucleases would prevent bacterial proliferation and would be less likely to affect human mRNA turnover processes. Accordingly, Table 1 provides a comparison of the known RNases for each of the ESKAPE bacterial pathogens. Separated by Gram-stain categorization, the table compares the amino acid conservation of each RNase across pathogens (percent identity listed in parentheses), its essentiality (if known), as well as the percent predicted amino acid identity of each ribonuclease to orthologous human enzymes. A survey of these data brings to light several observations. First, there is no RNase antibiotic-development target that is essential, highly conserved across each of the ESKAPE pathogens, and also lacks similarity to human enzymes. Second, subdividing the ESKAPE pathogens based on very granular evolutionary boundaries, such as cell wall composition (Gram-staining), provides several putative RNase therapeutic targets that are essential, have low similarity to human proteins, and are well conserved across Gram-negative (Table 1A) or Gram-positive (Table 1B) organisms. Thus, one could ostensibly develop antimicrobials targeting these RNases; such agents may not be broad spectrum in the strictest sense, rather they would likely be efficacious across pathogenic species belonging to a given Gram-stain-defined boundary. There may be advantages to this approach, as the concept of targeting a subset of bacteria has been predicted to more beneficial than “broad-spectrum” antibiotics. For instance, narrow-spectrum agents may spare the host’s normal bacterial flora and reduce selective pressure, thereby minimizing the development of resistance4. In that regard, antimicrobial agents that target essential RNases may provide a perfect blend of broadly exhibiting efficacy against a Gram-stain-specific set of bacterial pathogens and avoiding the side effects of truly “broad spectrum” agents. Table 1 indicates that three essential RNases are conserved across ESKAPE pathogens belonging to a given Gram-stain designation, each of which also exhibits limited or no sequence and/or functional conservation to members of the human RNA turnover machinery. These RNases may represent excellent antibiotic targets and include the Gram-negative RNase E, the Gram-positive RNase J1, and the protein component of RNase P, RnpA, found in both Gram types.
Table 1.
ESKAPE pathogen ribonucleases.
| A) Gram-negative* | ||||||
|---|---|---|---|---|---|---|
| Enzyme |
Escherichia coli24, 25, 79–90 |
Acinetobacter baumannii91a |
Klebsiella pneumoniae |
Pseudomonas aeruginosa91 |
Enterobacter cloacae |
Human |
| Oligoribonuclease | E | E (63%) | P (93%) | NE (67%) | P (96%) | REX2 (49%) |
| PNPase | NE | E (61%) | P (96%) | E (65%) | P (96%) | Exosome coreb (25–33%) |
| RNase I | NE | P (14%) | P (70%) | - | P (79%) | RNase T2 (25%) |
| RNase II | NE | - | P (90%) | - | P (80%) | Dis3 (26%) Dis3L (25%) |
| RNase III | NE | NE (58%) | P (97%) | E (57%) | P (98%) | Drosha (30%) RNase III (30%) |
| RNase BN/Z | NE | P (31%) | P (69%) | NEc (29%) | P (77%) | tRNase Z/ELAC1 (35%) |
| RNase D | NE | NE (25%) | P (79%) | NE (38%) | P (82%) | PM-Scl100/Rrp6 (26%) |
| RNase E | E | E (61%) | P (74%) | NE (63%) | P (77%) | No known ortholog |
| RNase G | NE | P (54%) | P (97%) | NE (58%) | P (96%) | No known ortholog |
| RNase HI | NE | Ed (49%) | P (91%) | E (62%) | P (92%) | RNase H1 (37%) |
| RNase HII | NE | NE (65%) | P (89%) | NE (71%) | P (91%) | RNase H2, subunit A (30%) |
| RNase P (RnpA) | E | P (39%) | P (97%) | NE (47%) | P (97%) | No known orthologe |
| RNase PH | NE | E (67%) | P (96%) | NE (72%) | P (96%) | Exosome coreb (25–28%) |
| RNase R | NE | NE (42%) | P (93%) | NE (53%) | P (93%) | Dis3 (29%) Dis3L (26%) |
| RNase T | NE | NE (54%) | P (86%) | E (63%) | P (89%) | No known ortholog |
| B) Gram-positive† | ||||||
| Enzyme |
Staphylococcus aureus38, 69 |
Enterococcus faecium92 |
Human |
|||
| PNPase | NE | P (63%) | Exosome coreb (25–33%) | |||
| RNase III | NE | P (53%) | Drosha (36%) RNase III (31%) |
|||
| RNase BN/Z | NE | P (53%) | tRNase Z/ELAC1 (38%) | |||
| RNase HI | NE | P (35%) | RNase H1 (35%) | |||
| RNase HII | NE | P (48%) | RNase H2, subunit A (26%) | |||
| RNase HIII | NE | P (41%) | RNase H2, subunit A (22%) | |||
| RNase J1 | E | Pf (63%) | No known ortholog | |||
| RNase J2 | E | NEf (35%) | No known ortholog | |||
| RNase M5 | NE | P (51%) | No known ortholog | |||
| RNase P (RnpA) | E | P (47%) | No known orthologe | |||
| RNase R | E | P (48%) | Dis3 (31%) Dis3L (26%) |
|||
| RNase Y | NE | P (63%) | No known ortholog | |||
Protein percent identity calculated in comparison to E. coli RNase
Protein percent identity calculated in comparison to S. aureus RNase
E = essential
NE = non-essential
P = present, essentiality unknown
Deletion studies performed in closely related Acinetobacter baylyi
Exosome core components Rrp41, Rrp42, Mtr3, OIP2/Rrp43, Rrp46, and PM-Scl75/Rrp45
Putative metal-dependent hydrolase
DnaQ
Human RNase P subunits Rpp14, Rpp20, Rpp21, Rpp25, Rpp29, Rpp30, Rpp38, Rpp40, Pop1, and Pop5 have no significant similarity to E. coli or S. aureus RnpA proteins
Metallo-beta-lactamase superfamily protein
As stated above, RNase E is thought to play a key role in mediating Gram-negative bacterial mRNA degradation. As the central component of the degradosome, it is essential for mRNA turnover, yet it is also required for rRNA and tRNA processing12, 47, 48. The enzyme is also well conserved across Gram-negative bacteria (Table 1A). Furthermore, when comparing E. coli RNase E to the human genome, there is no significant amino acid homology to human proteins. Thus a small molecule inhibitor of RNase E would presumably exhibit antimicrobial activity against a repertoire of Gram-negative bacterial pathogens with no predicted human toxicity.
Two Gram-positive RNases with considerable potential as antimicrobial development targets are RNase J1 and the protein component of RNase P (RnpA), both of which are essential components of the S. aureus degradosome-like complex (Table 1B). RNase J1 is hypothesized to be the functional analog to the E. coli RNase E37, which in addition to its endonucleolytic activity, is also a 5’→3’ exoribonuclease able to degrade down to single nucleotides49, 50. RNase J1 is similar to RNase J2 in sequence and activity, however the essentiality of RNase J2 varies among bacterial species, suggesting that RNase J1 is the better target for antimicrobial development. RNase P is a ubiquitous ribonucleoprotein whose composition differs between host and pathogen35. Bacterial RNase P is composed of one protein (RnpA) and one RNA subunit (RnpB), whereas human nuclear RNase P contains an RNA component and up to ten different protein subunits (Rpp14, Rpp20, Rpp21, Rpp25, Rpp29, Rpp30, Rpp38, Rpp40, Pop1, and Pop5) that do not share significant amino acid similarity to bacterial RnpA51. In bacteria, both RnpB and RnpA are essential and have historically been considered to work in concert to aid in tRNA maturation. As elaborated below, RnpA has also been shown to contribute to S. aureus cellular mRNA turnover, and small molecule inhibitors of this process have been shown to have considerable therapeutic potential as antimicrobials36. Due to sequence divergence, it is unlikely that those small molecules will exhibit efficacy against Gram-negative bacteria, however, RnpA is also essential in many Gram-negatives of considerable healthcare concern and could be considered a target for antibiotic development in those species as well (Table 1A).
Target-based Screening for RNase Inhibitors
Once an appropriate enzyme has been selected for target-based antimicrobial drug development, the protein is typically purified and an in vitro functional assay is developed to accurately measure the protein’s activity. This assay must be sensitive enough to detect a partial loss of enzyme activity, yet robust enough to be repeated thousands of times in the presence of individual members of chemical libraries. In addition, the assay should also be amenable to miniaturization and ideally be relatively simple in format so that it can be automated. Using such assays, literally thousands of compounds can rapidly be screened for their ability to limit the protein’s activity in a high-throughput manner. Resulting inhibitory agents would represent a starting point for antimicrobial development.
One of biggest challenges in designing a high-throughput assay is to ensure that the functional assay best recapitulates the protein’s essential cellular function. For example, a high-throughput screening campaign targeting “RNA elongation” failed to identify inhibitors of S. aureus or E. coli RNase activity, albeit the functional assay was not disclosed52. This failure was attributed to the predicted inadequacy in the structural diversity of the compound library, which consisted of 260,000–530,000 members52. However, a pilot screen using only the protein component of S. aureus RNase identified small molecule inhibitors of RnpA-mediated RNA degradation with tremendous antimicrobial therapeutic promise using a chemical library of only 29,066 compounds36. As proof of principle, one of those molecules, RNPA1000, exhibited antimicrobial activity against several pathogenic Gram-positive bacteria, limited human cytotoxicity, and prevented disease within an acute-lethal murine model of S. aureus infection36. Thus, in this example, it is probably not the number of screened compounds that determined success; rather it is likely contingent upon developing a functional assay that best simulates the protein’s essential cellular function.
In retrospect, this theme has been reiterated over and over again in target-based screens for antimicrobials and is arguably the predominant reason genomics has failed to deliver a single novel antibiotic. The late 1990’s marked the beginning of the so-called genomic era, when bacterial genomic sequences became available and were immediately mined for essential genes with predicted functions. Enzyme screening campaigns were frequently performed based on identifying inhibitors of a conveniently measurable predicted activity of a given target, such as ATP binding, without knowledge of whether the measured activity actually accounted for the enzyme’s essential cellular function. Consequently, assay inhibitors did not necessarily translate to molecules that exhibited antimicrobial efficacy. Only now do we fully appreciate that a complete understanding of given target’s cellular function should be a prerequisite to beginning a high-throughput screening campaign. As indicated above, much is known about bacterial cellular RNase functions, and with this information in hand, we argue that their essential functions can be easily assessed in a high-throughput manner.
The aforementioned S. aureus RnpA-mediated RNA degradation assay was performed using total or in vitro-transcribed bacterial RNA substrate molecules and incubating with RnpA in the presence of individual members of a small compound library. Enzyme inhibition was measured as the amount of intact RNA following the addition of RiboGreen, which fluoresces when bound to intact RNA species. A secondary gel-based RNA-degradation assay was used to distinguish bona fide RnpA inhibitors from high-throughput screening artifacts36. This technique has been used successfully for S. aureus RnpA, and we believe that a similar assay design has the potential to identify inhibitors of other essential endoribonucleases, such as Gram-positive RNase J1, Gram-negative RNase E, and other bacterial RnpA proteins. The lead compounds uncovered by these screens can be further investigated for their bacteriostatic or bactericidal activity, inhibitor specificity, potency, and human cytotoxicity. This information can then be used by medicinal chemists to create more effective and potent analogs for further drug development.
An Alternative Approach: Utilizing Pathogenic RNases for Drug Discovery
When considering targets for antibiotics, we have focused on those bacterial RNases that are essential for the organism’s survival; however, some investigators propose that blocking virulence factor function and/or inhibiting genes required for in vivo survival within the host can also prevent infection. In that regard, most virulence factors and/or regulatory cascades are specific to each bacterial species, thus such inhibitory agents would demonstrate very narrow-spectrum activity, with the advantages of preserving the native flora and potentially preventing resistance mechanisms from developing53. Many virulence factors have been identified in bacteria, but only recently has it been appreciated that RNases control their expression. Thus, we will introduce the reader to RNases that have been shown to affect bacterial pathogenesis and discuss their likelihood as possible alternative antimicrobial targets.
The 3’→5’ exoribonuclease RNase R was first characterized as vacB (virulence associated locus B) in E. coli and Shigella flexneri, and mutants demonstrated decreased virulence factor expression, epithelial cell invasion, and hemolytic activity54, 55. Further studies in Aeromonas hydrophila showed that RNase R mutants were significantly less virulent in a mouse model of infection56. Thus, the enzyme could be exploited as a target for reducing bacterial pathogenesis. However, it should be noted that the enzyme apparently does not contribute to the pathogenesis of all bacteria, as Brucella abortus RNase R mutant showed no difference in pathogenesis, bacterial burden, or viability in a mouse model of infection57. The functional variability of RNase R combined with its homology to members of the human RNA degradation machinery (Table 1), suggest that RNase R would not be an ideal target. Like RNase R, PNPase is a 3’→5’ exoribonuclease that is present in both Gram-positive and –negative bacterial species. E. coli PNPase contributes to the regulation of outer-membrane proteins that are important for virulence, and PNPase mutants in Yersinia species resulted in decreased cytotoxicity in cell culture, as well as decreased virulence in a mouse model of infection58–60. However, inactivation of S. enterica PNPase resulted in increased intracellular replication and invasion, and in S. pyogenes, PNPase appears to degrade virulence factor transcripts61, 62. Furthermore, both Gram-positive and –negative PNPase exhibit significant amino acid similarity to members of the human exosome core. Discrepancies in the contribution of PNPase to pathogenesis and its homology to human proteins limit enthusiasm for this RNase as an optimal target.
One of the known modulators of bacterial small regulatory RNA/mRNA complexes is RNase III, which cleaves non-coding RNA-bound transcripts63–65. Both Gram-positive and –negative bacteria encode RNase III, however its role in virulence has been limited to studies in S. aureus. In an RNase III-deletion mutant of S. aureus, secretion of virulence factors was inhibited, the supernatants of the mutant were less toxic to human cells, and the mutant was attenuated in a peritonitis mouse model of infection66. Despite showing promise as an anti-pathogenesis target, limited knowledge of RNase III function across many different bacterial species, combined with its amino acid similarity to human components of the RNA degradation machinery, allow us to conclude that targeting RNase III would be problematic.
The endonuclease RNase Y is a member of the Gram-positive degradosome and is essential in B. subtilis but not essential for S. aureus38, 39. Nonetheless, S. aureus RNase Y mutant strains are highly attenuated in a silkworm model of infection and demonstrate decreased hemolysin production67. Likewise, the Gram-positive pathogen Streptococcus pyogenes RNase Y enzyme is also non-essential and is involved in virulence factor expression, adaptation to nutrient stress, and contributes to the organism’s pathogenesis in several animal models of infection68, 69. Amino acid homology searches indicate that bacterial RNase Y does not exhibit significant sequence similarity to human proteins, and thus may be an attractive target for developing agents that prevent or reduce Gram-positive bacterial pathogenesis.
It is important to note that when considering non-essential RNases as targets for attenuating bacterial pathogenesis (or any other non-essential regulatory molecule for that matter) several additional considerations must be taken into account. First, different types of infections (i.e. abscess vs. endocarditis) are likely to involve unique subsets of virulence factors. Thus, the efficacy of agents that inhibit a particular virulence factor regulatory enzyme will likely vary widely, depending on infection type. Second, many non-essential RNases exhibit redundant activities, and as a result, high potential exists for resistance to develop, as one RNase may compensate for another RNase. Third, agents designed against virulence factors prevent pathogenesis but do not aid in eliminating the bacteria, thus these molecules would not be effective in immunocompromised patients. Fourth, implementing standard in vitro techniques to assess the agents’ potential, such as determining the minimum inhibitory concentration (MIC), would prove difficult and may complicate inhibitor optimization.
Successful RNase Drug Development: Promise for Antibacterials
Although the focus of this review is to target RNA degradation as a means for developing antimicrobials, this concept extends beyond the bacterial realm into successful application to antiviral and anticancer drug development. For instance, one current antiviral effort is to target the human immunodeficiency virus (HIV) reverse transcriptase (RT)-mediated RNase H activity70. During HIV replication, the RT-associated polymerase function synthesizes a DNA copy of the viral RNA genome, and RT-associated RNase H activity subsequently degrades the parental RNA copy. As reviewed by Tramontano and Di Santo, both functions are required for viral replication, however all known RT inhibitors selectively block the polymerase function70. As HIV mutates at a rapid frequency, the RNase H function has been an attractive target for novel anti-retroviral development70. In fact, a recent screen for inhibitors of RT-associated RNase H activity was performed, and several small molecules were found to bind and inhibit RNase H activity, demonstrating little to no cytotoxicity to human cells71.
The dysregulation of several RNases has been implicated in the development of cancer72. Of these, Angiogenin (Ang) RNase activity is essential for angiogenesis73. Several laboratories have successfully targeted Ang using a wide variety of agents including Ang-specific monocolonal antibiodies, antisense oligonucleotides, small inhibitor peptides, as well as small molecule inhibitors that block Ang RNase activity, and all have demonstrated antitumor activity in vitro and in vivo74–78. These examples of developing agents that block RNases outside of the prokaryotic kingdom show that inhibition is tangible and emphasize the need for advances in developing antibacterials along the same premise.
Conclusion
At the end of the day, the number of classes of antibacterial drugs must be expanded to include new targets in order to combat highly drug-resistant bacteria, most importantly the ESKAPE pathogens. Interfering with RNA-metabolizing processes has successfully produced many antibiotics (RNA polymerase—rifampicin; ribosome—macrolides, tetracyclines, and aminoglycosides; isoleucyl-tRNA synthetase—mupirocin). Yet one aspect of bacterial RNA physiology that has not yet been exploited for antimicrobial chemotherapy is the essential process of RNA turnover via RNases. In that regard, we propose that small molecule inhibitors of essential bacterial RNases with little homology to human proteins will sabotage cellular global mRNA homeostasis and in turn limit prokaryotic proliferation and/or pathogenesis. As proof of principle of this concept, the successful identification and therapeutic potential of inhibitors of S. aureus RnpA, suggest that the approach is with merit and that additional small molecule inhibitors of bacterial ribonucleases can be identified36. Indeed, we have shown that RnpA inhibitors exhibit “broad spectrum” antimicrobial activity toward Gram-positive pathogens of immediate healthcare concern, efficacy in animal models of infection and limit biofilm-associated bacteria to a level that meets or exceeds currently available antibiotics36. Accordingly, the intent of this review is to highlight additional putative RNase antimicrobial targets and provide strategies for their exploitation, and consequently, their development as agents that will be useful for the therapeutic intervention of bacterial infections.
Acknowledgements
P.M.D. and C.R. are supported by NIH/NIAID award 1R01 AI073780-01 and T.M.E. is supported, in part, by a University of Nebraska Medical Center Student Assistantship.
References
- 1.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spellberg B, Blaser M, Guidos RJ, Boucher HW, Bradley JS, Eisenstein BI, Gerding D, Lynfield R, Reller LB, Rex J, et al. Combating antimicrobial resistance: policy recommendations to save lives. Clin Infect Dis. 2011;52(Suppl 5):S397–S428. doi: 10.1093/cid/cir153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nathan C. Antibiotics at the crossroads. Nature. 2004;431:899–902. doi: 10.1038/431899a. [DOI] [PubMed] [Google Scholar]
- 5.Kushner SR. mRNA decay in prokaryotes and eukaryotes: different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- 6.Tomecki R, Dziembowski A. Novel endoribonucleases as central players in various pathways of eukaryotic RNA metabolism. RNA. 16:1692–1724. doi: 10.1261/rna.2237610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condon C. Maturation and degradation of RNA in bacteria. Curr Opin Microbiol. 2007;10:271–278. doi: 10.1016/j.mib.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 8.Anderson KL, Dunman PM. Messenger RNA Turnover Processes in Escherichia coli, Bacillus subtilis, and Emerging Studies in Staphylococcus aureus. Int J Microbiol. 2009;2009:525491. doi: 10.1155/2009/525491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 10.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 11.Morrison JM, Dunman PM. The modulation of Staphylococcus aureus mRNA turnover. Future Microbiol. 2011;6:1141–1150. doi: 10.2217/fmb.11.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carpousis AJ. The Escherichia coli RNA degradosome: structure, function and relationship in other ribonucleolytic multienzyme complexes. Biochem Soc Trans. 2002;30:150–155. [PubMed] [Google Scholar]
- 13.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 14.Carpousis AJ, Van Houwe G, Ehretsmann C, Krisch HM. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 15.Gupta RS, Kasai T, Schlessinger D. Purification and some novel properties of Escherichia coli RNase II. J Biol Chem. 1977;252:8945–8949. [PubMed] [Google Scholar]
- 16.Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 17.Ito R, Ohnishi Y. The roles of RNA polymerase and RNase I in stable RNA degradation in Escherichia coli carrying the srnB+ gene. Biochim Biophys Acta. 1983;739:27–34. doi: 10.1016/0167-4781(83)90040-4. [DOI] [PubMed] [Google Scholar]
- 18.Umitsuki G, Wachi M, Takada A, Hikichi T, Nagai K. Involvement of RNase G in in vivo mRNA metabolism in Escherichia coli. Genes Cells. 2001;6:403–410. doi: 10.1046/j.1365-2443.2001.00430.x. [DOI] [PubMed] [Google Scholar]
- 19.Drider D, Condon C. The continuing story of endoribonuclease III. J Mol Microbiol Biotechnol. 2004;8:195–200. doi: 10.1159/000086700. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Zhu L, Deutscher MP. Oligoribonuclease is encoded by a highly conserved gene in the 3′-5′ exonuclease superfamily. J Bacteriol. 1998;180:2779–2781. doi: 10.1128/jb.180.10.2779-2781.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altman S, Wesolowski D, Guerrier-Takada C, Li Y. RNase P cleaves transient structures in some riboswitches. Proc Natl Acad Sci U S A. 2005;102:11284–11289. doi: 10.1073/pnas.0505271102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Altman S. A specific endoribonuclease, RNase P, affects gene expression of polycistronic operon mRNAs. Proc Natl Acad Sci U S A. 2003;100:13213–13218. doi: 10.1073/pnas.2235589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alifano P, Rivellini F, Piscitelli C, Arraiano CM, Bruni CB, Carlomagno MS. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev. 1994;8:3021–3031. doi: 10.1101/gad.8.24.3021. [DOI] [PubMed] [Google Scholar]
- 24.Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci U S A. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghora BK, Apirion D. Structural analysis and in vitro processing to p5 rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli. Cell. 1978;15:1055–1066. doi: 10.1016/0092-8674(78)90289-1. [DOI] [PubMed] [Google Scholar]
- 26.Ow MC, Kushner SR. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev. 2002;16:1102–1115. doi: 10.1101/gad.983502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Commichau FM, Rothe FM, Herzberg C, Wagner E, Hellwig D, Lehnik-Habrink M, Hammer E, Volker U, Stulke J. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics. 2009;8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roux CM, Demuth JP, Dunman PM. Characterization of components of the Staphylococcus aureus messenger RNA degradosome holoenzyme-like complex. J Bacteriol. 2011;193:5520–5526. doi: 10.1128/JB.05485-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathy N, Hebert A, Mervelet P, Benard L, Dorleans A, Li de la Sierra-Gallay I, Noirot P, Putzer H, Condon C. Bacillus subtilis ribonucleases J1 and J2 form a complex with altered enzyme behaviour. Mol Microbiol. 75:489–498. doi: 10.1111/j.1365-2958.2009.07004.x. [DOI] [PubMed] [Google Scholar]
- 30.Richards J, Liu Q, Pellegrini O, Celesnik H, Yao S, Bechhofer DH, Condon C, Belasco JG. An RNA Pyrophosphohydrolase Triggers 5′-Exonucleolytic Degradation of mRNA in Bacillus subtilis. Mol Cell. 43:940–949. doi: 10.1016/j.molcel.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shahbabian K, Jamalli A, Zig L, Putzer H. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 2009;28:3523–3533. doi: 10.1038/emboj.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehnik-Habrink M, Newman J, Rothe FM, Solovyova AS, Rodrigues C, Herzberg C, Commichau FM, Lewis RJ, Stulke J. RNase Y in Bacillus subtilis: a Natively Disordered Protein That Is the Functional Equivalent of RNase E from Escherichia coli. J Bacteriol. 2011;193:5431–5441. doi: 10.1128/JB.05500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Condon C, Bechhofer DH. Regulated RNA stability in the Gram positives. Curr Opin Microbiol. 2011;14:148–154. doi: 10.1016/j.mib.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seif E, Altman S. RNase P cleaves the adenine riboswitch and stabilizes pbuE mRNA in Bacillus subtilis. RNA. 2008;14:1237–1243. doi: 10.1261/rna.833408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Condon C, Putzer H. The phylogenetic distribution of bacterial ribonucleases. Nucleic Acids Res. 2002;30:5339–5346. doi: 10.1093/nar/gkf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olson PD, Kuechenmeister LJ, Anderson KL, Daily S, Beenken KE, Roux CM, Reniere ML, Lewis TL, Weiss WJ, Pulse M, et al. Small molecule inhibitors of Staphylococcus aureus RnpA alter cellular mRNA turnover, exhibit antimicrobial activity, and attenuate pathogenesis. PLoS Pathog. 2011;7:e1001287. doi: 10.1371/journal.ppat.1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Even S, Pellegrini O, Zig L, Labas V, Vinh J, Brechemmier-Baey D, Putzer H. Ribonucleases J1 and J2: two novel endoribonucleases in B.subtilis with functional homology to E.coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, et al. Comprehensive identification of essential Staphylococcus aureus genes using Transposon-Mediated Differential Hybridisation (TMDH) BMC Genomics. 2009;10:291. doi: 10.1186/1471-2164-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci U S A. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Dijk EL, Schilders G, Pruijn GJ. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Allmang C, Petfalski E, Podtelejnikov A, Mann M, Tollervey D, Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ --> 5′ exonucleases. Genes Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raijmakers R, Egberts WV, van Venrooij WJ, Pruijn GJ. Protein-protein interactions between human exosome components support the assembly of RNase PH-type subunits into a six-membered PNPase-like ring. J Mol Biol. 2002;323:653–663. doi: 10.1016/s0022-2836(02)00947-6. [DOI] [PubMed] [Google Scholar]
- 43.Midtgaard SF, Assenholt J, Jonstrup AT, Van LB, Jensen TH, Brodersen DE. Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain. Proc Natl Acad Sci U S A. 2006;103:11898–11903. doi: 10.1073/pnas.0604731103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJ, Raijmakers R, Pruijn GJ. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J. 29:2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burkard KT, Butler JS. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol. 2000;20:604–616. doi: 10.1128/mcb.20.2.604-616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H. Complexes of tRNA and maturation enzymes: shaping up for translation. Curr Opin Struct Biol. 2007;17:293–301. doi: 10.1016/j.sbi.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 49.Mathy N, Benard L, Pellegrini O, Daou R, Wen T, Condon C. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. [DOI] [PubMed] [Google Scholar]
- 50.Fang M, Zeisberg WM, Condon C, Ogryzko V, Danchin A, Mechold U. Degradation of nanoRNA is performed by multiple redundant RNases in Bacillus subtilis. Nucleic Acids Res. 2009;37:5114–5125. doi: 10.1093/nar/gkp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jarrous N, Reiner R. Human RNase P: a tRNA-processing enzyme and transcription factor. Nucleic Acids Res. 2007;35:3519–3524. doi: 10.1093/nar/gkm071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 53.Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541–548. doi: 10.1038/nchembio.2007.24. [DOI] [PubMed] [Google Scholar]
- 54.Tobe T, Sasakawa C, Okada N, Honma Y, Yoshikawa M. vacB, a novel chromosomal gene required for expression of virulence genes on the large plasmid of Shigella flexneri. J Bacteriol. 1992;174:6359–6367. doi: 10.1128/jb.174.20.6359-6367.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheng ZF, Zuo Y, Li Z, Rudd KE, Deutscher MP. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J Biol Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- 56.Erova TE, Kosykh VG, Fadl AA, Sha J, Horneman AJ, Chopra AK. Cold shock exoribonuclease R (VacB) is involved in Aeromonas hydrophila pathogenesis. J Bacteriol. 2008;190:3467–3474. doi: 10.1128/JB.00075-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyoshi A, Rosinha GM, Camargo IL, Trant CM, Cardoso FC, Azevedo V, Oliveira SC. The role of the vacB gene in the pathogenesis of Brucella abortus. Microbes Infect. 2007;9:375–381. doi: 10.1016/j.micinf.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 58.Andrade JM, Arraiano CM. PNPase is a key player in the regulation of small RNAs that control the expression of outer membrane proteins. RNA. 2008;14:543–551. doi: 10.1261/rna.683308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosenzweig JA, Weltman G, Plano GV, Schesser K. Modulation of yersinia type three secretion system by the S1 domain of polynucleotide phosphorylase. J Biol Chem. 2005;280:156–163. doi: 10.1074/jbc.M405662200. [DOI] [PubMed] [Google Scholar]
- 60.Rosenzweig JA, Chromy B, Echeverry A, Yang J, Adkins B, Plano GV, McCutchen-Maloney S, Schesser K. Polynucleotide phosphorylase independently controls virulence factor expression levels and export in Yersinia spp. FEMS Microbiol Lett. 2007;270:255–264. doi: 10.1111/j.1574-6968.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- 61.Clements MO, Eriksson S, Thompson A, Lucchini S, Hinton JC, Normark S, Rhen M. Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc Natl Acad Sci U S A. 2002;99:8784–8789. doi: 10.1073/pnas.132047099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barnett TC, Bugrysheva JV, Scott JR. Role of mRNA stability in growth phase regulation of gene expression in the group A streptococcus. J Bacteriol. 2007;189:1866–1873. doi: 10.1128/JB.01658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagner EG, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- 64.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y, Dong J, Wu N, Gao Y, Zhang X, Mu C, Shao N, Fan M, Yang G. The production of extracellular proteins is regulated by ribonuclease III via two different pathways in Staphylococcus aureus. PLoS One. 6:e20554. doi: 10.1371/journal.pone.0020554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nagata M, Kaito C, Sekimizu K. Phosphodiesterase activity of CvfA is required for virulence in Staphylococcus aureus. J Biol Chem. 2008;283:2176–2184. doi: 10.1074/jbc.M705309200. [DOI] [PubMed] [Google Scholar]
- 68.Kang SO, Caparon MG, Cho KH. Virulence gene regulation by CvfA, a putative RNase: the CvfA-enolase complex in Streptococcus pyogenes links nutritional stress, growth-phase control, and virulence gene expression. Infect Immun. 2010;78:2754–2767. doi: 10.1128/IAI.01370-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kaito C, Kurokawa K, Matsumoto Y, Terao Y, Kawabata S, Hamada S, Sekimizu K. Silkworm pathogenic bacteria infection model for identification of novel virulence genes. Mol Microbiol. 2005;56:934–944. doi: 10.1111/j.1365-2958.2005.04596.x. [DOI] [PubMed] [Google Scholar]
- 70.Tramontano E, Di Santo R. HIV-1 RT-associated RNase H function inhibitors: Recent advances in drug development. Curr Med Chem. 17:2837–2853. doi: 10.2174/092986710792065045. [DOI] [PubMed] [Google Scholar]
- 71.Yanagita H, Urano E, Matsumoto K, Ichikawa R, Takaesu Y, Ogata M, Murakami T, Wu H, Chiba J, Komano J, Hoshino T. Structural and biochemical study on the inhibitory activity of derivatives of 5-nitro-furan-2-carboxylic acid for RNase H function of HIV-1 reverse transcriptase. Bioorg Med Chem. 19:816–825. doi: 10.1016/j.bmc.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 72.Kim WC, Lee CH. The role of mammalian ribonucleases (RNases) in cancer. Biochim Biophys Acta. 2009;1796:99–113. doi: 10.1016/j.bbcan.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 73.Curran TP, Shapiro R, Riordan JF. Alteration of the enzymatic specificity of human angiogenin by site-directed mutagenesis. Biochemistry. 1993;32:2307–2313. doi: 10.1021/bi00060a023. [DOI] [PubMed] [Google Scholar]
- 74.Olson KA, Fett JW, French TC, Key ME, Vallee BL. Angiogenin antagonists prevent tumor growth in vivo. Proc Natl Acad Sci U S A. 1995;92:442–446. doi: 10.1073/pnas.92.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olson KA, Byers HR, Key ME, Fett JW. Inhibition of prostate carcinoma establishment and metastatic growth in mice by an antiangiogenin monoclonal antibody. Int J Cancer. 2002;98:923–929. doi: 10.1002/ijc.10282. [DOI] [PubMed] [Google Scholar]
- 76.Olson KA, Byers HR, Key ME, Fett JW. Prevention of human prostate tumor metastasis in athymic mice by antisense targeting of human angiogenin. Clin Cancer Res. 2001;7:3598–3605. [PubMed] [Google Scholar]
- 77.Gho YS, Yoon WH, Chae CB. Antiplasmin activity of a peptide that binds to the receptor-binding site of angiogenin. J Biol Chem. 2002;277:9690–9694. doi: 10.1074/jbc.M105526200. [DOI] [PubMed] [Google Scholar]
- 78.Kao RY, Jenkins JL, Olson KA, Key ME, Fett JW, Shapiro R. A small-molecule inhibitor of the ribonucleolytic activity of human angiogenin that possesses antitumor activity. Proc Natl Acad Sci U S A. 2002;99:10066–10071. doi: 10.1073/pnas.152342999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gesteland RF. Isolation and characterization of ribonuclease I mutants of Escherichia coli. J Mol Biol. 1966;16:67–84. doi: 10.1016/s0022-2836(66)80263-2. [DOI] [PubMed] [Google Scholar]
- 80.Apirion D, Watson N. Mapping and characterization of a mutation in Escherichia coli that reduces the level of ribonuclease III specific for double-stranded ribonucleic acid. J Bacteriol. 1975;124:317–324. doi: 10.1128/jb.124.1.317-324.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee K, Bernstein JA, Cohen SN. RNase G complementation of rne null mutation identifies functional interrelationships with RNase E in Escherichia coli. Mol Microbiol. 2002;43:1445–1456. doi: 10.1046/j.1365-2958.2002.02848.x. [DOI] [PubMed] [Google Scholar]
- 82.Horiuchi T, Maki H, Sekiguchi M. RNase H-defective mutants of Escherichia coli: a possible discriminatory role of RNase H in initiation of DNA replication. Mol Gen Genet. 1984;195:17–22. doi: 10.1007/BF00332717. [DOI] [PubMed] [Google Scholar]
- 83.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schedl P, Primakoff P. Mutants of Escherichia coli thermosensitive for the synthesis of transfer RNA. Proc Natl Acad Sci U S A. 1973;70:2091–2095. doi: 10.1073/pnas.70.7.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Asha PK, Deutscher MP. Escherichia coli CAN lacks a tRNA-processing nuclease. J Bacteriol. 1983;156:419–420. doi: 10.1128/jb.156.1.419-420.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reiner AM. Isolation and mapping of polynucleotide phosphorylase mutants of Escherichia coli. J Bacteriol. 1969;97:1431–1436. doi: 10.1128/jb.97.3.1431-1436.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nikolaev N, Folsom V, Schlessinger D. Escherichia coli mutants deficient in exoribonucleases. Biochem Biophys Res Commun. 1976;70:920–924. doi: 10.1016/0006-291x(76)90679-3. [DOI] [PubMed] [Google Scholar]
- 88.Blouin RT, Zaniewski R, Deutscher MP. Ribonuclease D is not essential for the normal growth of Escherichia coli or bacteriophage T4 or for the biosynthesis of a T4 suppressor tRNA. J Biol Chem. 1983;258:1423–1426. [PubMed] [Google Scholar]
- 89.Craven MG, Henner DJ, Alessi D, Schauer AT, Ost KA, Deutscher MP, Friedman DI. Identification of the rph (RNase PH) gene of Bacillus subtilis: evidence for suppression of cold-sensitive mutations in Escherichia coli. J Bacteriol. 1992;174:4727–4735. doi: 10.1128/jb.174.14.4727-4735.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Padmanabha KP, Deutscher MP. RNase T affects Escherichia coli growth and recovery from metabolic stress. J Bacteriol. 1991;173:1376–1381. doi: 10.1128/jb.173.4.1376-1381.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Berardinis V, Vallenet D, Castelli V, Besnard M, Pinet A, Cruaud C, Samair S, Lechaplais C, Gyapay G, Richez C, et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gao P, Pinkston KL, Nallapareddy SR, van Hoof A, Murray BE, Harvey BR. Enterococcus faecalis rnjB is required for pilin gene expression and biofilm formation. J Bacteriol. 2010;192:5489–5498. doi: 10.1128/JB.00725-10. [DOI] [PMC free article] [PubMed] [Google Scholar]