SUMMARY
Regulation of self-renewal and differentiation of neural stem cells is still poorly understood. Here we investigate the role of a developmentally expressed protein, Botch, which blocks Notch, in neocortical development. Downregulation of Botch in vivo leads to cellular retention in the ventricular and subventricular zones, whereas overexpression of Botch drives neural stem cells into the intermediate zone and cortical plate. In vitro neurosphere and differentiation assays indicate that Botch regulates neurogenesis by promoting neuronal differentiation. Botch prevents cell surface presentation of Notch by inhibiting the S1 furin-like cleavage of Notch, maintaining Notch in the immature full-length form. Understanding the function of Botch expands our knowledge regarding both the regulation of Notch signaling and the complex signaling mediating neuronal development.
INTRODUCTION
Among the proteins and signal cascades that participate in building the complex architecture of the brain from neural progenitors cells (Ayala et al., 2007; Doe, 2008) Notch signaling is prominent (Kopan and Ilagan, 2009; Louvi and Artavanis-Tsakonas, 2006). Notch maintains cells in the neuronal progenitor fates by inhibiting neuronal differentiation and promoting gliogenesis. Notch is a highly evolutionary conserved signaling pathway controlling cell fate decisions, differentiation, proliferation, and apoptosis both during development and in adult tissues (Artavanis-Tsakonas et al., 1999; Kopan and Ilagan, 2009). Due to the emerging role of Notch signaling in health and disease, understanding the regulation and actions of the Notch pathway are developing clinical interest and importance.
Although regulating functionally diverse physiologic outcomes, the canonical core Notch pathway remains constant (Selkoe and Kopan, 2003). In mammals the Notch pathway consists of Notch 1-4, Delta-like ligands (Dll) 1, 3 and 4 and Jagged ligands (J1, J2). Immature Notch is processed by cleavage by a furin-type protease to form a mature heterodimeric receptor. Notch is localized to the surface of cells where it interacts with its ligands, Dll 1, 3, 4, J1 and J2. On binding of ligands, Notch is rendered susceptible to a series of proteolytic steps, first by the ADAM family metalloproteases then an intramembrance cleavage by the γ-secretase complex (Ilagan and Kopan, 2007). These ligand-dependent cleavage events, eventually promotes the release of the Notch intracellular domain (NICD) from the plasma membrane. NICD translocates to the nucleus, where it converts the C-promoter binding factor-1 (CBF-1) complex from a transcriptional repressor to a transcriptional activator resulting in expression of Notch target genes. This represents the ‘canonical’ or core signaling pathway. However, recent work is defining additional, non-canonical, points of regulation of Notch activity.
Many developmental processes are extremely sensitive to the dosage of Notch signaling (Donoviel et al., 1999; Duarte et al., 2004; Gale et al., 2004; Krebs et al., 2004; McCright et al., 2002). Thus, Notch signaling pathways must be precisely modulated and regulated in order for Notch to choreograph the complex events of development. Here we describe and characterize, neuroprotective gene 7 (NPG7), which we rename Botch (Blocks Notch), that promotes neurogenesis by downregulating the Notch signaling pathway. The mechanism by which Botch inhibits Notch1 is through interfering with its processing and trafficking.
RESULTS
Botch (NPG7) is developmentally expressed
NPG7 (EF688602), Botch was identified in functional screen for neuroprotective proteins (Dai et al., 2010). Botch is a protein of unknown function. Sequence analysis reveals Botch homologues in drosophila, C. elegans, chickens, rodents and man (Figure S1). It is a 24.5 kDa protein of 223 amino acids in mouse (NM_026929) and human (NM_024111) and 222 amino acids in rat (NM_001173437). Botch has no known mammalian protein domains and no closely related mammalian homolog. Botch is distantly related to ChaC, a protein that is thought to be associated with the putative ChaA Ca2+/H+ cation transport protein in Escherichia coli. The gene is currently annotated as ChAC1, although there is no evidence for such a function in mammalian cells. It is induced by the unfolded protein response and is thought to play a role in cell survival (Mungrue et al., 2009).
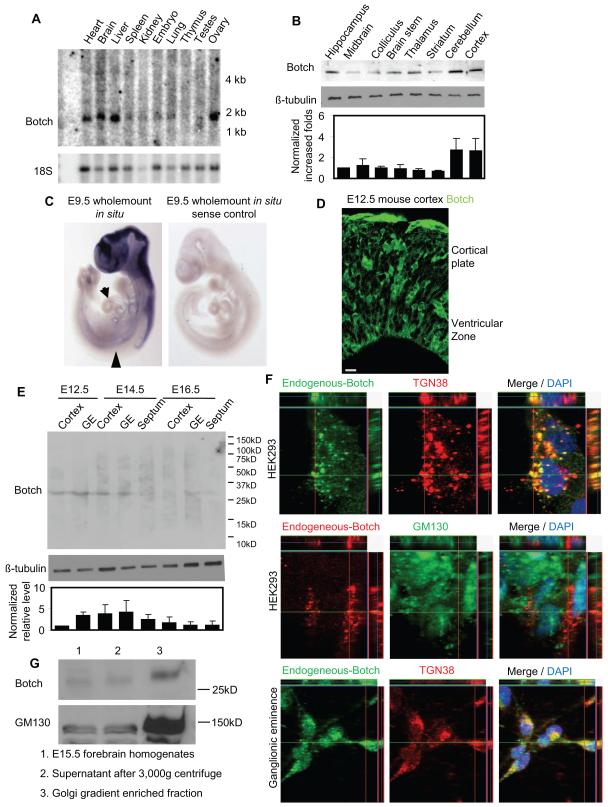
By Northern blot analysis Botch message is about 1.8 kb in mouse (Figure 1A) and it is widely expressed in multiple organs including the brain (Figure 1A). A monoclonal antibody was raised to Botch (NeuroMab, Davis CA) that recognizes a single band on immunoblot, the signal of which is reduced following shRNA mediated knockdown of Botch (Figure S2A, S2B, S2D, and S2E) indicating that the antibody is specific for Botch. By immunoblot analysis Botch is heterogeneously distributed throughout the adult mouse brain (Figure 1B). Whole mount in situ hybridization at E9.5 reveals Botch is highly expressed in mouse forebrain and anterior spinal cord and moderately expressed in the dorsal aorta and heart (Figure 1C). Endogenous Botch is detected in E12.5 mouse forebrain cortex by immunohistochemistry (Figure 1D). Expression of Botch message in adult brain by in situ hybridization is much lower than expression during development therefore immunoblot analysis was performed at different embryonic stages (E12.5, E14.5, E16.5) and the relative expression levels of Botch was compared in the cortex, ganglionic eminence and septum (Figure 1E). E14.5 cortex expresses the high levels of Botch and Botch is also expressed at relatively high levels in the germinal zones including the ganglionic eminence and septum at E14.5. Levels in these areas decrease at E16.5. High-resolution Z-stack confocal immunohistochemistry of cultured HEK293 cells or primary neuronal precursor cultures from E14.5 ganglionic eminence indicates Botch co-localizes with the trans-Golgi marker (TGN38) but not the cis-Golgi marker (GM130), indicating that in the Golgi, Botch resides in the trans-Golgi (Figure 1F). Botch doesn’t co-localize with the endocytic markers Caveolin-1, Clathrin, EEA1 or Rab5 (data not shown). Subcellular fractionation by sucrose gradient for the Golgi fractionation shows Botch enrichment in the Golgi fraction similar to the Golgi marker, GM130 (Figure 1G). We cannot exclude the possibility that Botch may be localized to other subcellular cytoplasmic compartments, but these results indicate that Botch is localized to the trans-Golgi where it could mediate it biological actions.
Figure 1. Botch is developmentally expressed and localized to the Golgi.
(A) Northern analysis of Botch in adult mouse tissues.
(B) Immunoblot analysis of mouse brain with the Botch monoclonal antibody. Optical densitometry measurements were normalized to β-tubulin (bottom panel).
(C) Whole mount Botch in situ hybridization of E9.5 mouse. Arrow: dorsal aorta; arrowhead: heart.
(D) Immunostaining with anti-Botch antibody for endogenous Botch in E12.5 mouse forebrain cortex. Scale bar: 10 μM.
(E) Immunoblot analysis of regional mouse brain tissue at E12.5, E14.5, and E16.5 with the Botch monoclonal antibody. Optical densitometry measurements were normalized to β-tubulin (bottom panel).
(F) Immunohistochemical subcellular localization of Botch with the Botch monoclonal antibody compared to the trans-Golgi marker (TGN38) and the cis-Golgi marker (GM130) in HEK293 or primary ganglionic eminence cultures. Co-localization is represented by yellow in the Merge panels with Z-stack. (C-G) All experiments were repeated 3 times.
(G) Immunoblot analysis of sucrose gradient Golgi fractions from E15.5 mouse forebrains. GM130 was used as a Golgi marker as sucrose gradient enrichment doesn’t separate cis-Golgi from trans-Golgi.
Botch regulates embryonic neurogenesis in vivo
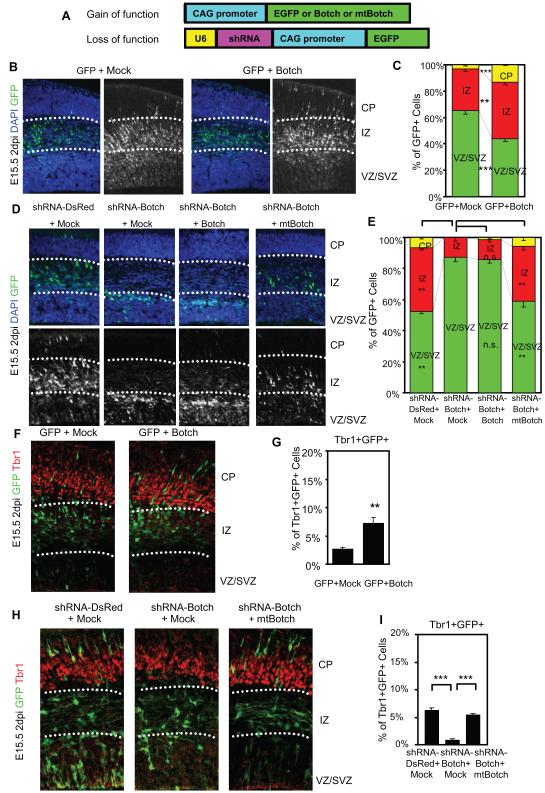
Since Botch is expressed at high levels during development and is enriched in the germinal zone of the forebrain, gain and loss of function studies were performed to explore the role of Botch on neocortical development. In utero co-electroporation of pCAG-EGFP and pCAG-Botch into E13.5 CD1 mouse brains was performed (Figure 2A-C). Embryos were harvested at E15.5 and immunostained for GFP to identify Botch overexpressing cells and counterstained with DAPI to identify all cells. Botch overexpression resulted in fewer GFP+ cells in the ventricular (VZ) and subventricular zones (SVZ) and more cells in the cortical plate (CP) and intermediate zone (IZ) when compared to co-electroporation of pCAG-EGFP with Mock control (pCAG empty vector) (Figure 2A-C), suggesting that Botch could promote neurogenesis.
Figure 2. Botch regulates embryonic neurogenesis in vivo.
(A) A schematic diagram of pCAG constructs for overexpression (gain of function) or knockdown (loss of function) for in utero injection and electroporation.
(B-I): Distribution of GFP+ cells 2 days after in utero injection and electroporation. (B) Representative confocal images of cortex immunostained for GFP with and without the DAPI channel with Botch expression. Abbreviations: CP, cortical plate; IZ, intermediate zone; VZ, ventricular zone; SVZ, subvetricular zone.
(C) Quantification of distribution of GFP+ cells in (B). Values represent the mean ± SEM (n = 4; ** p < 0.01; *** p < 0.001, Student’s t-test).
(D) Representative confocal images of cortex immunostained for GFP with and without the DAPI channel following knockdown of Botch and rescue with mtBotch.
(E) Quantification of distribution of GFP+ cells in (D). Values represent the mean ± SEM (n ≥ 3; ** p < 0.01; n.s. p > 0.05; one-way ANOVA, post-test: Tukey’s multiple comparison test).
(F) Representative confocal images of GFP+ (green) and Tbr1+ (red) in cortex following Botch expression. The box represents the source of the high power image (Magnification 45 X).
(G) Quantification of GFP+Tbr1+ cells in (F). Values represent the mean ± SEM (n = 4, ** p < 0.01, Student’s t-test).
(H) Representative confocal images of GFP+ (green) and Tbr1+ (red) double positive cells in cortex with the Botch knockdown and rescue. The box represents the source of the high power image (Magnification 45 X).
(I) Quantification of GFP+Tbr1+ cells in (H). Values represent the mean ± SEM (n ≥ 3; *** p < 0.001; Student’s t-test).
To explore loss of function a shRNA was designed to knockdown expression of Botch. Expression constructs were generated from a bicistronic construct pCAG-EGFP (Hitoshi et al., 1991) with U6 driving shRNA-Botch or shRNA-DsRed and CAG promoter driving EGFP (Figure 2A). The shRNA was targeted towards a region of Botch conserved between mouse and rat (Figure S2A). A shRNA resistant Botch was also generated (mtBotch) (Figure S2A). shRNA-Botch is effective in knocking down transiently expressed wild-type Botch in HEK293 cells (Figure S2B), whereas it is ineffective in knocking down mtBotch (Figure S2C). In ganglionic eminence cultures shRNA-Botch knocks down endogenous Botch (Figure S2D-F). shRNA to DsRed (Duan et al., 2007), which engages the RISC complex, serves as an additional control and has no effect on overexpressed or endogenously expressed Botch (Figure S2B-E).
To explore the role of Botch in neurogenesis in utero electroporation of shRNA-DsRed, shRNA-Botch, shRNA-Botch and Botch, or shRNA-Botch and mtBotch into E13.5 CD1 mouse brains was performed and embryos were harvested at E15.5 (Figure 2D-E). Knockdown of Botch greatly increases the percentage of cells in the VZ and SVZ while significantly decreasing the percentage of GFP+ cells in the CP and IZ (Figure 2D-E). Co-expression of wild-type Botch, which is sensitive to shRNA-Botch, does not alter the shRNA-Botch phenotype but co-expression of mtBotch, which is not susceptible to knockdown by shRNA-Botch, rescues the knockdown phenotype (Figure 2D-E).
The number of Tbr1+ (T-box brain 1) cells (an indicator of neuronal differentiation) were assessed following in utero electroporation of Botch or shRNA-Botch or rescue of shRNA-Botch with mtBotch (Figure 2F-I) to evaluate Botch regulation of neurogenesis. Overexpression of Botch increases the number of Tbr1+ and GFP+ cells implying regulation of neurogenesis rather than mis-positioned progenitor cells from altered migration (Figure 2F-G). Conversely, knockdown of Botch results in a loss of Tbr1+ expression in GFP+ cells indicating suppression of neurogenesis, which can be rescued by expression of mtBotch (Figure 2H-I). Furthermore, GFP+ cells are either Pax6+ in the VZ (Figure S2G) or Tbr2+ in the SVZ (Figure S2H) but are Tbr1- in both cases (Figure 2H). There is no alteration in the glial marker GFAP (Figure S2I). Nestin immunohistochemistry reveals intact radial-glial scaffolds in neocortex following electroporation with shRNA-Botch (Figure S2J). Active caspase-3 immunohistochemistry confirms that the altered cellular distribution pattern is not due to apoptosis, following knockdown of Botch (Figure S2K-L). Taken together, these data indicate Botch regulates neurogenesis, not migration or altered cell viability.
Embryos were harvested at E17.5 to observe the role of Botch on a later stage of neuronal development. Knockdown of Botch greatly increases the number of cells in the VZ, SVZ and IZ while significantly decreasing the number of GFP+ cells in the CP. mtBotch rescues the knockdown phenotype (Figure S2M-N). Thus Botch appears to regulate neocortical neurogenesis and may direct the proliferating-zone exit of neural stem cells (NSC).
Botch inhibits the Notch signaling pathway
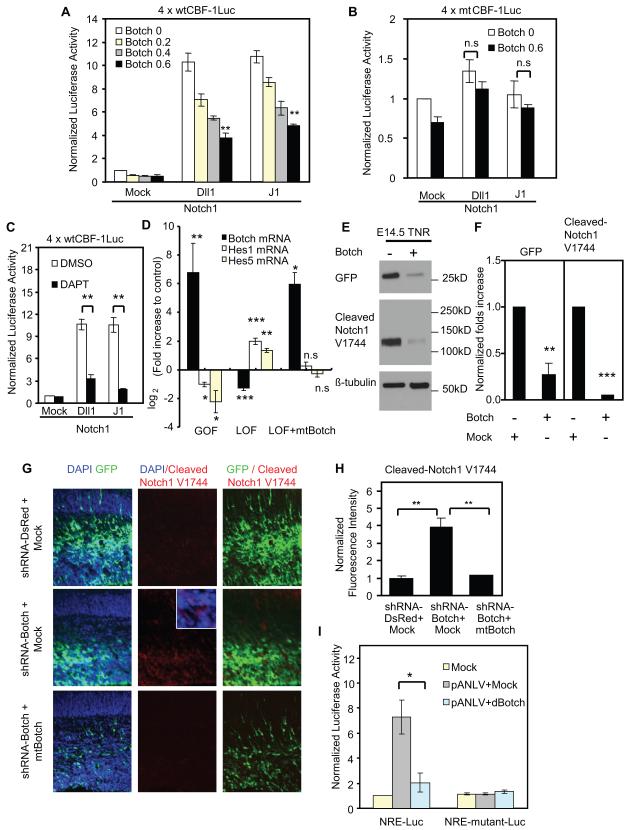
Notch activation results in observations that are similar to the effects of shRNA-Botch suggesting a potential interaction (Mizutani and Saito, 2005; Mizutani et al., 2007). Thus, HeLa cells were transfected with full length Notch1, and 4 × wtCBF-1-response-element luciferase reporter, β-galatacosidase reporter (β-gal), and different concentrations of a Botch expression construct (Hsieh et al., 1996). 48 hours later, HeLa cells were co-cultured with NIH 3T3 cells expressing the Notch ligand Dll1 or J1, or mock control. 24 hours after co-culture, β-gal and luciferase activity was determined. Botch significantly inhibits the activation of CBF-1-dependent Notch1 activity in a dose-dependant manner (Figure 3A). To evaluate the specificity of this interaction, a CBF-1 transactivation inactive mutant reporter was used (Hsieh et al., 1996) with no difference observed between Botch and the controls (Figure 3B). HeLa cells transfected with full length Notch2, Notch3 or Notch4 and 4 × wtCBF-1-response-element luciferase reporter, β-galatacosidase reporter (β-gal), and different concentrations of Botch expression constructs (Figure S3A-C) display a dose-dependent botch inhibition of CBF-1-dependent Notch2, Notch3 or Notch4 activity (Figure S3A-C). The γ-secretase inhibitor, DAPT, inhibits CBF-1 dependent Notch signaling serving as a positive control for the specificity of this Notch reporter assay (Figure 3C).
Figure 3. Botch inhibits the Notch signaling pathway.
(A) wtCBF-1 luciferase reporter assays of HeLa cells expressing Notch1 and different concentrations of Botch co-cultured with NIH 3T3 cells expressing Notch ligands. wtCBF-1 luciferase activity was normalized to β-galatacosidase activity and then to the Mock group without Botch. Values represent the mean ± SEM (n ≥ 3; for Dll1 group, p < 0.001, one-way ANOVA; for J1 group, p < 0.001, one-way ANOVA, post-test: Tukey’s multiple comparison test).
(B) mtCBF-1 luciferase reporter assays of HeLa cells expressing Notch1 and Botch co-cultured with NIH 3T3 cells expressing Notch ligands. mtCBF-1 luciferase activity was normalized to β-galatacosidase activity and then to the Mock group without Botch. Values represent the mean ± SEM (n ≥ 3, n.s. p >0.05 Student’s t-test).
(C) wtCBF-1 luciferase reporter assays of HeLa cells expressing Notch1 co-cultured with NIH 3T3 cells expressing Notch ligands in the presence of DMSO or DAPT. wtCBF-1 luciferase activity was normalized to β-galatacosidase activity and then to the Mock group with DMSO. Values represent the mean ± SEM (n ≥ 3; ** p<0.01, Student’s t-test).
(D) RT-PCR results of Botch, Hes1 and Hes5 mRNA from E14.5 ganglionic eminence cultures following gain-of-function (GOF) with Botch overexpression, loss-of-function group (LOF) with shRNA-Botch knockdown and rescue with mtBotch. For GOF the control is empty pCAG vector and for LOF is shRNA-DsRed. Values represent the mean ± SEM (n ≥ 3; * p < 0.05; ** p < 0.01; *** p < 0.001, one-way ANOVA, post-test: Tukey’s multiple comparison test).
(E) Immunoblots of GFP signal driven by CBF1 in the TNR mouse and V1744 from ganglionic eminence cultures following overexpression of Botch.
(F) Optical densitometry quantification normalized to β-tubulin in (E). Values represent the mean ± SEM (n ≥ 3; ** p < 0.01; *** p < 0.001, Student’s t-test).
(G) Representative confocal images of cortex immunostained for GFP with DAPI or for cleaved Notch1 (V1744).
(H) Quantification of normalized fluorescence intensity in (G) Values represent the mean ± SEM (n = 3; *** p < 0.001, one-way ANOVA, post-test: Tukey’s multiple comparison test).
(I) β-gal and luciferase activity in S2 cells transfected with Drosophila Notch-VP16 (pANLV), NRE luciferase reporter or NRE-mutant luciferase reporter, β-galatacosidase reporter (β-gal), or Drosophila Botch (dBotch). Luciferase activities were normalized to β-galatacosidase activity and then to the Mock group. Values represent the mean ± SEM (n = 4; * p<0.05, Student’s t-test).
The effect of gain or loss of function of Botch on the expression of Notch target genes, Hes1 and Hes5, were evaluated in E14.5 ganglionic eminence neural precursor cultures by quantitative RT-PCR analysis (Tatsuya Iso, 2003). Overexpression of Botch leads to the downregulation of Hes1 and Hes5 mRNA. Knockdown of Botch has the opposite effect leading to an upregulation of Hes1 and Hes5 mRNA (Figure 3D) an effect that is rescued by expression of mtBotch. (Figure 3D). In ganglionic eminence cultures from the transgenic Notch reporter (TNR) mice (Mizutani et al., 2007), overexpression of Botch leads to a decrease of endogenous GFP signal indicating decreased Notch signaling (Figure 3E-F). Cleaved Notch1 is also decreased as detected by an antibody recognizing γ-secretase-cleaved Notch1 (V1744, Cell Signaling Technology, Beverly, MA, USA) (Figure 3E-F). These results indicate that Botch through inhibiting Notch signaling, can regulate the expression of Notch target genes.
Knockdown of Botch affects endogenous Notch signaling as there is a 4-fold increase of cleaved Notch1 immunoreactivity (V1744) in shRNA-Botch cells compared with shRNA-DsRed cells in vivo. This effect is rescued by expression of mtBotch (Figure 3G-H). To determine whether Botch influences Notch signaling in simple organisms, S2 Drosophila cells were transfected with full length Drosophila Notch-VP16 (pANLV) (Saj et al., 2010), NRE luciferase reporter or NRE-mutant luciferase reporter (Furriols and Bray, 2001), β-galatacosidase reporter (β-gal), and a Drosophila Botch expression construct. 24 hours later, β-gal and luciferase activity was measured. In pANLV transfected S2 cells, Botch significantly inhibits the activation of Notch activity (Figure 3I). These data indicate that Botch inhibition of Notch signaling might be evolutionarily conserved.
Botch and Notch are expressed in a similar temporal and spatial pattern
For Botch to regulate Notch signaling during development, both proteins would need to be expressed in a similar temporal and spatial pattern. In situ hybridization reveals an overlapping expression pattern for Botch and Notch1 at E12.5, E14.5 and E16.5 (Figure S4A). By immunohistochemical analysis there is a partial intracellular co-localization of Botch with Notch in HEK293 cells, HeLa cells, primary cultures of ganglionic eminence and primary cultures of hippocampus (Figure S4B-E). Thus, it is spatially and temporally possible for Botch to regulate Notch signaling.
Botch interacts with the Notch1 extracellular domain
To evaluate whether Botch can physically interact with Notch in vivo, a co-immunoprecipitation was performed from E14.5 ganglionic eminence lysates. Surprisingly, Botch immunoprecipitation pulls down more of the uncleaved immature full-length form of Notch1 than the Transmembrane and Intracellular (TMIC) domain, which is the S1-cleaved mature form of Notch and also the dominant form of Notch in the input (Figure 4A). This interaction is specific as Notch1 immunoreactivity is absent in the IgG immunoprecipitation control and following immunoabsorption with excess Botch (Figure 4A). Furthermore, Botch does not co-immunoprecipitate the EGF-repeat containing protein Dll1, nor does it bind to other membrane-receptors including EGFR or FGFR2 (Figure 4B). Botch is a 25 kD protein, and it migrates closely with the antibody light chain and thus is not readily discriminated from the light chain on immunoblot analysis or by coomassie staining. To show that Botch immunoprecipitation indeed immunoprecipitates Botch, overexpressed Botch-myc was immunoprecipitated by the Botch antibody and immunoblot analysis indicates that the Botch antibody is capable of immunoprecipitating Botch (Figure S4F).
Figure 4. Botch preferentially interacts with the Notch1 extracellular domain.
(A) Immunoblot of co-immunoprecipitation of endogenous uncleaved immature full-length Notch1 by an anti-Botch polyclonal antibody (Botch-PoAb) from E14.5 ganglionic eminence. An unrelated rabbit IgG antibody and preabsorption of the Botch-PoAb with recombinant Botch-GST are negative controls. Abbreviations: IP, immunoprecipitation; FL, Full Length; TMIC, TransMembrane and IntraCellular domain.
(B) Immunoblot of co-immunoprecipitation with Botch-PoAb and Dll1, EGFR or FGRF2 from E14.5 ganglionic eminence.
(C and D) Immunoblots of Botch co-immunoprecipitation with the Notch ExtraCellular Domain (NECD1) or the Notch IntraCellular Domain (NICD1).
(E) Immunoblot for GFP of Botch co-immunoprecipitation with four fragments of the NECD1; EGF repeats 1-12, 11-24, 22-33 and 32-36 with Lin12/Notch repeats (LNR) that encode rat Notch1 amino acids 1-493, 482-910, 832-1310 and 1224-1723 with cDNA encoding the 20 amino-acid signal peptide from Notch1 at the N-terminal for proper subcellular localization.
(F) Immunoblot for GFP of Botch co-immunoprecipitation with fragments containing EGF repeats 32-36 and LNR divided into EGF repeats 32-36 (rat Notch1 amino acids 1224-1448) or the Lin12/Notch repeats (LNR) (rat Notch1 amino acids 1449-1723) with cDNA encoding the 20 amino-acid signal peptide from Notch1 at the N-terminal for proper subcellular localization.
(G) Co-immunoprecipitation of Botch with Notch1 (Flag-N1-Gal4) or Notch Loop Out (Flag-N1-Gal4-LO) which lacks the S1-cleavage sites, visualized by immunoblot. Flag-N1-Gal4-LO lacks of amino acid sequence 1624-1670 in human Notch1.
(H) Scatchard plot of quantitative binding of Botch-N-AP to Notch1 immobilized by anti-Notch1 on protein G sepharose beads.
A-H, Experiments were repeated three times with similar results.
Co-immunoprecipitation experiments show Botch interacts with the extracellular domain of Notch1 (NECD1) (Figure 4C), but not the intracellular domain (NICD1) (Figure 4D). To further refine the binding site on Notch1, NECD1 was evenly divided into four fragments containing EGF repeats 1-12, 11-24, 22-33 and 32-36 with Lin12/Notch repeats (LNR), respectively, with each fragment also expressing the 20 amino-acid signal peptide from Notch1 for proper subcellular localization. These four constructs encode rat Notch1 amino acids 1-493, 482-910, 832-1310 and 1224-1723, respectively. Botch binds to the construct containing EGF repeats 32-36 with LNR (Figure 4E). The EGF repeats 32-36 with LNR was further subdivided. Botch binds to the LNR-GFP containing the LNR and HD domain (heterodimerization domain) (rat Notch1 amino acids 1449-1723) but not to the fragment containing EGF repeats 32-36 (rat Notch1 amino acids 1224-1448) (Figure 4F). Because LNR-GFP also contains the S1-cleavage sites the ability of Botch to bind to Notch1 versus the S1-cleavage resistant Notch1 (Notch1-Loopout, Flag-N1-Gal4-LO) (Gordon et al., 2009) was investigated. Botch binds to Notch1 but not to Notch1-Loopout (Figure 4G). Thus, Botch does not bind to either EGF-like repeats or the LNR, but it specifically binds to the S1 furin-like cleavage site.
The binding properties of Botch to Notch were ascertained by using an alkaline phosphatase tagged Botch (Botch-N-AP) (Chapman et al., 2006; Flanagan and Cheng, 2000). Notch1 was transiently transfected into HEK293 cells and lysates were immunoprecipitated with anti-Notch1 antibody (Santa Cruz) and protein G beads. Binding affinity was determined by incubating different concentrations of Botch-N-AP fusion protein with an equal volume of Notch1 binding protein G beads. Botch binds to Notch1 in a saturable manner with a binding affinity of 3.3 nM (Figure 4H). Botch also binds with varying affinity to Notch2 (8.8 nM), Notch3 (3.3 nM) and Notch4 (6.3 nM) (Figure S4G-I). These results in combination indicate that Botch binds to Notch with the strongest interaction with Notch1 and Notch3.
Botch interferes with processing of Notch1 to its mature form
Whole cell and cell surface expression of Botch and Notch were monitored in neural precursor cells during differentiation into neuronal cultures. Botch expression increases and Notch surface expression decreases during differentiation (Figure 5A-B). To further investigate the actions of Botch on Notch, Botch and Flag-Notch1-GFP constructs were overexpressed in HEK293 cells to monitor Notch1 processing. The effects of Botch were compared to the furin inhibitor, DEC-RVKR-CMK. Overexpression of Botch and the furin inhibitor, DEC-RVKR-CMK leads to an approximately 2-fold increase in the level of unprocessed full-length Notch1 (Notch1-FL) with a significant decrease in processed Notch1 extracellular domain (Notch1-ECD) (Figure 5C-D). Biotin surface labeling of Notch1 under identical conditions shows an 80% reduction of surface Notch1 (Figure 5C-D). By FACS analysis binding of the Fc-tagged Notch1 ligand J1-Fc was significantly reduced with expression of Botch (Figure 5E). To confirm that Botch requires full length Notch1 to mediate its inhibitory effects, the Notch intracellular domain (NICD1) and Notch1 extracellular domain deleted form (Notch1-DeltaE-GFP), as the constitutively active forms of Notch1, were overexpressed in HeLa cells and Notch1 activity was monitored by the CBF-1 luciferase reporter assay (Figure 5F-G). Botch has no effects on the abilities of NICD1 or Notch1-DeltaE to activate the CBF-1 luciferase reporter indicating the actions of Botch on Notch1 occur before Notch1 cleavage induced by ligand-receptor binding and NICD1 signaling.
Figure 5. Botch interferes with processing of Notch1 to its mature form.
(A) Immunoblot of Notch1, Tuj1 or Botch in whole cell lysates, and surface expression of Notch1 in mouse ganglionic eminence cultures over time (day 0 to day 6) during differentiation in to neuronal cultures.
(B) Linear regression analysis of normalized surface Notch1 expression to normalized Botch expression. Densitometry quantification of (A) normalized to β-tubulin. Values represent the mean ± SEM (n = 3; * p < 0.05, one-way ANOVA, post-test: Tukey’s multiple comparison test).
(C) Immunoblot analysis of Notch in whole cell lysate and surface expression following overexpression of Botch and Flag-Notch1-GFP with a furin inhibitor. Abbreviations: FL, Full Length; ECD, Extracellular domain.
(D) Optical densitometry quantification of (C) normalized to β-tubulin. Values represent the mean ± SEM (n ≥ 3; * p < 0.05; *** p < 0.001, Student’s t-test).
(E) Cell-membrane-bound Jagged-1-Fc (J1-Fc) quantified by FACS analysis. Values are normalized with Fc control in Mock group and represent the mean ± SEM (n = 4; * p < 0.05; *** p < 0.001, Student’s t-test).
(F and G) wtCBF-1 luciferase reporter assays of HeLa cells expressing NICD1 or Notch1-DeltaE-GFP with or without Botch. wtCBF-1 luciferase activity was normalized to β-galatacosidase activity and then to the Mock group. Values represent the mean ± SEM (n = 3; n.s. p > 0.05, Student’s t-test).
(H) Immunoblots of Notch cleavage by furin and inhibition of furin cleavage by the inhibitor, DEC-RVKR-CMK, or by Botch-AP. The experiment was repeated 3 times with similar results.
Since furin is required for Notch1 S1 processing (Kopan and Ilagan, 2009; Louvi and Artavanis-Tsakonas, 2006), an in vitro furin cleavage assay was performed to test whether Botch regulates S1 cleavage (Figure 5H). Immunoprecipitated Flag-Notch1-GFP was evenly divided into three groups. The first group was pre-treated with AP, and then treated with furin and DMSO; the second group was pre-treated with AP, and then treated with furin in the presence of furin inhibitor, DEC-RVKR-CMK (Enzo LifeSciences); the last group was pre-treated with Botch-AP, and then treated with furin and DMSO. Botch blocks furin cleavage in a manner similar to the furin inhibitor, DEC-RVKR-CMK (Figure 5H). Thus, Botch maintains Notch1 in an immature form primarily by blocking the S1 furin-like cleavage of Notch, although we cannot exclude the possibility that Botch also retains full-length Notch in the Golgi preventing its trafficking.
HEK293 cells were co-transfected with the EGF repeat-containing protein Dll1-GFP and Botch-myc followed by co-immunoprecipitation. Botch does not interact with Dll1 (Figure S5A). Binding studies indicate there is no interaction between Botch and Dll1 (Figure S5B) and analysis of surface expression of Dll1 shows that Botch does not alter the expression of Dll1 (Figure S5C-D). Additionally, Botch does not alter the surface expression of the unrelated glutamate receptor protein subunits, GluR2 or NR2A as well as the Notch1 ligand Jagged1 (Figure S5E-J). Taken together these data indicate that Botch is not binding indiscriminately to any EGF repeat containing protein or receptor protein, but Botch shows selectivity for Notch proteins. Moreover, Botch’s affects on Notch1 surface expression does not appear to be due to a generalized defect in the trans-Golgi network since Botch has no effect on the surface expression of other membrane receptors such as the glutamate NR2A or GluR1 receptors, nor does it have any effect on the trafficking of Delta1 or Jagged 1.
Botch promotes C2C12 differentiation by inhibiting the Notch signaling pathway
The role of Botch in antagonizing Notch1 was explored in a classic model of Notch signaling of C2C12 cell differentiation to myotubes (Lindsell et al., 1995). C2C12 cells stably expressing Notch1 (C2C12-N1) (Chapman et al., 2006), were transiently transfected with an 8 × wtCBF-1-response-element luciferase reporter, aβ-galatacosidase reporter (β-gal) and a Botch expression construct. 48 hours later, C2C12-N1 cells were co-cultured with NIH 3T3 cells expressing the Notch ligands Dll1 or J1, or mock control. In addition, the effects of Botch on co-cultured wild-type C2C12 and C2C12-N1 cells were also monitored. Botch significantly inhibits Notch activity (Figure 6A). Myotube differentiation was monitored by myosin heavy chain (MHC) immunostaining in C2C12 cells overexpressing Botch, and compared to control C2C12 cells overexpressing GFP (Figure 6B) and quantified (Figure 6C) (Chapman et al., 2006). Botch leads to an approximately 3-fold increase of MHC+ cells in wild-type, C2C12, or C2C12-N1 cells (Figure 6C). Co-culture of C2C12-N1 cells with NIH 3T3 expressing Dll1 or J1, almost completely inhibits the expression of MHC. However, overexpression of Botch reverses the inhibition of ligand-dependent Notch activation, leading to an increase in MHC+ cells (Figure 6C). The γ-secretase inhibitor, DAPT, which blocks Notch signaling, increases the number of MHC+ cells in a manner similar to Botch (Figure 6D). There is no additional effect with overexpression of Botch indicating that both DAPT and Botch are acting in the same pathway. Overexpressing NICD1 overrides the Botch inhibitory effects on Notch signaling consistent with the notion that Botch is acting upstream of ligand-receptor binding (Figure 6E). At day 3 there is increased expression of the early differentiation marker myogenin (Chapman et al., 2006; Wright et al., 1989) following overexpression of Botch (Figure 6F) indicating differentiation in the presence of Botch, an action opposite that of Notch. Loss of function experiments cannot be conducted in C2C12 cells as this cell line does not express Botch. Therefore in C2C12 cells, overexpression of Botch antagonizes the differentiation inhibition induced by Notch signaling.
Figure 6. Botch promotes C2C12 differentiation and embryonic neurogenesis by inhibiting the Notch signaling pathway.
(A) wtCBF-1 luciferase reporter assays with Mock or Botch transfected C2C12 wild-type cells, C2C12-Notch1 cells and C2C12-Notch1 cells co-cultured with NIH 3T3 cells expressing Notch ligands. wtCBF-1 luciferase activity was normalized to β-galatacosidase activity and to the Mock. Values represent the mean ± SEM (n ≥ 3; ** p < 0.01, Student’s t-test).
(B) Representative confocal images of either GFP or Botch transfected C2C12 cells immunostained for myosin heavy chain (MHC), GFP or Botch and counterstained for DAPI.
(C) Differentiation assays in either GFP or Botch transfected wild-type C2C12 cells, C2C12-Notch1 cells or C2C12-Notch1 cells co-cultured with NIH 3T3 cells expressing different Notch ligands. Percentages of MHC positive cells are shown. Values represent the mean ± SEM (n ≥ 3; * p < 0.05; ** p < 0.01; *** p < 0.001, Student’s t-test).
(D) Differentiation assays in GFP or Botch transfected C2C12 wild-type cells in the presence of DMSO or gamma-secretase inhibitor DAPT. Percentages of MHC positive cells are shown. Values represent the mean ± SEM (n ≥ 3; n.s. p > 0.05, Student’s t-test).
(E) Differentiation assays in GFP or Botch transfected C2C12 wild-type cells in the presence of Mock or NICD1. Percentages of MHC positive cells are shown. Values represent the mean ± SEM (n ≥ 3; *** p < 0.001, Student’s t-test).
(F) Immunoblot analysis for Myogenin, Botch and β-tubulin following expression of Botch or GFP in C2C12.
(G) A schematic diagram of pCAG constructs for dominant-negative coactivator MAML DN-MAML-GFP or shRNA-Botch-mCherry.
(H) Distribution of GFP+ cells following Botch overexpression with NICD or DN-MAML-EGFP.
(I) Quantification of (H). Values represent the mean ± SEM (n ≥ 3; n.s. p > 0.05, Student’s t-test) Quantification of GFP with Mock alone is the same as Figure 2C which is displayed for comparison purposes.
(J) Distribution of GFP+ and mCherry+ cells following expression of DN-MAML-GFP or shRNA-Botch-mCherry (red).
(K) Quantification of (J). Values represent the mean ± SEM (n ≥ 5; *** p < 0.01; n.s. p > 0.05, Student’s t-test).
Botch promotes embryonic neurogenesis by inhibiting the Notch signaling pathway
NICD or the dominant-negative version of the coactivator MAML (DN-MAML-EGFP) (Figure 6G), which forms a transcriptionally inactive complex with NICD to produce loss of Notch signaling (Maillard et al., 2004), were expressed with Botch in utero. NICD blocks Botch repression of Notch signaling (Figure 6H-I). In the presence of DN-MAML-EGFP, Botch has no effect (Figure 6H-I). Knockdown of Botch greatly increases the percentage of cells in the VZ and SVZ while significantly decreasing the percentage of GFP+ cells in the CP and IZ (Figure 6J) an effect that is reversed by reduction of Notch signaling through expression of DN-MAML-EGFP (Figure 6J-K) in which normal cellular distribution is restored. Additionally, knockdown of Botch and inhibition of Notch signaling by expression of DN-MAML are not synergistic when compared with inhibition of Notch signaling alone (Figure 6J-K). Taken together, these results suggest that Botch and Notch are acting in the same pathway to regulate embryonic neurogenesis.
Botch promotes neurogenesis by regulating cell-fate choices in vitro
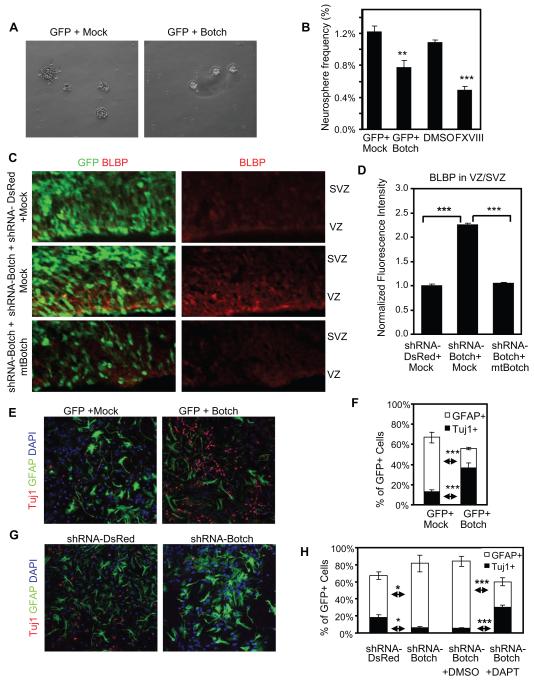
An important feature of neural precursor cells is the capacity for self-renewal that is mediated in part by Notch signaling therefore the actions of Botch were evaluated in neurosphere cultures established from the lateral and medial ganglionic eminences of E14.5 CD-1 mouse embryos. Botch expression decreases neurosphere frequency as compared to empty vector (Figure 7A-B). The γ-secretase inhibitor factor eighteen (GSI FXVIII) that blocks Notch signaling (Dang et al., 2006), also results in decreased neurosphere frequency similar to Botch (Figure 7B). In vivo shRNA-Botch knockdown results in an increase in brain lipid binding protein (BLBP) expression, a marker for early radial glia, which is rescued by expression of mtBotch (Figure 7C-D). These data are consistent with our previous report that Notch signaling induces BLBP expression and that BLBP is a Notch target (Anthony et al., 2005; Gaiano et al., 2000).
Figure 7. Botch promotes neurogenesis by regulating cell-fate choices.
(A) Representative images of neurosphere cultures.
(B) Quantification of neurosphere frequencies following expression of Botch or exposure to FXVIII. Values represent the mean ± SEM (n ≥ 3; ** p < 0.01; *** p < 0.001, Student’s t-test).
(C) Representative confocal images of cortex immunostained for GFP with DAPI and immunostained for BLBP following shRNA knockdown of Botch and expression of mtBotch.
(D) Quantification of normalized fluorescence intensity in (C). Values represent the mean ± SEM (n = 3; *** p < 0.001, Student’s t-test).
(E) Representative confocal images of DAPI and immunostaining for GFAP and Tuj1 in cultured ganglionic eminence following expression of Botch and shRNA-Botch.
(F) Quantification of (E). Values represent the mean ± SEM (n = 6; *** p < 0.001, Student’s t-test).
(G) Representative confocal images of DAPI and immunostaining for GFAP and Tuj1 in ganglionic eminence cultures with expression of shRNA-Botch in the presence or absence of DAPT. (H) Quantification of (G). Values represent the mean ± SEM (n = 6; * p < 0.05; *** p < 0.001, Student’s t-test).
The developmental potential of neural precursor cells (Mizutani et al., 2007) in adherent cultures from E14.5 CD-1 mouse ganglionic eminences were evaluated by immunostaining for Tuj1 to identify neuronal fate choices and GFAP for glial ones. Gain and loss of function experiments of Botch were conducted. Botch overexpression leads to an approximately 3-fold increase in the number of Tuj1+ cells and a corresponding reduction in the GFAP+ cells (Figure 7E-F). In contrast, knockdown of Botch by shRNA leads to a 3-fold reduction in Tuj1+ cells and a significant increase in the number of GFAP+ cells (Figure 7G-H). The γ-secretase inhibitor DAPT is effective in reversing the effects of Botch knockdown on glial fate choice, indicating that the enhanced glial cell fate following shRNA knockdown of Botch is due to Notch signaling (Figure 7H). These experiments taken together indicate that Botch promotes embryonic neuronal differentiation by inhibiting Notch signaling.
DISCUSSION
The major finding of this study is the characterization of a previously unknown developmentally expressed protein, Botch, which inhibits Notch1 signaling, thereby promoting embryonic neurogenesis. In both mammals and fruit flies Notch signaling plays a critical role in regulating self-renewal and differentiation of neural stem cells (Louvi and Artavanis-Tsakonas, 2006). NICD1 keeps neural stem cells in the proliferation zones (Mizutani and Saito, 2005; Mizutani et al., 2007), while knockdown of CBF-1 drives neural stem cells into the IZ and CP (Mizutani et al., 2007). In contrast Botch drives more cells to the IZ and CP, whereas downregulation of Botch tends to keep cells in the proliferating zones indicating Botch is acting in opposition to Notch.
The neurogenic phenotypes induced by Botch are primarily mediated by changes in Notch signaling. The cellular retention in the VZ and SVZ induced by Botch knockdown is prevented when Notch signaling is inhibited by DN-MAML indicating knockdown of Botch up-regulates Notch signaling. In cells overexpressing DN-MAML, shRNA-Botch does not restore normal cellular distribution. These results support the idea that shRNA-Botch acts upstream of the CBF-1 complex. If Botch and Notch were regulating the same process but not each other, we would have expected a partial rescue of the DN-MAML phenotype. Botch overexpression has no effect in cells in which Notch was already inhibited by DN-MAML. If Botch and Notch were regulating the same process but not each other, we would have expected a certain degree of synergistic effects between Botch and DN-MAML. Overexpressing NICD1 ablates the Botch overexpression phenotype consistent with the notion that the actions of Botch are on the immature full-length form of Notch1 upstream of the S3 cleavage that generates NICD. Botch decreases neurosphere frequency similar to the γ-secretase inhibitor FXVIII, which prevents Notch signaling and an in vitro differentiation assay indicates that overexpression of Botch promotes neuronal differentiation. Knockdown of Botch increases radial glia and increases glial differentiation, which can be rescued by γ-secretase inhibitor, DAPT. Taken together these results imply that Botch regulates neural stem cell self-renewal and differentiation, and promotes embryonic neurogenesis in opposition to Notch since all the effects of Botch overexpression are opposite to Notch signaling and knockdown of Botch causes similar effects as Notch activation (Grandbarbe et al., 2003; Tanigaki et al., 2001; Yamamoto et al., 2001; Yoon et al., 2004).
Botch inhibits Notch by regulating Notch processing
Botch is localized, in part, to the trans-Golgi where Notch is processed by S1 cleavage by a furin-like protease (Logeat et al., 1998). This S1 cleavage of the Notch receptor is thought be required for Notch receptor maturation where Notch is ultimately cleaved into a TMIC domain and a NECD to form a functional receptor. Our findings indicate that Botch may interfere with Notch maturation by maintaining Notch in its full-length immature form. Evidence includes the observation that overexpression of Botch leads to accumulation of unprocessed immature full-length Notch and a corresponding decrease in the S1 cleaved ECD domain with decreased surface Notch1. Consistent with the possibility that Botch interferes with the S1 furin-like cleavage of Notch is the observation that Botch blocks the furin cleavage of Notch in a similar manner as the furin inhibitor DEC-RVKR-CMK. This ultimately leads to a corresponding decrease in functional canonical Notch signaling. The failure to observe defects in the trafficking of other membrane receptors such as the glutamate NR2A or GluR1 receptors and the failure to observe defects in trafficking of Delta1 or Jagged 1 indicates that Botch has a rather specific role in regulating Notch processing. It appears Botch selectively regulates Notch trafficking to the cell surface via interference with the S1 furin-like cleavage of Notch. The exact role of Botch in this aspect of Notch signaling requires further investigation and identification of Botch’s biochemical mechanism of action. As recently reviewed by Kopan and Ilagan(Kopan and Ilagan, 2009), this aspect of Notch signalling is poorly understood. Further study of how Botch regulates the intracellular processing of Notch holds tremendous promise for understanding this aspect of Notch biology.
EXPERIMENTAL PROCEDURES
Plasmid constructs and Vector-based shRNA constructs
In situ hybridization
DIG-labeled anti-sense and sense RNA Botch probes were generated by T7 or T3 RNA polymerases using a template containing the mouse Botch cDNA clone (Image number 4483043) as described (Blackshaw and Snyder, 1997). Whole-mount and sectioned embryos of specified stages were subjected to alkaline phosphatase conjugated anti-DIG antibodies (Roche) to detect hybridized probes. Wholemount images were acquired with a Zeiss Axiocam digital camera on a Zeiss Stemi SV11 microscope. For sections images were taking using a Zeiss Axioskop compound microscope.
Production of antibodies
In utero injection and electroporation
DNA transfer into E13.5 CD1 mouse brains in utero was performed as previously described (Mizutani et al., 2007) using a Nepagene CUY21EDIT electroporator.
Cell line cultures and transfection
HEK293, HeLa, C2C12, S2 and NIH 3T3 cells were obtained from the American Type Culture Collections. C2C12 cells stably expressing Notch1 (C2C12-N1) was provided by U. Lendahl (Karolinska Institute, Sweden) and have been described previously (Chapman et al., 2006). For C2C12 differentiation, 10% FBS was replaced with 2% horse serum (HS) in the culture medium for 4 days before immunostaining.
CBF-1RE and NRE Luciferase reporter assay
The co-culture reporter assay was performed as described (Lindsell et al., 1995).
Mouse neuroprogenitor cultures and transfection
Neurosphere and adherent progenitor cultures were established from E14.5 CD-1 mouse lateral and medial ganglionic eminences as described (Yoon et al., 2004).
Quantitative Real-time PCR (RT–PCR)
Immunoprecipitation assays
Notch1-Botch-N-AP binding assays
As described (Flanagan and Cheng, 2000) Botch-N-AP constructs were transiently transfected into HEK293 cells. Varied concentrations of Botch-N-AP fusion protein were incubated with an equal volume of Notch binding protein G beads at room temperature and assayed for AP activity.
Cell surface biotinylation labeling and Notch1-Jagged1 binding assays
Biotin was used to label and isolate cell surface protein as described (Ladi et al., 2005).
Immunoblot analysis and immunostaining
Golgi sucrose gradient enrichment
A Golgi isolation kit (Sigma) was used to isolate the Golgi fraction from E15.5 mouse forebrain by following the manufacturer’s instruction.
Furin cleavage assay
HEK293 cells were transfected with Flag-Notch1-GFP construct. 24 hours later, the cell lysates were subjected to anti-GFP antibody immunoprecipitation with protein G sepharose beads. The Flag-Notch1-GFP protein binding on protein G beads was first pre-treated with AP or Botch-AP, and then treated with furin (New England Biolabs) and DMSO or furin with 50μM DEC-RVKR-CMK (Enzo LifeSciences). Flag-Notch1-GFP protein at different time points of furin treatment was subject to immunoblot analysis with anti-Flag antibody. Furin cleavage was carried out at room temperature in total volume of 1 ml using 20 Units of recombinant furin in 100mM HEPES 7.5, 0.5% Triton and 1 mM CaCl2 suggested by manufacturer.
Statistical analysis
Statistical analysis was performed using Prism or Excel software and specific tests are noted in the text and figure legends. Unless otherwise noted, all error bars represent ± SEM. and significance was assessed as p < 0.05.
Supplementary Material
HIGHLIGHTS.
Botch inhibits the S1 furin-like cleavage of Notch.
Botch likely interacts with Notch in the Golgi leading to decreased Notch processing.
Botch inhibits Notch function and promotes neurogenesis in vitro and in vivo.
ACKNOWLEDGMENTS
This work was supported by an American Heart Association Predoctoral Award to Z.C., USPHS FNS068010A to S.B., McKnight Foundation Neuroscience of Brain Disorders Award, USPHS NS40809, DA00266, and MSCRFII-0429 to V.L.D. T.M.D. is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Anthony TE, Mason HA, Gridley T, Fishell G, Heintz N. Brain lipid-binding protein is a direct target of Notch signaling in radial glial cells. Genes & Development. 2005;19:1028–1033. doi: 10.1101/gad.1302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Ayala R, Shu T, Tsai LH. Trekking across the brain: the journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- Blackshaw S, Snyder SH. Parapinopsin, a Novel Catfish Opsin Localized to the Parapineal Organ, Defines a New Gene Family. J Neurosci. 1997;17:8083–8092. doi: 10.1523/JNEUROSCI.17-21-08083.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman G, Liu L, Sahlgren C, Dahlqvist C, Lendahl U. High levels of Notch signaling down-regulate Numb and Numblike. J Cell Biol. 2006;175:535–540. doi: 10.1083/jcb.200602009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Liang D, Li H, Sasaki M, Dawson TM, Dawson VL. Functional Identification of Neuroprotective Molecules. PLoS ONE. 2010;5:e15008. doi: 10.1371/journal.pone.0015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, Yoon K, Wang M, Gaiano N. Notch3 signaling promotes radial glial/progenitor character in the mammalian telencephalon. Dev Neurosci. 2006;28:58–69. doi: 10.1159/000090753. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Donoviel DB, Hadjantonakis A-K, Ikeda M, Zheng H, Hyslop PSG, Bernstein A. Mice lacking both presenilin genes exhibitearlyembryonic patterningdefects. Genes Dev. 1999;13:2801–2810. doi: 10.1101/gad.13.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Chang JH, Ge S, Faulkner RL, Kim JY, Kitabatake Y, Liu XB, Yang CH, Jordan JD, Ma DK, et al. Disrupted-In-Schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell. 2007;130:1146–1158. doi: 10.1016/j.cell.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duarte A, Hirashima M, Benedito R, Trindade A, Diniz P, Bekman E, Costa L, Henrique D, Rossant J. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 2004;18:2474–2478. doi: 10.1101/gad.1239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan JG, Cheng HJ. Alkaline phosphatase fusion proteins for molecular characterization and cloning of receptors and their ligands. Methods Enzymol. 2000;327:198–210. doi: 10.1016/s0076-6879(00)27277-7. [DOI] [PubMed] [Google Scholar]
- Furriols M, Bray S. A model Notch response element detects Suppressor of Hairless-dependent molecular switch. Curr Biol. 2001;11:60–64. doi: 10.1016/s0960-9822(00)00044-0. [DOI] [PubMed] [Google Scholar]
- Gaiano N, Nye JS, Fishell G. Radial glial identity is promoted by Notch1 signaling in the murine forebrain. Neuron. 2000;26:395–404. doi: 10.1016/s0896-6273(00)81172-1. [DOI] [PubMed] [Google Scholar]
- Gale NW, Dominguez MG, Noguera I, Pan L, Hughes V, Valenzuela DM, Murphy AJ, Adams NC, Lin HC, Holash J, et al. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15949–15954. doi: 10.1073/pnas.0407290101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon WR, Vardar-Ulu D, L’Heureux S, Ashworth T, Malecki MJ, Sanchez-Irizarry C, McArthur DG, Histen G, Mitchell JL, Aster JC, et al. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS One. 2009;4:e6613. doi: 10.1371/journal.pone.0006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbarbe L, Bouissac J, Rand M, Hrabe de Angelis M, Artavanis-Tsakonas S, Mohier E. Delta-Notch signaling controls the generation of neurons/glia from neural stem cells in a stepwise process. Development. 2003;130:1391–1402. doi: 10.1242/dev.00374. [DOI] [PubMed] [Google Scholar]
- Hitoshi N, Ken-ichi Y, Jun-ichi M. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Hsieh JJ, Henkel T, Salmon P, Robey E, Peterson MG, Hayward SD. Truncated mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan MXG, Kopan R. SnapShot: Notch Signaling Pathway. Cell. 2007;128 doi: 10.1016/j.cell.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs LT, Shutter JR, Tanigaki K, Honjo T, Stark KL, Gridley T. Haploinsufficient lethality and formation of arteriovenous malformations in Notch pathway mutants. Genes Dev. 2004;18:2469–2473. doi: 10.1101/gad.1239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladi E, Nichols JT, Ge W, Miyamoto A, Yao C, Yang LT, Boulter J, Sun YE, Kintner C, Weinmaster G. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J Cell Biol. 2005;170:983–992. doi: 10.1083/jcb.200503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Isra毛l A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Maillard I, Weng AP, Carpenter AC, Rodriguez CG, Sai H, Xu L, Allman D, Aster JC, Pear WS. Mastermind critically regulates Notch-mediated lymphoid cell fate decisions. Blood. 2004;104:1696–1702. doi: 10.1182/blood-2004-02-0514. [DOI] [PubMed] [Google Scholar]
- McCright B, Lozier J, Gridley T. A mouse model of Alagille syndrome: Notch2 as a genetic modifier of Jag1 haploinsufficiency. Development. 2002;129:1075–1082. doi: 10.1242/dev.129.4.1075. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Saito T. Progenitors resume generating neurons after temporary inhibition of neurogenesis by Notch activation in the mammalian cerebral cortex. Development. 2005;132:1295–1304. doi: 10.1242/dev.01693. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Mungrue IN, Pagnon J, Kohannim O, Gargalovic PS, Lusis AJ. CHAC1/MGC4504 is a novel proapoptotic component of the unfolded protein response, downstream of the ATF4-ATF3-CHOP cascade. J Immunol. 2009;182:466–476. doi: 10.4049/jimmunol.182.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saj A, Arziman Z, Stempfle D, van Belle W, Sauder U, Horn T, Durrenberger M, Paro R, Boutros M, Merdes G. A combined ex vivo and in vivo RNAi screen for notch regulators in Drosophila reveals an extensive notch interaction network. Dev Cell. 2010;18:862–876. doi: 10.1016/j.devcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- Tanigaki K, Nogaki F, Takahashi J, Tashiro K, Kurooka H, Honjo T. Notch1 and Notch3 instructively restrict bFGF-responsive multipotent neural progenitor cells to an astroglial fate. Neuron. 2001;29:45–55. doi: 10.1016/s0896-6273(01)00179-9. [DOI] [PubMed] [Google Scholar]
- Tatsuya Iso LKYH. HES and HERP families: Multiple effectors of the notch signaling pathway. Journal of Cellular Physiology. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Nagao M, Sugimori M, Kosako H, Nakatomi H, Yamamoto N, Takebayashi H, Nabeshima Y, Kitamura T, Weinmaster G, et al. Transcription factor expression and notch-dependent regulation of neural progenitors in the adult rat spinal cord. Journal Of Neuroscience. 2001;21:9814–9823. doi: 10.1523/JNEUROSCI.21-24-09814.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K, Nery S, Rutlin ML, Radtke F, Fishell G, Gaiano N. Fibroblast Growth Factor Receptor Signaling Promotes Radial Glial Identity and Interacts with Notch1 Signaling in Telencephalic Progenitors. J Neurosci. 2004;24:9497–9506. doi: 10.1523/JNEUROSCI.0993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.