Abstract
Alagille syndrome (ALGS, OMIM #118450) is an autosomal dominant disorder that affects multiple organ systems including the liver, heart, eyes, vertebrae, and face. ALGS is caused by mutations in one of two genes in the Notch Signaling Pathway, JAGGED1 or NOTCH2. In this study, analysis of 21 Vietnamese ALGS individuals led to the identification of 19 different mutations (18 JAGGED1 and 1 NOTCH2), 17 of which are novel, including the third reported NOTCH2 mutation in Alagille Syndrome. The spectrum of JAGGED1 mutations in the Vietnamese patients is similar to that previously reported, including nine frameshift, three missense, two splice site, one nonsense, two whole gene, and onw partial gene deletion. The missense mutations are all likely to be disease causing, as two are loss of cysteines (C22R and C78G) and the third creates a cryptic splice site in exon 9 (G386R). No correlation between genotype and phenotype was observed. Assessment of clinical phenotype revealed that skeletal manifestations occur with a higher frequency than in previously reported Alagille cohorts. Facial features were difficult to assess and a Vietnamese pediatric gastroenterologist was only able to identify the facial phenotype in 61% of the cohort. To assess the agreement among North American dysmorphologists at detecting the presence of ALGS facial features in the Vietnamese patients, 37 clinical dysmorphologists evaluated a photographic panel of 20 Vietnamese children with and without ALGS. The dysmorphologists were unable to identify the individuals with ALGS in the majority of cases, suggesting that evaluation of facial features should not be used in the diagnosis of ALGS in this population. This is the first report of mutations and phenotypic spectrum of ALGS in a Vietnamese population.
Keywords: Alagille Syndrome, JAGGED1, NOTCH2, JAGGED1 missense mutation
INTRODUCTION
Alagille syndrome (ALGS, OMIM# 118450) is a complex, multisystem, autosomal dominant disorder characterized by variable expressivity of developmental abnormalities in several organs including the liver, heart, eyes, vertebrae, kidneys, and face [Alagille et al., 1975; Alagille et al., 1987]. ALGS is the second most common cause of intrahepatic cholestasis in infancy and occurs with an estimated frequency of 1:30,000 [Kamath et al., 2003]. Traditionally, the diagnosis of ALGS is based on a combination of the presence of intrahepatic bile duct paucity on liver biopsy in association with at least three of the major clinical features. Major clinical features include: chronic cholestasis, cardiac disease, skeletal abnormalities, ocular abnormalities, renal abnormalities, vascular anomalies, and characteristic facial features [Elmslie et al. 1995; Emerick et al., 1999; Krantz et al., 1999].
ALGS is caused by mutations in or deletions of either the JAGGED1 (JAG1) or NOTCH2 genes [Oda et al., 1997; Li et al., 1997; McDaniell et al., 2006]. Both genes are involved in the Notch signaling pathway (NSP), an intercellular signaling mechanism, which plays a key role in cell fate determination. NOTCH2 encodes one of the four Notch receptors and JAG1 encodes a ligand that binds to the Notch receptors. This binding results in the release of the Notch intracellular domain, allowing for activation of transcription factors that play a role in cell differentiation and morphogenesis [Artavanis-Tsakonas et al., 1999]. Mutations in JAG1, which is found on chromosome 20, can be identified in over 90% of clinically diagnosed individuals with ALGS [Warthen et al., 2006]. Currently, 377 different JAG1 gene mutations have been identified in ALGS patients [Krantz et al., 1998; Yuan et al., 1998, 2001; Crosnier et al., 1999, 2000; Onouchi et al., 1999; Pilia et al., 1999; Heritage et al., 2000, 2002; Colliton et al., 2001; Giannakudis et al., 2001; Ropke et al., 2002; Jurkiewicz et al., 2005]. More recently, mutations in NOTCH2 were found to cause ALGS and two NOTCH2 mutations have been reported to date [McDaniell et al. 2006].
The identification of the genetic basis of ALGS and the availability of molecular testing has led to a proposed revision in the diagnostic criteria, suggesting that the presence of a disease causing mutation in addition to only one major clinical feature is enough for the diagnosis of ALGS [Kamath et al., 2003]. Of the major clinical features, the characteristic facial dysmorphism is one of the most penetrant findings and is the only one that is dependent on actual clinician assessment instead of diagnostic testing. Characteristic facial features of ALGS consist of a prominent forehead, deep-set eyes that are hyperteloric, a straight nose with a flattened tip, and a prominent, pointed chin. Early data suggested that these facial features were associated with cholestasis and not unique to ALGS [Sokol et al., 1983]. However, a study by Kamath et al. [2002] demonstrated that the diagnostic specificity of ALGS facial features in a Caucasian cohort was above 80%, as determined by a group of clinical dysmorphologists. The hypothesis that the facial features of ALGS are unique to this syndrome is further supported by the expression of JAG1 in the developing mouse facial bones [Kamath et al., 2002]. Thus, it has recently been widely accepted that ALGS facies are unique to this syndrome. The prevalence of the facial phenotype in JAG1-mutation positive individuals is over 90% [Emerick et al. 1999] and a similar prevalence has been seen in population-based studies reporting unique JAG1 mutations in Italian, Australian, and Polish populations [Pilia et al., 1999; Heritage et al., 2000, 2002; Ropke et al., 2002; Jurkiewicz et al., 2005]. There have also been reports of ALGS in Asian populations, including Japanese, Korean, and Chinese [Yuan et al., 1998, 2001; Onouchi et al., 1999; Kasahara et al., 2003; Kim et al., 2005; Wang et al., 2008] and all of these described characteristic ALGS facial features, although only one study specifically mentioned confirmation of the facies by a clinical dysmorphologist. In this study we present the results of JAG1 and NOTCH2 mutation analysis of 21 Vietnamese patients with ALGS and a systematic assessment of the agreement among North American dysmorphologists at detecting the presence of ALGS facial features in this population.
MATERIALS AND METHODS
Patients
Twenty-one unrelated probands were referred from the Department of Gastroenterology at Children’s Hospital #1 in Ho Chi Minh City, Vietnam under a protocol of informed consent and enrolled into our IRB approved study at The Children’s Hospital of Philadelphia. Patients were enrolled from April 2007 to October 2010. Most of the patients in this study were initially referred to the Department of Gastroenterology at Children’s Hospital #1 in Ho Chi Minh City, Vietnam with the chief complaint of jaundice. A few were referred for cardiovascular or renal disease plus hepatitis. Initial screening for ALGS consisted of routine blood work to assess liver function and a clinical exam looking for classic ALGS facial features, hepatosplenomegaly, and the presence of a heart murmur. Presence of any clinical features or an elevated GGT led to additional analysis consisting of an echocardiogram, x-ray of the spine, and an abdominal ultrasound. If a patient had three or more of the major clinical features of ALGS, the patient was referred for genetic testing. All patients met the standard clinical diagnostic criteria for ALGS [Alagille et al., 1987], although not all individuals had a liver biopsy performed. Blood samples were collected from each patient and using standard procedures, genomic DNA was extracted from peripheral blood leukocytes, and a lymphoblastoid cell line was also created.
Mutation Detection
The 26 exons of the JAG1 gene were amplified by polymerase chain reactions (PCR) as previously described [Krantz et al., 1998; Colliton et al., 2001; Warthen et al., 2006]. PCR products of different exons were purified with ExoSAP-IT (USB Corporation) and then sequenced with the BigDye Terminator v3.1 Cycle Sequencing Kit on an ABI 3730 DNA Analyzer (Applied Biosystems) in the Nucleic Acid and Protein Core Facility at the Children’s Hospital of Philadelphia. Sequences of analyzed fragments were compared with the JAG1 cDNA sequence (JAG1 mRNA GenBank RefSeq: NM_000214.1). All mutations were numbered based on the cDNA reference sequence, with position +1 corresponding to the A of the ATG translation initiation codon. If no mutations in JAG1 were detected, large deletion screening was performed using either a single-nucleotide polymorphism (SNP) array or multiplex ligationdependent probe amplification (MLPA) to detect exonic deletions as previously described [Kamath et al., 2009; Cho et al., 2009]. SNP array analysis was carried out using the Illumina Infinium SNP genotyping platform (HumanHap610 chips, BeadStation Scanner, and BeadStudio analysis software). If no large-scale JAG1 deletions were detected, PCR reactions were carried out with 36 primer pairs required to amplify the 34 exons of the NOTCH2 gene. Sequencing and determination of mutations in NOTCH2 followed the same protocol as for JAG1 screening (NOTCH2 mRNA GenBank RefSeq: NM_024408).
Facial Features Questionnaire
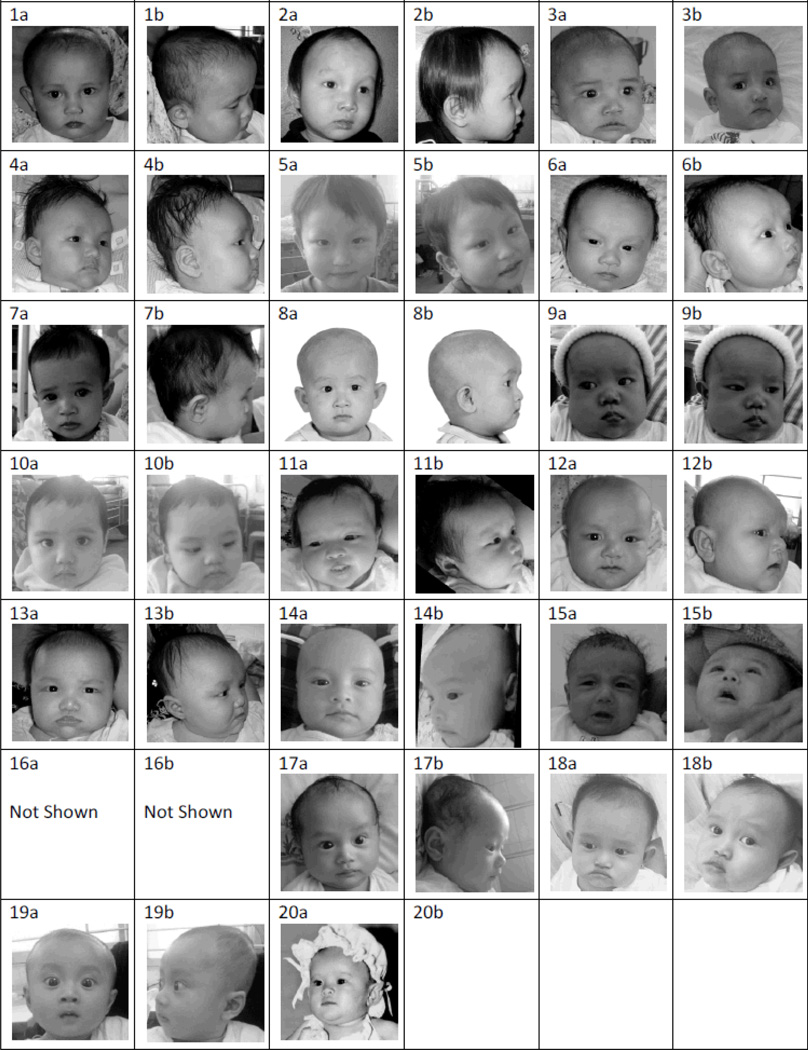
To evaluate the ability of North American dysmorphologists to assess ALGS facial features in the Vietnamese patients, a photographic panel was created of 20 Vietnamese children, which included 16 children with ALGS and 4 children without ALGS who had no known clinical liver disease. All ALGS subjects met the classic clinical diagnostic criteria for ALGS [Alagille et al., 1987]. Fourteen of the 16 ALGS subjects had mutations (13 JAG1 mutations and 1 NOTCH2 mutation), as described in the results section of this paper. The panel included ALGS children of both genders and of various ages. The non-ALGS subjects were matched by representative gender and ages and were randomly selected from patients seen at the Children’s Hospital #1 in Ho Chi Minh City, Vietnam. All patients with the exception of one were photographed in two views: full face and profile (Fig 1). A profile view was not available from one of the subjects (Photographic Panel Subject #20). Informed consent was obtained for use of the photographs in the study.
Figure 1.
Vietnamese Photographic Panel - Full-face and profile views of the individuals in the photographic panel distributed to the dysmorphologists participating in the study. Original dimensions of all images were 7 × 7 cm and all photographs were printed in gray-scale. Proband number 5, 10, 14, and 19 do not have ALGS. All other probands have ALGS.
A photographic panel consisting of two 7cm by 7cm black-and white pictures of each individual (full face and profile) along with a score sheet were distributed to participants of the 2010 David W. Smith Annual Workshop on Malformations and Morphogenesis (Supplemental Figure 1). Participants analyzed the photographs on the basis of facial features to determine if the individuals in the photographs had features consistent with ALGS. All participants had no prior knowledge of the patients or their actual diagnoses. Participant responses were compared with each other to determine the percentage agreement on analysis of facial features.
RESULTS
Mutations in JAG1 or NOTCH2 were found in 19 out of the 21 individuals (90%) with the clinical diagnosis of ALGS (Table I). A total of 18 JAG1 mutations (86%) and 1 NOTCH2 mutation (5%) were found, 17 of which are novel. Sixteen of the JAG1 mutations and the NOTCH2 mutation have not been previously described. The NOTCH2 mutation identified is a nonsense mutation with a substitution C to T at position 6007 (exon 33, codon 2003) and is predicted to lead to the creation of a protein which is truncated within the ankyrin repeats located in the intracellular domain of the protein.
Table I.
Gene Mutations in Vietnamese ALGS Patients
| Patient No. |
Gene | Location | Mutation Position a, b | Predicted Consequence c |
Protein Domain d |
Origin e | Mutation Type |
|---|---|---|---|---|---|---|---|
| 1 | JAG1 | Exon 1 | c.64T→C | p.Cys22Arg | SP | ND | Missense |
| 2 | JAG1 | Exon 2 | c.232T→G | p.Cys78Gly | NL | de novo | Missense |
| 3 | JAG1 | Exon 2 | c.338insC | p.Arg113fs | 1 | de novo | Frame Shift |
| 4 | JAG1 | Exon 3 | c.337_338+3delTTCGT | p.Val146fs | 1 | de novo | Frame Shift |
| 5 | JAG1 | Exon 6 | c.839G→A | p.Trp280X | EGF 2 | de novo | Nonsense |
| 6 | JAG1 | Exon 8 | c.1019_1022delGCCT | p.Cys340fs | EGF4 | ND | Frame Shift |
| 7 | JAG1 | Exon 9 | c.1156G→A | p.Gly386Arg | EGF 5 | de novo | Missense f |
| 8 | JAG1 | Exon 9 | c.1205delC | p.Pro402fs | EGF 5 | de novo | Frame Shift |
| 9 | JAG1 | Exon 13 | c.1678delT | p.Cys560fs | EGF 9 | de novo | Frame Shift |
| 10 | JAG1 | Exon 17 | c.2122_2125delCAGT | p. Gln708fs | EGF 13 | de novo | Frame Shift |
| 11 | JAG1 | Exon 18 | c.2269_2270dupGG | p.Gly757fs | EGF 14 | ND | Frame Shift |
| 12 | JAG1 | Exon 20 | c.2458+1delG | r. spl? | EGF 15-16 | de novo | Splice Site |
| 13 | JAG1 | Exon 21 | c.2572+1 G→T | r. spl? | EGF 16-CR | ND | Splice Site |
| 14 | JAG1 | Exon 22 | c.2587dupT | p.Cys863fs | CR | ND | Frame Shift |
| 15 | JAG1 | Exon 23 | c.2820_2826delTCTCCAG | p.Ser940fs | 2 | ND | Frame Shift |
| 16 | JAG1 | Exons 9-12 | Exons 9-12 | EGF 5-8 | ND | Deletion | |
| 17 | JAG1 | Full gene | Del20: 2.84Mb, 8,647,519-11,490,289 | Gene Deletion | All | ND | Deletion |
| 18 | JAG1 | Full gene | Del20: 839 SNPs, 2.96 Mb, 9,251,107-12,214,763 | Gene Deletion | All | ND | Deletion |
| 19 | NOTCH2 | Exon 33 | c.6007C→T | p.Arg2003X | Ankyrin Repeat | ND | Nonsense |
Novel mutations appear in boldface type
JAG1 cDNA sequence according to the GenBank RefSeq: NM_000214.1, position +1 corresponds to the A of the ATG translation initiation codon in the reference sequence.
ins – insertion, del – deletion, dup – duplication, Mb – megabases, SNP – single nucleotide polymorphism
JAG1 protein sequence according to the GenBank RefSeq: NP_000205.1, the translation initiator methionine is numbered as +1; r.spl? – the change is expected to affect splicing, not experimentally analyzed
SP – signal peptide, NL – Notch ligand, 1 – Unknown protein domain 1, DSL – Delta/Serrate/Lag-2 domain, EGF – epidermal growth factor, CR – cysteine rich region, 2 – Unknown protein domain 2
ND – not determined, parents’ samples not available
Possible Cryptic Splice Site
The eighteen different JAG1 mutations included nine frameshift mutations (50%), 3 missense mutations (16.6%); two splice site mutations (11.1%), one nonsense mutation (5.6%), 2 whole gene deletions (11.1%), and one partial gene deletion (5.6%). The partial gene deletion was noted by MLPA and involves exons 9–12 of JAG1. The two whole gene deletions were detected by SNP array analysis. The chromosome 20 deletion for Patient 17 is predicted to span at least 2.84Mb from position 8,647,519 to 11,490,289. The deletion for Patient 18 is also predicted to span at least 2.96Mb from position 9,251,107 to 12,214,763 on chromosome 20. In addition, 15 different JAG1 polymorphisms, six of which have been previously reported, were identified in the Vietnamese cohort (Supplemental Table I).
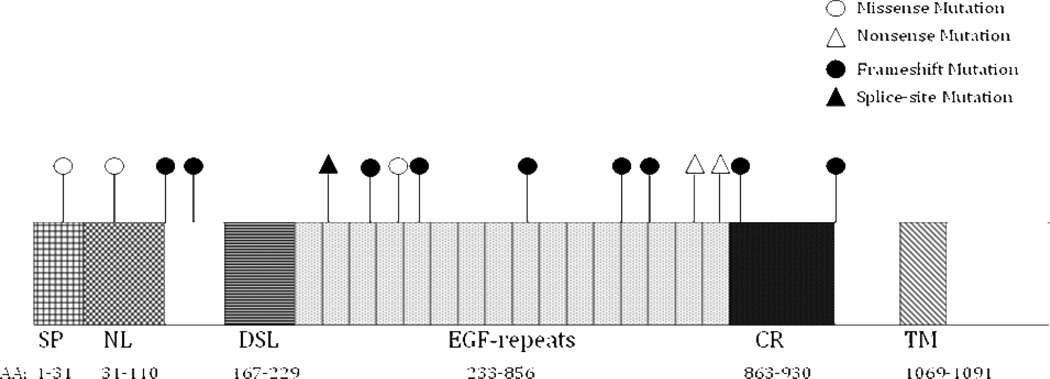
Figure 2 provides a diagram of the amino acid positions compared to the domains of the JAG1 protein. All of the JAG1 mutations map into the extracellular domain of JAG1 protein and the mutations appear to be distributed throughout the JAG1 gene. Fifty-six percent (10/18) of mutations occurred in epidermal growth factor (EGF) repeat regions. Over half of the mutations (nine frameshift and one nonsense) are predicted to lead to premature termination codons. There are two mutations that are located in consensus splice site sequences that interfere with splicing. Patient 13 had a G to T substitution at position 2572, changing the GT to TT at the donor site of intron 21. Patient 12 had a deletion of a single G at the donor site of intron 20. The missense mutations were C22R, C78G and G386R. It is predicted that the loss of cysteine in C22R and C78G can lead to changes in protein function, although no confirmatory studies were performed. G386R, seen in Patient 7, has previously been reported and results in a G to A substitution at position 1156 in exon 9. This substitution results in a CGG to CAG within exon 9, which we predicted could cause the formation of a GT-AG cryptic splice site. Sequencing of cDNA from this JAG1 mutation confirmed this cryptic splice site, as the resultant cDNA was deleted for 37 nucleotides of exon 9, with initiation of the altered cDNA immediately following the new splice site (Supplemental Figure 2).
Figure 2.
JAG1 Protein Map and Mutations
Map of JAG1 mutations of Vietnamese ALGS patients on the JAG1 protein. AA – Amino Acid. Conserved regions of JAG1 include: SP – signal peptide, NL – Notch ligand, DSL – Delta/Serrate/Lag-2 domain, EGF – epidermal growth factor, CR – cysteine rich region, TM – transmembrane domain
Assessment of the ALGS clinical phenotype consisted of evaluation for the five major clinical manifestations as well as looking for renal involvement. ALGS clinical manifestations of the 18 patients with JAG1 mutations are summarized in Table II. Not all patients had full clinical evaluation. Of the 18 patients, the prevalence of the clinical phenotype based on available clinical evaluation data is as follows: liver 100% (18/18), cardiac 94.4% (17/18), ophthalmologic 100% (2/2), skeletal 94.1% (16/17), and renal 27.3% (3/11). The facial phenotype was unable to be accurately determined.
Table II.
Clinical Features of ALGS by Mutation
| Patient No. |
Gene | Mutation | Clinical Features | |||||
|---|---|---|---|---|---|---|---|---|
| Liver | Cardiac | Eyed | Skeletale | Faciesf | Renal | |||
| 1 | JAG1 | c.63T→C | + | ++++ b | ND | + | − | + g |
| 2 | JAG1 | c.232T→G | ++ | − | + | − | ND g | − |
| 3 | JAG1 | c.337insC | + | ++++ | ND | + | − | ND |
| 4 | JAG1 | c.337_338+3delTTCGT | + | ++ c | ND | + | − g | ND |
| 5 | JAG1 | c.8393G→A | + | +++ | ND | + | + g | − |
| 6 | JAG1 | c.1019_1022delGCCT | + | ++ | ND | ND | − g | − |
| 7 | JAG1 | c.1156G→A | + | + b | ND | + | − g | − |
| 8 | JAG1 | c.1205delC | + | + b | ND | + | − | ND |
| 9 | JAG1 | c.1678delT | + | + | ND | + | + g | ND |
| 10 | JAG1 | c.2122_2125delCAGT | + | +++ | ND | + | − | − |
| 11 | JAG1 | c.2269_2270dupGG | ++ a | +++ | ND | + | + g | − |
| 12 | JAG1 | c.2458+1delG | + a | + | ND | + | − g | − |
| 13 | JAG1 | c.2572+1 G→T | + | +++ | ND | + | − g | ND |
| 14 | JAG1 | c.2587dupT | + | ++ | + | + | + | + h |
| 15 | JAG1 | c.2820_2826delTCTCCAG | ++ | +++ | ND | + | − | − |
| 16 | JAG1 | Partial gene deletion | + | +++ | ND | ND | − g | ND |
| 17 | JAG1 | Full gene deletion | + a | +++ | ND | + | − | ND |
| 18 | JAG1 | Full gene deletion | ++ a | + c | ND | + | − g | + i |
| 19 | NOTCH2 | c.6007C→T | ++ a | + | ND | + | + g | + g |
(+) Indicates the presence of a feature; (−) Indicates a feature is not present; ND indicates that this feature was not defined.
In the liver and heart columns the additional (+)’s indicates an increase in the severity of the clinical features as follows:
Liver = (+) cholestasis/jaundice; (++) pruritis
Heart – (+) murmur only, (++) peripheral pulmonary stenosis (PPS), (+++) PPS plus septal abnormality (ASD/VSD), (++++) tetralogy of fallot
Had liver biopsy
Pulmonary atresia
Aortic Stenosis
Posterior Embryotoxon
Butterfly-like vertebrae
Facial phenotype as per our North American Dysmorphologist
Facial phenotype per Vietnamese Pediatric Gastroenterologist
Renal hyperechogenicity
Glomerulopathy
Renal hypoplasia or solitary kidney
We compared the frequency of clinical features in JAG1 mutation-positive Vietnamese individuals with JAG1 mutation-positive ALGS probands in our cohort at The Children’s Hospital of Philadelphia (Table III). Our cohort is primarily Caucasian (75%) and has been partially reported by Emerick et al. [1999] and Warthen et al. [2006]. Since 2006, approximately 100 new individuals have been enrolled. The frequency of clinical phenotypes in the patients we have studied is comparable to the clinical phenotypes seen in previously reported studies, although we find a slighter lower frequency of the ophthalmologic (68.3% to 80%) and the renal phenotype (30.5% to 44%) [Alagille et al., 1987; Depretere et al., 1987; Hoffenberg et al., 1995; Emerick et al., 1999; Quiros-Tejeira et al., 1999; reviewed in Kamath et al., 2007].
Table III.
Phenotype Comparison in JAG1 + Cohort versus Vietnamese JAG1+ Cohort
| Phenotype Presentation in JAG1 (+) Cohort and Vietnamese JAG1(+) Cohort | |||||||
|---|---|---|---|---|---|---|---|
| Type of Proband | Total # of Probands |
Phenotype | |||||
| Liver | Cardiac | Eye | Skeletal | Facies | Renal | ||
| Composite of Prior Studiesa | 268 | 95% | 94% | 80% | 61% | 92% | 44% |
| Cohort partially reported in Warthen et al. [2006] | 326 | 97.3% | 97.9% | 68.3% | 57.9% | 96% | 30.5% |
| JAG1 Vietnamese Probands (Excludes Unknowns) | 18 | 100% (18/18) | 94.4% (17/18) | 100% (2/2) | 94.1% (16/17) | NDb | 27.3% (3/11) |
Data from Kamath BM, Spinner NB, “Piccoli DA. Alagille Syndrome. In: Suchy FJ, Sokol RJ, Balistreri WF, eds. Liver disease in children, 3rd ed. Cambridge: Cambridge University Press, 2007:326–345.
Facial phenotype frequency was not assessed
In comparing the clinical phenotype of the Vietnamese cohort with JAG1 mutations to our existing ALGS cohort, the liver and cardiac phenotypes occur in over 94% in both populations. Notably, bile duct paucity was found in all four children who had liver biopsies. Renal involvement occurs with a similar frequency of 27.3% in the Vietnamese cohort compared to 30.5% in our cohort. Only two children had ophthalmologic exams so the frequency of the ophthalmologic phenotype is difficult to determine. Differences in clinical phenotype frequency are seen with the skeletal phenotype. The Vietnamese population has an increased frequency of skeletal involvement (93.3% compared to 57.9%). The facial phenotype was not assessed by a Vietnamese dysmorphologists, making it difficult to determine the true frequency of the ALGS facial dysmorphism. Prior studies have reported the frequency of facial dysmorphism in ALGS to be 92% and the facial phenotype occurs in 96% of the patients in our ALGS cohort. Facial phenotype assessment by our clinical dysmorphologist noted that ALGS facies in only 23.5% (4/17) of the Vietnamese cohort with JAG1 mutations. By comparison, Dr. Phuc, the referring pediatric gastroenterologist in Vietnam felt that 61.1% (11/18) of the cohort had characteristic ALGS facies.
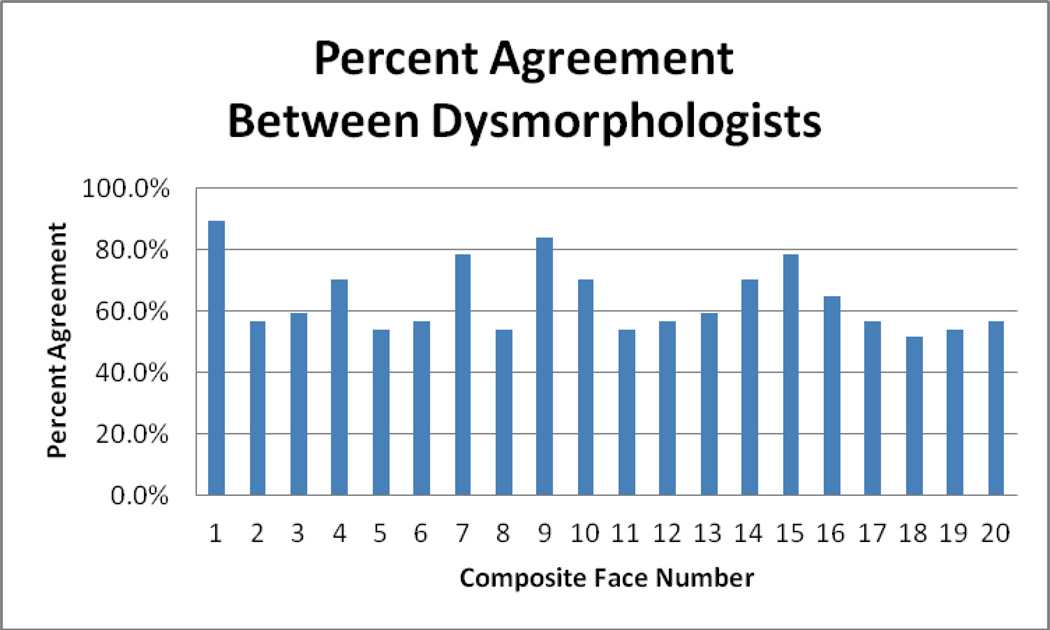
Recognizing the limitations of identifying the ALGS facies in this cohort, we conducted a survey to evaluate the ability of North American dysmorphologists to assess facial dysmorphism and the presence of the characteristic ALGS facial phenotype in the Vietnamese population. A total of 37 respondents with varying levels of experience actively working in clinical genetics with experience in facial feature identification participated in the study. For each individual in the photographic panel, respondents were asked to identify ALGS versus non-ALGS facial phenotype. For each facial composite, the percentage agreement was calculated by dividing the numerical value of the most frequent response by the total number of respondents for that particular facies. The percentage agreement between respondents ranged from 51.4% to 89.2% for each facial composite (Fig 3). Dysmorphologists had over 75% agreement of the facial phenotype for only four of the individuals in the photographic panel. Overall, the percentage agreement of the presence or absence of the ALGS facial phenotype was 51.8% among the 37 respondents. Professional grade and experience were not correlated with improved percent agreement (Table IV).
Figure 3.
Percentage Agreement Among Dysmorphologists for Each Face Reviewed
Table IV.
Percent Agreement of Facial Feature Analysis by Professional Group
| Professional Group | Number in Group | Percent Agreement |
|---|---|---|
| Fellows | 5 | 51% |
| Assistant Professor | 7 | 50% |
| Associate Professor | 10 | 59% |
| Professor | 14 | 51.4% |
| Other a | 1 | N/A |
| Total | 37 | 51.8% |
Clinical geneticist
DISCUSSION
In this study of 21 clinically defined ALGS patients from Vietnam, we report a JAG1/NOTCH2 mutation rate of 90%, which is similar to the previously reported rate in other clinically defined patient populations [Warthen et al., 2006]. Of the 21 Vietnamese patients, screening of the JAG1 and NOTCH2 genes identified 19 different mutations, 17 of which are novel. The type and distribution of the JAG1 mutations in Vietnamese ALGS patients are similar to previous reports [Krantz et al., 1998; Warthen et al., 2006]. The low frequency of NOTCH2 as compared to JAG1 mutations in this cohort (1/21 patients) is also similar to that seen previously, and this is only the third reported NOTCH2 mutation [McDaniell et al., 2006]. The NOTCH2 mutation falls within exon 33 and leads to truncation of the NOTCH2 protein within the intracellular domain, specifically within the ankyrin repeats.
While the prevalence of cardiac (94%) and liver involvement (100%) in this Vietnamese cohort was similar compared to prior series, the frequency of vertebral anomalies was markedly different. Ninety-three percent of the Vietnamese cohort had skeletal involvement, suggesting an increased frequency of vertebral anomalies in the Vietnamese population. To compare, in a review of 92 patients with ALGS, Emerick et al. [1999] report the frequency of butterfly vertebrae at 51%. The distribution of the type of skeletal involvement was similar to previous observations; all skeletal involvement in the Vietnamese cohort consisted of butterfly vertebrae.
In our survey of North American dysmorphologists with experience in facial feature recognition, the assessment of the Vietnamese facies proved difficult. The overall percent agreement among respondents was only 51.8%. Having a percentage agreement close to 50% suggests definite disagreement among dysmorphologists as to the presence of absence of ALGS facial features in the Vietnamese cohort. This difficulty in recognizing the ALGS facial phenotype is further highlighted by the fact that experience did not play a role in improvement of agreement of the facial phenotype. It is possible that the North American dysmorphologists, who do not evaluate facial dysmorphism in a population they do not routinely treat, may have had intrinsic difficulty recognizing the ALGS facies. A similar cohort of reviewers (many of whom were the same) from the 1998 David W. Smith Annual Workshop on Malformations and Morphogenesis completed a similar survey for the facial phenotype study by Kamath et al. [2002] and performed more successfully. In that study of a Caucasian cohort of differing ages, the calculated sensitivity and specificity of facies as a diagnostic tool for ALGS was over 75%.
Previous experience indicates that the effect of JAG1 mutations on facial features is similar among different ethnic groups. In a review of the ALGS probands enrolled in our studies (data not reported), 96% (23/ 24) of African-American individuals, 97% (32/33) of Hispanics and 92% (12/13) of Asians had characteristic facies. However, the lack of agreement among North American dysmorphologists suggests that clinicians should consider other organ system involvement in lieu of relying on the characteristic facies in the evaluation of a Vietnamese patient for suspicion of ALGS. Notably, the referring pediatric gastroenterologist, who sees Vietnamese patients daily, but is not clinically trained as a dysmorphologist, was only able to identify ALGS facial features in 61.1% of the patients. Another consideration for the level of disagreement among the North American dysmorphologists is that the facial dysmorphisms have been reported to evolve, with the ALGS facies becoming more prominent with age [Kamath et al., 2002]. The Vietnamese individuals in the photographic panel were all young children under the age of 10 years, so it is possible that analysis of this same cohort at a later age may yield different results. While familial data was unavailable, looking at familial cases of ALGS could help clarify the prevalence of the Vietnamese facial phenotype. Ultimately, to effectively determine the frequency of the facial phenotype in the Vietnamese cohort, assessment by a Vietnamese dysmorphologist is needed
Limitations of this study include the small size of the study cohort, leading to difficulty in making generalizations about ALGS in the Vietnamese population. Out of the novel JAG1 mutations, three are missense mutations. We cannot completely exclude these mutations as polymorphisms in the Vietnamese population in the absence of screening population-matched controls for polymorphisms or performing JAG1 functional protein studies on these missense mutations. However, we note that two of the mutations are loss of a cysteine residue in the extracellular domain, which has a high index of suspicion based for pathogenicity, and the third mutation has been shown to produce an abnormal mRNA. Prior studies have already cited the challenges of trying to assess facial morphology from two-dimensional photographs, as facial features may be more noticeable in person. While the survey respondents were blinded to the actual condition of each individual in the photograph panel, there may be some response bias, with the expectation of identification of the ALGS facial phenotype.
In conclusion, this study reports novel JAG1 and NOTCH2 mutations in a Vietnamese ALGS cohort. The Vietnamese cohort shares a similar distribution of the major clinical phenotypes with the exception of the skeletal and facial phenotype. While the facial phenotype in ALGS is specific to the syndrome, this study demonstrates a decreased frequency of the facial phenotype amongst a Vietnamese cohort, lowering its value as a diagnostic tool in this population.
Supplementary Material
ACKNOWLEDGMENTS
We thank all of the families that participated in the study. Supported by the National Institutes of Health DK081702 (to NBS).
REFERENCES
- Alagille D, Odievre M, Gautier M, Dommergues JP. Hepatic ductular hypoplasia associated with characteristic facies, vertebral malformations, retarded physical, mental, and sexual development, and cardiac murmur. J Pediatr. 1975;86:63–71. doi: 10.1016/s0022-3476(75)80706-2. [DOI] [PubMed] [Google Scholar]
- Alagille D, Estrada A, Hadchouel M, Gautier M, Odievre M, Dommergues JP. Syndromic paucity of interlobular bile ducts. J Pediatr. 1987;110:195–200. doi: 10.1016/s0022-3476(87)80153-1. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Cho EH, Park BY, Cho JH, Kang YS. Comparing two diagnostic laboratory tests for several microdeletions causing mental retardation syndromes: multiplex ligation-dependent amplification vs fluorescent in situ hybridization. Korean J Lab Med. 2009;29:71–76. doi: 10.3343/kjlm.2009.29.1.71. [DOI] [PubMed] [Google Scholar]
- Colliton RP, Bason L, Lu FM, Piccoli DA, Krantz ID, Spinner NB. Mutation analysis of Jagged1 (JAG1) in Alagille syndrome patients. Mutation in Brief #397. Online. Hum Mutat. 2001;17:151–152. doi: 10.1002/1098-1004(200102)17:2<151::AID-HUMU8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Crosnier C, Driancourt C, Raynaud N, Dhorne-Pollet S, Pollet N, Bernard O, Hadchouel M, Meunier-Rotival M. Mutations in JAGGED1 gene are predominantly sporadic in Alagille syndrome. Gastroenterology. 1999;116:1141–1148. doi: 10.1016/s0016-5085(99)70017-x. [DOI] [PubMed] [Google Scholar]
- Crosnier C, Driancourt C, Raynaud N, Hadchouel M, Meunier-Rotival M. Fifteen novel mutations in the JAGGED1 gene of patients with Alagille syndrome. Mutation in Brief #385. Online. Hum Mutat. 2000;17:72–73. doi: 10.1002/1098-1004(2001)17:1<72::AID-HUMU11>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Deprettere A, Portmann B, Mowat AP. Syndromic paucity of the intrahepatic bile ducts: diagnostic difficulty; severe morbidity throughout early childhood. J Pediatr Gastroenterol Nutr. 1987;6:865–871. doi: 10.1097/00005176-198711000-00008. [DOI] [PubMed] [Google Scholar]
- Elmslie FV, Vivian AJ, Gardiner H, Hall C, Mowat AP, Winter RM. Alagille syndrome: family studies. J Med Genet. 1995;32:264–268. doi: 10.1136/jmg.32.4.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerick KM, Rand EB, Goldmuntz E, Krantz ID, Spinner NB, Piccoli DA. Features of Alagille syndrome in 92 patients: frequency and relation to prognosis. Hepatology. 1999;29:822–829. doi: 10.1002/hep.510290331. [DOI] [PubMed] [Google Scholar]
- Giannakudis J, Ropke A, Kujat A, Krajewska-Walasek M, Hughes H, Fryns JP, Bankier A, Amor D, Schlicker M, Hansmann I. Parental mosaicism of JAG1 mutations in families with Alagille syndrome. Eur J Hum Genet. 2001;9:559. doi: 10.1038/sj.ejhg.5200613. [DOI] [PubMed] [Google Scholar]
- Heritage ML, MacMillan JC, Colliton RP, Genin A, Spinner NB, Anderson GJ. Jagged1 (JAG1) mutation detection in an Australian Alagille syndrome population. Hum Mutat. 2000;16:408–416. doi: 10.1002/1098-1004(200011)16:5<408::AID-HUMU5>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Heritage ML, MacMillan JC, Anderson GJ. DHPLC mutation analysis of Jagged1 (JAG1) reveals six novel mutations in Australian Alagille syndrome patients. Mutation in Brief #566. Online. Hum Mutat. 2002;20:481. doi: 10.1002/humu.9095. [DOI] [PubMed] [Google Scholar]
- Hoffenberg EJ, Narkewicz MR, Sondheimer JM, Smith DJ, Silverman A, Sokol RJ. Outcome of syndromic paucity of interlobular bile ducts (Alagille syndrome) with onset of cholestasis in infancy. J Pediatr. 1995;127:220–224. doi: 10.1016/s0022-3476(95)70298-9. [DOI] [PubMed] [Google Scholar]
- Jurkiewicz D, Popowska E, Glaser C, Hansmann I, Krajewska-Walasek M. Twelve Novel JAG1 Gene Mutations in Polish Alagille Syndrome Patients. Mutation in Brief #784. Online. Hum Mutat. 25:321. doi: 10.1002/humu.9313. [DOI] [PubMed] [Google Scholar]
- Kamath BM, Loomes KM, Oakey FJ, Emerick KE, Conversano T, Spinner NB, Piccoli DA, Krantz DA. Facial Features in Alagille Syndrome: Specific or Cholestasis Facies? American Journal of Medical Genetics. 2002;112:163–170. doi: 10.1002/ajmg.10579. [DOI] [PubMed] [Google Scholar]
- Kamath BM, Enson L, Piccoli DA, Krantz ID, Spinner NB. Consequences of JAG1 mutations. J Med Genet. 2003;40:891–895. doi: 10.1136/jmg.40.12.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath BM, Spinner NB, Piccoli DA. Alagille Syndrome. In: Suchy FJ, Sokol RJ, Balistreri WF, editors. Liver disease in children. 3e. Cambridge: Cambridge University Press; 2007. pp. 326–345. [Google Scholar]
- Kamath BM, Thiel BD, Gai X, Conlin LK, Munoz PS, Glessner J, Clark D, Warthen DM, Shaikh TH, Mihci E, Piccoli DA, Grant SF, Hakonarson H, Krantz ID, Spinner NB. SNP array mapping of 20p deletions: Genotypes, Phenotypes and Copy Number Variation. Hum Mutat. 2009;30:371–378. doi: 10.1002/humu.20863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, Kiuchi T, Inomata Y, Uryuhara K, Sakamoto S, Ito T, Fujimoto Y, Ogura Y, Oike F, Tanaka K. Living-related liver transplantation for Alagille syndrome. Transplantation. 2003;75:2147–2150. doi: 10.1097/01.TP.0000066804.33006.17. [DOI] [PubMed] [Google Scholar]
- Kim B, Park SH, Yang HR, Seo JK, Kim WS, Chi JG. Hepatocellular carcinoma occurring in Alagille syndrome. Pathol Res Pract. 2005;201:55–60. doi: 10.1016/j.prp.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Krantz ID, Colliton RP, Genin A, Rand EB, Li L, Piccoli DA, Spinner NB. Spectrum and frequency of Jagged1 (JAG1) Mutations in Alagille syndrome patients and their families. Am J Hum Genet. 1998;62:361-13. doi: 10.1086/301875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz ID, Piccoli DA, Spinner NB. Clinical and molecular genetics of Alagille syndrome. Curr Op Pediatr. 1999a;11:558–564. doi: 10.1097/00008480-199912000-00015. [DOI] [PubMed] [Google Scholar]
- Li L, Krantz ID, Deng Y, Genin A, Banta AB, Collins CC, Qi M, Trask BJ, Kuo WL, Cochran J, Costa T, Pierpont MEM, Rand EB, Piccoli DA, Hood L, Spinner NB. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- McDaniell R, Warthen DM, Sanchez-Lara PA, Pai A, Krantz ID, Piccoli DA, Spinner NB. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the Notch signaling pathway. Am J Hum Genet. 2006;79:169–171. doi: 10.1086/505332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Elkahlous AG, Pike BL, Okajima K, Krantz ID, Genin A, Piccoli DA, Meltzer PM, Spinner NB, Collins FS, Chandrasekharappa SC. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- Onouchi Y, Kurahashi H, Tajiri H, Ida S, Okada S, Nakamura Y. Genetic alterations in the JAG1 gene in Japanese patients with Alagille syndrome. J Hum Genet. 1999;44:235–239. doi: 10.1007/s100380050150. [DOI] [PubMed] [Google Scholar]
- Pilia G, Uda M, Macis D, Frau F, Crisponi L, Balli F, Barbera C, Colombo C, Frediani T, Gatti R, Iorio R, Marazzi MG, Marcellini M, Musumeci S, Nebbia G, Vajro P, Ruffa G, Zancan L, Cao A, DeVirgilis S. Jagged-1 Mutation Analysis in Italian Alagille Syndrome Patients. Hum Mutat. 1999;14:394.400. doi: 10.1002/(SICI)1098-1004(199911)14:5<394::AID-HUMU5>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Quiros-Tejeira RE, Ament ME, Heyman MB, Martin MG, Rosenthal P, Hall TR, Vargas JH. Variable morbidity in Alagille syndrome: a review of 43 cases. J Pediatr Gastroenterol Nutr. 1999;29:431–437. doi: 10.1097/00005176-199910000-00011. [DOI] [PubMed] [Google Scholar]
- Ropke A, Kujat A, Graber M, Giannakudis J, Hansmann I. Identification of 36 novel Jagged 1 (JAG1) mutations in patients with Alagille syndrome. Mutation in Brief #573. Online. Hum Mutat. 2002;21:100. doi: 10.1002/humu.9102. [DOI] [PubMed] [Google Scholar]
- Sokol RJ, Heubi JE, Balistreri WF. Intrahepatic “cholestasis facies”: is it specific for Alagille syndrome? J Pediatr. 1983;103:205–208. doi: 10.1016/s0022-3476(83)80345-x. [DOI] [PubMed] [Google Scholar]
- Wang JS, Wang XH, Zhu QR, Wang ZL, Hu XQ, Zheng S. Clinical and pathological characteristics of Alagille syndrome in Chinese children. World J Pediatr. 2008;4:283–288. doi: 10.1007/s12519-008-0051-5. [DOI] [PubMed] [Google Scholar]
- Warthen DM, Moore EC, Kamath BM, Morrissette JJD, Sanchez P, Piccoli DA, Krantz ID, Spinner NB. Jagged1 (JAG1) mutations in Alagille syndrome: increasing the mutation detection rate. Hum Mutat. 2006;27:436–443. doi: 10.1002/humu.20310. [DOI] [PubMed] [Google Scholar]
- Yuan ZR, Kohsaka T, Ikegaya T, Suzuki T, Okano S, Abe J, Kobayashi N, Yamada M. Mutational analysis of the Jagged 1 gene in Alagille syndrome families. Hum Mol Genet. 1998;7:1363–1369. doi: 10.1093/hmg/7.9.1363. [DOI] [PubMed] [Google Scholar]
- Yuan ZR, Okaniwa M, Nagata I, Tazawa Y, Ito M, Kawarazaki H, Inomata Y, Okano S, Yoshida T, Kobayashi N, Kohsaka T. The DSL domain in mutant JAG1 ligand is essential for the severity of the liver defect in Alagille syndrome. Clin Genet. 2001;59:330–337. doi: 10.1034/j.1399-0004.2001.590506.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.