Abstract
Recent data suggests that PAS kinase acts as a signal integrator to adjust metabolic behavior in response to nutrient conditions. Specifically, PAS kinase controls the partitioning of nutrient resources between the myriad of possible fates. In this capacity, PAS kinase elicits a pro-growth program, which includes both signaling and metabolic control, both in yeast and in mammals. We propose that, like other kinases possessing these properties—AMPK and TOR, PAS kinase might be target for therapy of diabetes, obesity and cancer.
Every organism must strike a balance between energy consumption, as required for growth and reproduction, and energy preservation for survival during starvation. Organismal growth requires the coordinated execution of biochemical processes, linked to nutrient and energy availability, by which organisms increase cell size and/or cell number through the biosynthesis of new cellular components. Equally important is the ability to rapidly suppress these synthetic processes to conserve energy and survive under depleted nutrient conditions. Suppression of biosynthetic pathways must be coordinated with mobilization of energy stores (carbohydrate and lipid), recycling of proteins and organelles through autophagy and the cessation of cell proliferation[1]. The combination of these stress responses prevents death under conditions of fasting or starvation.
The ability to coordinately up-regulate biosynthetic cellular processes in response to available nutrients is largely dependent upon activation of target-of-rapamycin (TOR) signaling[2] and inhibition of AMP-activated protein kinase (AMPK) signaling[3]. However, the converse occurs rapidly upon nutrient depletion, AMPK is activated in response to an increased AMP:ATP ratio, and TOR is inactivated[4]. The capacity to respond rapidly and appropriately to a dynamic nutrient environment can determine the fate of the organism, whether it be survival and reproduction or death. In order to synthesize the myriad of signals from both within and without the cell, TOR and AMPK integrate with a wide variety of signaling molecules and pathways[5–7]. One such protein is PAS kinase, which is a highly conserved serine/threonine kinase containing an N-terminal PAS domain[8]. Deletion of PAS kinase, both in yeast and mice, leads to abnormal responses to perturbations in nutrient availability and aberrant partitioning of those nutrients[9, 10].
STRUCTURAL INSIGHTS INTO PAS KINASE REGULATION AND FUNCTION
Insight into the regulation and function of PAS kinase has come from structural studies. PAS kinase contains an N-terminal Per-Arnt-Sim (PAS) domain. PAS domains serve as versatile sensory domains responsive to a variety of intracellular cues, including light, oxygen, redox state and many others[11]. PAS domains are not highly conserved at the primary amino acid sequence level (~ 20% identity), but they adopt a characteristic core fold consisting of an alternating five strand β-sheet that is flanked by a varying number of α-helices. PAS domains are often regulated by the binding of a diverse group of small ligands, including ATP, heme or flavins, within the hydrophobic pocket at the core of the domain[12–15]. As with other PAS domains, the PAS domain of PAS kinase adopts this characteristic fold and can bind small organic molecules within its hydrophic core[16]. Unlike other PAS domains, however, the physiological ligand(s) for PAS kinase remain unknown. When the PAS domain is deleted, the kinase activity of PASK increases, but it is repressed by addition in trans of the PAS domain, which acts through direct binding to the kinase domain[8]. We hypothesize that allosteric regulation by the PAS domain enables PAS kinase to display ligand-responsive kinase activity in vivo. PAS domain binding could force PAS kinase into an inactive state until the PAS domain binds an endogenous ligand. The binding of ligand to the PAS domain could then, through altering the PAS domain conformation, disrupt the interaction with the kinase domain thereby derepressing kinase activity.
The C-terminus of PAS kinase contains a canonical serine/threonine kinase domain. Presumably kinase activity is the major mode by which PAS kinase mediates its cellular effects; this has been formally demonstrated in yeast[10, 17, 18]. Recently, we determined the structure of the kinase domain of PAS kinase[19]. Many protein kinases are converted from an inactive to an active conformation by phosphorylation of one or more residues in a conserved loop within the kinase domain, known as the activation loop[20]. Most PAS kinase orthologs possess a phosphorylatable threonine residue in the canonical activation loop site, but we made the surprising observation that the kinase domain adopted an active conformation in the absence of activation loop phosphorylation [19]. The evolutionary conservation of this threonine residue raises the possibility that, although phosphorylation on this residue is not required for basal activity, it may be important for modulating kinase activity in some situations. As a part of this study, we also determined the substrate preferences for PAS kinase. We found a strong preference for a basic residue at the -3 position relative to the site of phosphorylation [19], which is fairly common among kinases. We also found an unusual preference for histidine at the -5 position, which has only been previously described for the human LATS kinase and its yeast homolog Cbk1[21]. This unique preference for histidine may prove informative in determining substrates for PAS kinase, which has remained difficult in mammalian systems.
The unusual regulatory properties of PAS kinase might enable it to play a role in coordinating energy sensing with metabolic control. The PAS domain is poised to bind small metabolites and derepress kinase activity in response. In this manner PAS kinase could recognize certain aspects of the nutrient environment and then lead to appropriate metabolic adaptation via phosphorylation of downstream effectors. Thus, PAS kinase could respond to the metabolic status of the cell in order to elicit an appropriate metabolic response.
PAS KINASE AND NUTRIENT PARTITIONING
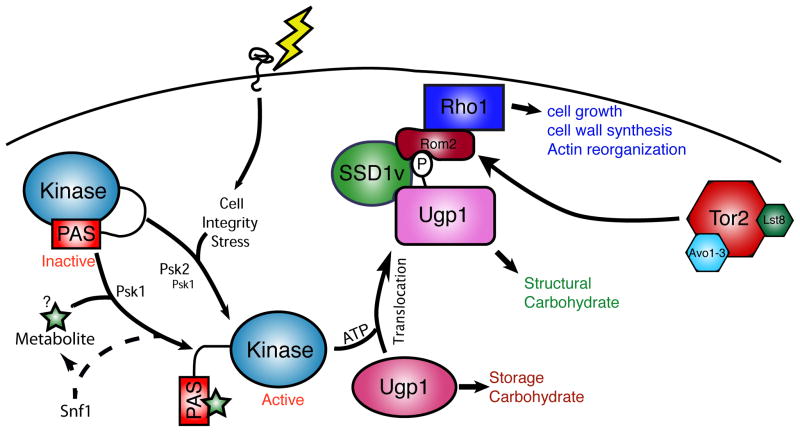
In S. cerevisiae, PAS kinase responds to environmental changes to regulate nutrient partitioning. The two PAS kinase paralogs in yeast, termed PSK1 and PSK2 (together referred to as PAS kinase), are redundant for all known functions[10]. PAS kinase is activated by growth in non-fermentable carbon sources[17], which lead to stimulated respiration and mitochondrial metabolism. Additionally, PAS kinase is activated by various forms of cell wall/cell membrane stress. Upon activation, PAS kinase directly phosphorylates UDP-glucose pyrophosphorylase (Ugp1) at serine-11[18]. Ugp1 catalyzes the formation of UDP-glucose, which is the glucose donor for the production of both glycogen and cell wall glucans, which are the major structural component of the yeast cell wall[22]. Under conditions of severe cell integrity stress, the cell increases the partitioning of glucose toward cell wall synthesis to stabilize this essential structure. One way this partitioning is enacted is through PAS kinase-dependent phosphorylation of Ugp1. Interestingly, PAS kinase-dependent phosphorylation of Ugp1 does not affect the enzymatic activity of Ugp1[10]. However, it induces a conformational change that can be detected either by limited proteolysis or by ion-exchange chromatography, and thereby enacts a metabolic switch. Unphosphorylated Ugp1 produces UDP-glucose that preferentially contributes to the synthesis of glycogen. Phosphorylated Ugp1 produces UDP-glucose that is preferentially incorporated into cell wall glucans at the expense of glycogen. We provided evidence that phosphorylated Ugp1 translocates from the cytoplasm to the cell periphery, which enables its UDP-glucose product to be preferentially used in cell wall synthesis, part of which happens there. The role of PAS kinase in glucose partitioning is completely dependent on phosphorylation of Ugp1. A psk1Δ psk2Δ strain shows a dramatic increase in glycogen production at the expense of cell wall production, which is completely phenocopied in a UGP1-S11A mutant[10, 17]. The shift in glucose utilization in the psk1Δ psk2Δ double mutant or in the UGP1-S11A mutant is associated with increased sensitivity to cell wall perturbing agents. Thus, PAS kinase activation allows the cell to respond to harsh extracellular conditions and continue to survive and grow.
Glucose is a finite resource and is the preferred carbon source for the single-cell eukaryote S. cerevisiae. Each cell must strike a balance between consumption of glucose for energy production, utilization of glucose as a building block and storage of glucose for future use. In order to determine where to partition its limited supply of glucose, the cell must continuously monitor its dynamic metabolic state. In glucose replete conditions, S. cerevisiae represses genes involved in mitochondrial biogenesis and oxidative phosphorylation, a process called glucose repression[23]. Under these conditions, glycolysis is the predominant source of ATP. However, as extracellular glucose is depleted, the cell must be able to utilize other energy sources in order to survive and grow. This requires reversal of glucose repression (derepression) and up-regulation of oxidative phosphorylation[24]. Multiple signaling mechanisms are involved in glucose derepression, the majority of which depend upon the yeast AMPK homolog Snf1[25–27]. Similar to mammalian AMPK, Snf1 is activated by an increase in AMP concentrations [27, 28]. Activation of Snf1 is associated with an increase in mitochondrial biogenesis, oxidative phosphorylation and expression of proteins involved in ATP production from non-glucose substrates[3]. Snf1 activation also leads to activation of PAS kinase and Ugp1 phosphorylation, which induces altered glucose partitioning from storage as glycogen to utilization in cell wall synthesis[17]. The mechanistic details of the relationship between PAS kinase and Snf1 remain unknown. It is possible that the altered metabolic environment present upon Snf1 activation causes PAS kinase activation. It is also possible, however, that Snf1-dependent PASK activation is a concerted response to force utilization of the available glucose for critical cellular processes (like cell wall synthesis) rather than for storage or energy production.
PAS KINASE AND GROWTH CONTROL
Given the fundamental importance of coordinating growth decisions with available energy resources, the mechanisms, proteins and pathways that manage this relationship are typically evolutionarily conserved. As a result, S. cerevisiae has proven to be a valuable tool for deciphering these signaling mechanisms. Because cell growth and division are very expensive processes energetically, the cell must constantly monitor the nutrient environment and react rapidly and appropriately to changes in nutrient availability. Cells have developed several mechanisms that allow them to sense nutrient availability and alter their growth rate. One such mechanism is dependent on the Target of Rapamycin (TOR) protein. From yeast to mammals, TOR has been shown to be a master regulator of cellular growth and division[2, 7, 29, 30]. TOR is activated by nutrients and promotes both temporal and spatial growth. The S. cerevisiae genome, as with PAS kinase, contains two paralogs of TOR: TOR1 and TOR2. The protein products of these two genes function in two distinct complexes termed TOR complex 1 (TORC1) and TOR complex 2 (TORC2)[29]. TORC1, which contains Tor1 or Tor2 as well as Kog1 (raptor), Tco89 and Lst8, regulates protein synthesis and cell size in response to nutrient availability[29, 31]. TORC1 function is sensitive to the inhibitor rapamycin. TORC2, which contains Tor2 (but not Tor1), Avo1, Avo2, Avo3 (Tsc11 or rictor), Bit61 and Lst8, is essential for cell division and cell cycle- dependent polarization of the actin cytoskeleton[32]. TORC2 function is not directly sensitive to rapamycin.
TORC1 promotes spatial growth (i.e. increases cell size) through an increase in protein translation[33–35]. TORC1 stabilizes the 5’-cap dependent initiation factor, eIF4G, and upregulates the translation of ribosomal proteins. Interestingly, a screen for PAS kinase substrates revealed direct phosphorylation of a number of proteins involved in translational regulation. One protein found to be a phosphorylation target of PAS kinase was Cap-Associated Factor 20 (Caf20) in complex with eukaryotic initiation factor 4E (eIF4E)[18]. Caf20 negatively regulates translation by blocking assembly of the translation initiation complex[36]. In higher organisms, the Caf20 homologs (4E-BPs) are directly phosphorylated by the TORC1 complex, causing increased eIF4E activity and protein synthesis[33]. PAS kinase can also phosphorylate Tif11, which is the yeast eukaryotic translation initiation factor 1A (eIF1A)[18]. eIF1A mediates Met-tRNA transfer to the 40S preinitiation complex[37]. These data indicate that PAS kinase may, like TOR, regulate protein synthesis through phosphorylation of downstream effectors, but the significance of its effect on translation has not been defined.
TOR not only regulates translation and spatial growth, but also controls cell division as a component of TORC2. TORC2 activates temporal growth through reorganization and polarization of the actin cytoskeleton by activating the small GTPase Rho1[38–40]. S. cerevisiae TOR2 is an essential gene given its obligate role in the TORC2 complex. In order to better understand the function of TOR2, a temperature sensitive allele of TOR2 (tor2ts) was isolated that is non-functional at the restrictive temperature (37°C)[30]. Therefore, a tor2ts mutant is inviable at 37°C, but it can be rescued by over-expression of Rho1 and its relatives, which function downstream of TORC2 [41]. Interestingly, we have found that the lethality of the tor2ts can also be suppressed by over-expression of either PSK1 or PSK2[42]. PAS kinase-dependent suppression of the tor2ts requires both Ugp1 phosphorylation and Rho1 activation. Our biochemical experiments indicate that phospho-Ugp1 nucleates the formation of a signaling complex that includes Rom2, a Rho1 activating guanine nucleotide exchange factor and the RNA binding protein Ssd1. The complex is able to induce activation of Rho1, presumably through the activity of Rom2. Active Rho1 then promotes cell growth and cell division through activation of the MAP kinase cascade and cell wall synthesis enzymes like the β-1,3-glucan synthase Fks1. Activation of the MAPK cascade in yeast leads to actin reorganization and transcriptional up-regulation of genes involved in cell wall biogenesis[43]. Rho1 also directly regulates cell wall biogenesis via binding and activation of Fks1[44].
These observations indicate that PAS kinase-dependent Ugp1phosphorylation generates a pro-growth signal while at the same time stimulating a pro-growth metabolic program. It increases cell wall biogenesis, which is required to accommodate the increase in cell size associated with cell division[10], and simultaneously nucleates the formation of a pro-growth signaling complex resulting in Rho1 activation[42]. It is not clear if these two functions are inter-dependent or if they operate in complete isolation from one another. For example, two independent populations of Ugp1 may be responsible for these two distinct outputs or Ugp1 may perform both activities simultaneously. It is also unclear what function Ssd1, which has been shown to regulate mRNA stability and translation, may have in the pro-growth signaling complex[45]. However, one mechanism of spatial growth control is dependent upon localized regulation of mRNA stability and translation[46]. It is therefore possible that the Ugp1-Rom2-Ssd1 complex also regulates some aspects of spatial growth through Ssd1-dependent control of RNA stability or translation. The coordination of spatial and temporal growth and their integration with the nutrient environment are critically important processes in all living organisms. PAS kinase appears to tie these stimuli and processes together in yeast. Based on the evolutionary conservation of PAS kinase sequence and structure, we predict that it would act similarly in mammals, but such processes are much more complicated in complex, multicellular mammals.
PAS KINASE FUNCTION IN MAMMALS
Defects in nutrient partitioning lie at the heart of the metabolic alterations that characterize many human pathologies, including obesity, diabetes and cancer. TOR and AMPK are master regulators of cellular nutrient utilization in mammals[47]. As in yeast, mammalian TOR functions in two distinct complexes termed mTORC1 and mTORC2[2]. The functions of the TOR complexes to control cell growth and cell division in response to environmental conditions is evolutionarily conserved throughout the eukaryotic kingdom. The role of mTORC1 in driving inappropriate cell proliferation is now well appreciated and is a high value therapeutic target in cancer. Small molecule inhibition of mTORC1 by rapamycin, as well as second generation rapamycin analogs (rapalogs)[48], have proven effective against certain types of cancer[49]. Some data also suggest that mTORC1 inhibition might be of therapeutic benefit for obesity and diabetes[50]. Pharmacological activation of AMPK by biguanide drugs like metformin, however, has been used as a treatment for type 2 diabetes for over 40 years[51]. AMPK and TOR exemplify the intimate connection between the pathways that monitor energy availability and the cellular decisions related to growth, division and nutrient consumption and storage. Given the commonalities in function, we postulate that PAS kinase might also be a part of the cellular decision-making apparatus concerning nutrient partitioning, thereby controlling cell growth and energy homeostasis.
We, and others, have found that mice lacking PAS kinase (PASK−/−) exhibit a number of tissue specific defects in metabolic control[9]. Specifically, PASK−/− mice exhibit impaired glucose-stimulated insulin secretion from pancreatic β-cells, altered triglyceride storage in the liver, and increased metabolic rate in skeletal muscle. The role of PASK in glucose-responsiveness in β-cells is the best understood and PASK appears to act through controlling activity of the promoter driving insulin gene expression [52–55]. In the liver, PASK is required for the normal expression of genes involved in fatty acid synthesis, uptake and storage, which could partially explain the protection from high fat diet-induced fatty liver observed in the PASK−/− mice[9]. The mechanism(s) by which PASK affects metabolic rate in skeletal muscle remains unknown. Mitochondrial morphology and number were unaffected by PASK deletion in soleus muscle, while oxygen consumption and ATP production were elevated. This increased metabolic rate is almost certainly part of the mechanism underlying the relatively lean phenotype of the PASK−/− mice on a high fat diet. We also found that the hypermetabolic phenotype observed in skeletal muscle was recapitulated in cultured cells. Silencing of PASK in cultured myoblasts resulted in increased glucose and palmitate oxidation[9]. These data strongly suggest that PASK is required for normal nutrient partitioning and energy homeostasis in the mouse and that this activity is primarily carried out in a cell-autonomous fashion.
As described above, PASK enacts a pro-growth (broadly defined) program both in yeast and mammals. The fact that PASK itself is regulated nutritionally places this protein kinase in the class of proteins, like AMPK and TOR, which monitor the cellular and organismal metabolic state and initiate or suppress growth signals. We speculate that, also like AMPK and TOR, PASK may be a viable drug target not only for obesity and diabetes, for which some evidence already exists, but for some cancers as well. This question awaits further studies, not only to define the mechanism(s) of metabolic control elicited by PASK, but also to explore the relevance of these mechanisms to human disease.
Figure 1. PAS kinase signaling in S. Cerevisiae.
PAS kinase is activated by cell intergrity stress and non-fermentable carbon sources. Activation of PAS kinase leads to phosphorylation of Ugp1. Phospho-Ugp1 translocates from the cytoplasm to the cell periphery. At the cell periphery Ugp1 produces UDP-glucose that is a substrate for cell wall synthesis. Phospho-Ugp1 also nucleates the formation of a pro-growth complex involved in Rho1 activation.
Highlights.
PAS kinase integrates nutrient availability to nutrient partitioning
Growth and nutrient availability are tightly coupled through intracellular signaling
PAS kinase is required for normal nutrient utilization
PAS kinase activates Rho1 to promote growth
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hardie DG. AMPK and autophagy get connected. Embo J. 2011;30:634–5. doi: 10.1038/emboj.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–85. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 4.Shaw RJ. LKB1 and AMP-activated protein kinase control of mTOR signalling and growth. Acta Physiol (Oxf) 2009;196:65–80. doi: 10.1111/j.1748-1716.2009.01972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardie DG. Roles of the AMP-activated/SNF1 protein kinase family in the response to cellular stress. Biochem Soc Symp. 1999;64:13–27. [PubMed] [Google Scholar]
- 6.Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1:15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–75. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 8.Rutter J, Michnoff CH, Harper SM, Gardner KH, McKnight SL. PAS kinase: an evolutionarily conserved PAS domain-regulated serine/threonine kinase. Proc Natl Acad Sci U S A. 2001;98:8991–6. doi: 10.1073/pnas.161284798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hao HX, Cardon CM, Swiatek W, Cooksey RC, Smith TL, Wilde J, et al. PAS kinase is required for normal cellular energy balance. Proc Natl Acad Sci U S A. 2007;104:15466–71. doi: 10.1073/pnas.0705407104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith TL, Rutter J. Regulation of glucose partitioning by PAS kinase and Ugp1 phosphorylation. Mol Cell. 2007;26:491–9. doi: 10.1016/j.molcel.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 11.Moglich A, Ayers RA, Moffat K. Structure and signaling mechanism of Per-ARNT-Sim domains. Structure. 2009;17:1282–94. doi: 10.1016/j.str.2009.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitanishi K, Igarashi J, Hayasaka K, Hikage N, Saiful I, Yamauchi S, et al. Heme-binding characteristics of the isolated PAS-A domain of mouse Per2, a transcriptional regulatory factor associated with circadian rhythms. Biochemistry. 2008;47:6157–68. doi: 10.1021/bi7023892. [DOI] [PubMed] [Google Scholar]
- 13.Stephenson K, Hoch JA. PAS-A domain of phosphorelay sensor kinase A: a catalytic ATP-binding domain involved in the initiation of development in Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98:15251–6. doi: 10.1073/pnas.251408398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soshilov A, Denison MS. Role of the Per/Arnt/Sim domains in ligand-dependent transformation of the aryl hydrocarbon receptor. J Biol Chem. 2008;283:32995–3005. doi: 10.1074/jbc.M802414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monson EK, Ditta GS, Helinski DR. The oxygen sensor protein, FixL, of Rhizobium meliloti. Role of histidine residues in heme binding, phosphorylation, and signal transduction. J Biol Chem. 1995;270:5243–50. doi: 10.1074/jbc.270.10.5243. [DOI] [PubMed] [Google Scholar]
- 16.Amezcua CA, Harper SM, Rutter J, Gardner KH. Structure and interactions of PAS kinase N-terminal PAS domain: model for intramolecular kinase regulation. Structure. 2002;10:1349–61. doi: 10.1016/s0969-2126(02)00857-2. [DOI] [PubMed] [Google Scholar]
- 17.Grose JH, Smith TL, Sabic H, Rutter J. Yeast PAS kinase coordinates glucose partitioning in response to metabolic and cell integrity signaling. Embo J. 2007;26:4824–30. doi: 10.1038/sj.emboj.7601914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rutter J, Probst BL, McKnight SL. Coordinate regulation of sugar flux and translation by PAS kinase. Cell. 2002;111:17–28. doi: 10.1016/s0092-8674(02)00974-1. [DOI] [PubMed] [Google Scholar]
- 19.Kikani CK, Antonysamy SA, Bonanno JB, Romero R, Zhang FF, Russell M, et al. Structural bases of PAS domain-regulated kinase (PASK) activation in the absence of activation loop phosphorylation. J Biol Chem. 2010;285:41034–43. doi: 10.1074/jbc.M110.157594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–58. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 21.Mazanka E, Alexander J, Yeh BJ, Charoenpong P, Lowery DM, Yaffe M, et al. The NDR/LATS family kinase Cbk1 directly controls transcriptional asymmetry. PLoS Biol. 2008;6:e203. doi: 10.1371/journal.pbio.0060203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daran JM, Dallies N, Thines-Sempoux D, Paquet V, Francois J. Genetic and biochemical characterization of the UGP1 gene encoding the UDP-glucose pyrophosphorylase from Saccharomyces cerevisiae. Eur J Biochem. 1995;233:520–30. doi: 10.1111/j.1432-1033.1995.520_2.x. [DOI] [PubMed] [Google Scholar]
- 23.Szekely E, Montgomery DL. Glucose represses transcription of Saccharomyces cerevisiae nuclear genes that encode mitochondrial components. Mol Cell Biol. 1984;4:939–46. doi: 10.1128/mcb.4.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parrou JL, Enjalbert B, Plourde L, Bauche A, Gonzalez B, Francois J. Dynamic responses of reserve carbohydrate metabolism under carbon and nitrogen limitations in Saccharomyces cerevisiae. Yeast. 1999;15:191–203. doi: 10.1002/(SICI)1097-0061(199902)15:3<191::AID-YEA358>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 25.Schuller HJ, Entian KD. Isolation and expression analysis of two yeast regulatory genes involved in the derepression of glucose-repressible enzymes. Mol Gen Genet. 1987;209:366–73. doi: 10.1007/BF00329667. [DOI] [PubMed] [Google Scholar]
- 26.Wright RM, Poyton RO. Release of two Saccharomyces cerevisiae cytochrome genes, COX6 and CYC1, from glucose repression requires the SNF1 and SSN6 gene products. Mol Cell Biol. 1990;10:1297–300. doi: 10.1128/mcb.10.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson WA, Hawley SA, Hardie DG. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996;6:1426–34. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- 28.Chandrashekarappa DG, McCartney RR, Schmidt MC. Subunit and domain requirements for adenylate-mediated protection of Snf1 activation loop from dephosphorylation. J Biol Chem. 2011 doi: 10.1074/jbc.M111.315895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–68. doi: 10.1016/s1097-2765(02)00636-6. [DOI] [PubMed] [Google Scholar]
- 30.Helliwell SB, Wagner P, Kunz J, Deuter-Reinhard M, Henriquez R, Hall MN. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol Biol Cell. 1994;5:105–18. doi: 10.1091/mbc.5.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature. 1999;402:689–92. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- 32.Kamada Y, Fujioka Y, Suzuki NN, Inagaki F, Wullschleger S, Loewith R, et al. Tor2 directly phosphorylates the AGC kinase Ypk2 to regulate actin polarization. Mol Cell Biol. 2005;25:7239–48. doi: 10.1128/MCB.25.16.7239-7248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beretta L, Gingras AC, Svitkin YV, Hall MN, Sonenberg N. Rapamycin blocks the phosphorylation of 4E-BP1 and inhibits cap-dependent initiation of translation. Embo J. 1996;15:658–64. [PMC free article] [PubMed] [Google Scholar]
- 34.Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell. 2004;119:969–79. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 35.Crespo JL, Hall MN. Elucidating TOR signaling and rapamycin action: lessons from Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2002;66:579–91. doi: 10.1128/MMBR.66.4.579-591.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altmann M, Schmitz N, Berset C, Trachsel H. A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. Embo J. 1997;16:1114–21. doi: 10.1093/emboj/16.5.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei CL, Kainuma M, Hershey JW. Characterization of yeast translation initiation factor 1A and cloning of its essential gene. J Biol Chem. 1995;270:22788–94. doi: 10.1074/jbc.270.39.22788. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt A, Kunz J, Hall MN. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc Natl Acad Sci U S A. 1996;93:13780–5. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell. 1993;73:585–96. doi: 10.1016/0092-8674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 40.Jacinto E, Loewith R, Schmidt A, Lin S, Ruegg MA, Hall A, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–8. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt A, Bickle M, Beck T, Hall MN. The yeast phosphatidylinositol kinase homolog TOR2 activates RHO1 and RHO2 via the exchange factor ROM2. Cell. 1997;88:531–42. doi: 10.1016/s0092-8674(00)81893-0. [DOI] [PubMed] [Google Scholar]
- 42.Cardon CM, Beck T, Hall MN, Rutter J. PAS kinase promotes cell survival and growth via activation of Rho1. Science Signaling. doi: 10.1126/scisignal.2002435. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Helliwell SB, Schmidt A, Ohya Y, Hall MN. The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr Biol. 1998;8:1211–4. doi: 10.1016/s0960-9822(07)00511-8. [DOI] [PubMed] [Google Scholar]
- 44.Qadota H, Python CP, Inoue SB, Arisawa M, Anraku Y, Zheng Y, et al. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science. 1996;272:279–81. doi: 10.1126/science.272.5259.279. [DOI] [PubMed] [Google Scholar]
- 45.Jansen JM, Wanless AG, Seidel CW, Weiss EL. Cbk1 regulation of the RNA-binding protein Ssd1 integrates cell fate with translational control. Curr Biol. 2009;19:2114–20. doi: 10.1016/j.cub.2009.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berset C, Trachsel H, Altmann M. The TOR (target of rapamycin) signal transduction pathway regulates the stability of translation initiation factor eIF4G in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1998;95:4264–9. doi: 10.1073/pnas.95.8.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hardie DG. AMPK and Raptor: matching cell growth to energy supply. Mol Cell. 2008;30:263–5. doi: 10.1016/j.molcel.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 48.Carew JS, Kelly KR, Nawrocki ST. Mechanisms of mTOR inhibitor resistance in cancer therapy. Target Oncol. 2011;6:17–27. doi: 10.1007/s11523-011-0167-8. [DOI] [PubMed] [Google Scholar]
- 49.Cloughesy TF, Yoshimoto K, Nghiemphu P, Brown K, Dang J, Zhu S, et al. Antitumor activity of rapamycin in a Phase I trial for patients with recurrent PTEN-deficient glioblastoma. PLoS Med. 2008;5:e8. doi: 10.1371/journal.pmed.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inoki K, Kim J, Guan KL. AMPK and mTOR in Cellular Energy Homeostasis and Drug Targets. Annu Rev Pharmacol Toxicol. 2011 doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- 51.Fogarty S, Hardie DG. Development of protein kinase activators: AMPK as a target in metabolic disorders and cancer. Biochim Biophys Acta. 2010;1804:581–91. doi: 10.1016/j.bbapap.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 52.da Silva Xavier G, Farhan H, Kim H, Caxaria S, Johnson P, Hughes S, et al. Per-arnt-sim (PAS) domain-containing protein kinase is downregulated in human islets in type 2 diabetes and regulates glucagon secretion. Diabetologia. 2011;54:819–27. doi: 10.1007/s00125-010-2010-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.da Silva Xavier G, Rutter J, Rutter GA. Involvement of Per-Arnt-Sim (PAS) kinase in the stimulation of preproinsulin and pancreatic duodenum homeobox 1 gene expression by glucose. Proc Natl Acad Sci U S A. 2004;101:8319–24. doi: 10.1073/pnas.0307737101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fontes G, Semache M, Hagman DK, Tremblay C, Shah R, Rhodes CJ, et al. Involvement of Per-Arnt-Sim Kinase and extracellular-regulated kinases-1/2 in palmitate inhibition of insulin gene expression in pancreatic beta-cells. Diabetes. 2009;58:2048–58. doi: 10.2337/db08-0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Semplici F, Vaxillaire M, Fogarty S, Semache M, Bonnefond A, Fontes G, et al. A human mutation within the per-ARNT-sim (PAS) domain-containing protein kinase (PASK) causes basal insulin hypersecretion. J Biol Chem. 2011 doi: 10.1074/jbc.M111.254995. [DOI] [PMC free article] [PubMed] [Google Scholar]