Abstract
We present results from machine classification of melanoma biopsies sectioned and stained with hematoxylin/eosin (H&E) on tissue micro-arrays (TMA). The four stages of melanoma progression were represented by seven tissue types, including benign nevus, primary tumors with radial and vertical growth patterns (stage I) and four secondary metastatic tumors: subcutaneous (stage II), lymph node (stage III), gastrointestinal and soft tissue (stage IV). Our experiment setup comprised 14,208 image samples based on 164 TMA cores. In our experiments we constructed an HE color space by digitally deconvolving the RGB images into separate H (hematoxylin) and E (eosin) channels. We also compared three different classifiers: Weighted Neighbor Distance (WND), Radial Basis Functions (RBF), and k-Nearest Neighbors (kNN). We found that the HE color space consistently outperformed other color spaces with all three classifiers, while the different classifiers did not have as large of an effect on accuracy. This showed that a more physiologically relevant representation of color can have a larger effect on correct image interpretation than downstream processing steps. We were able to correctly classify individual fields of view with an average of 96% accuracy when randomly splitting the dataset into training and test fields. We also obtained a classification accuracy of 100% when testing entire cores that were not previously used in training (four random trials with one test core for each of 7 classes, 28 tests total). Because each core corresponded to a different patient, this test more closely mimics a clinically relevant setting where new patients are evaluated based on training with previous cases. The analysis method used in this study contains no parameters or adjustments that are specific to melanoma morphology, suggesting it can be used for analyzing other tissues and phenotypes, as well as potentially different image modalities and contrast techniques.
Key Terms: Melanoma progression, histopathological image analysis, non-parametric image analysis, H&E data, tissue classification
Melanoma is the most aggressive form of skin cancer, affecting 1/75 people in the United States. Cutaneous Malignant Melanoma (CMM) starts and grows locally, initially spreading superficially across the surface of the skin (radial growth phase), and later invading into the dermis and subcutaneous fat (vertical growth phase). The vertical progression of melanoma is graded using “Clark’s level” measurement (1), where level I tumors are contained within epidermis, and level V tumors have invaded the subcutaneous tissue. Further, the melanoma may be carried via the lymph to the lymph nodes, and via the bloodstream, to distal sites such as the lungs, brain, and liver. The American Joint Committee on Cancer (AJCC) recommended a system with four stages (2). The first two stages indicate a localized tumor with a thickness less than 4 mm, corresponding to Clark’s levels I–III. Stage III refers to tumors thicker than 4 mm, potentially with presence of satellite metastases or involvement of regional lymph nodes. Stage IV indicates distal metastases, which are ultimately responsible for the death of melanoma patients. Understanding and identifying the factors that promote the progression of melanoma among these different stages is a key focus of the melanoma community. Studying melanoma progression, and identifying the stage of a patient’s tumor requires immunohistochemical analysis of the involved tissues. Such an analysis of CMM’s transition to secondary sites is essential in understanding mechanisms of metastasis and finding its relevant markers. The development of computer-aided image processing techniques capable of distinguishing these stages can help in identifying novel biomarkers of melanoma progression as well as potentially serve as diagnostic aides in clinical settings.
Existing Research on Automatic Classification of Histopathology Images
Over the course of last decade, automatic annotation of histopathological images has been established as a field (3–33). The existing research on automatic annotation of melanoma is mostly concerned with distinguishing malignant melanoma from healthy controls (binary classification problems) (18–23) rather than exploring patterns in primary and secondary sites as we do in this work. Using in vivo confocal laser-scanning microscopy (857 images total, 574 used for training) Gerger et al (19) were able to classify correctly 97.5% of melanoma images. A similar result (97%) was obtained by Handels et al (20), where they employed a laser profilometry technique using 44 images in their report. Principal components were used as features in 213 images by Iyatomi et al (23) in their acral volar skin data set, where they achieved 93% overall accuracy in this two-class problem. Similarly, Wilgten et al (22) obtained 92% on a histopathological set of 40 images using texture features. Cheng et al (21) reported 86% classification rate on their 285 image set employing relative color features. A large set of 5,389 skin images was studied by Ganster et al (18), where 574 images were used in training a kNN classifier. Using size, shape and color descriptors they reported classification accuracy of 92%.
Scope of the Study and Contribution Summary
The scope of this study is to investigate critical factors for accurate machine-based analysis of histopathology samples, using melanoma as a test-case. Specifically, our interest was to investigate the effectiveness of a non-parametric, segmentation-free approach to analyzing histopathology samples. In as far as the analysis does not rely on parameters specific to melanoma, our findings stand a greater chance of applying to the automated analysis of histopathology in other cancers and diseases.
A major source of error in machine-based analysis is variation in sample preparation. In this study, we specifically control for this by analyzing biopsies on a tissue micro-array (TMA). While TMAs are more prevalent in clinical studies than in clinical practice, standardization of sample preparation and staining techniques will facilitate machine-assisted analysis in real clinical samples where a slide typically contains a biopsy from a single case including normal and potentially diseased tissue.
A second source of error is the accuracy of manual, often subjective readings that are commonly used as the “gold standard” or “ground truth” in machine-assisted diagnosis studies. We avoided this source of error by focusing on melanoma progression through several disease stages, where the goal was to correctly identify the objectively known biopsy sites.
The interpretation of color information is a major consideration in machine-based analysis of stained histopathology samples. The sole source of contrast in these samples is the specific color profile of the stains used, so different color models are expected to have significant effects on the information content of the images in subsequent processing. We are able to make direct comparisons between the classification accuracies achievable using different color models because our analysis system is non-parametric and does not rely on segmentation. This allows it to be used in the same way with the same original data even though the source of contrast is significantly altered by the different color models. Similarly, different downstream processing steps can be compared for their effectiveness in preserving and interpreting the information present in these samples.
In this study, we show that non-parametric techniques can be used to achieve very high classification accuracy in this melanoma test-case. Up to 96% accuracy can be achieved for individual fields of view at 50× magnification, and up to 100% when using several fields for aggregate classifications of whole TMA cores. Importantly, these accuracies are achieved on new melanoma cases that were entirely left out of the training phase. We find that the major factor affecting overall accuracy is the color model used to represent the color information, where digitally separating the RGB data into stain-specific hematoxylin and eosin channels produces the best accuracy regardless of the downstream processing techniques used.
The image analysis software used in this study - WND-CHARM (34) - was specifically designed to be as free from user-supplied parameters as possible. A major source of these parameters are those used for segmentation to identify cells or subcellular structures used in downstream processing. Instead, unlike nearly all work on H&E-stained histopathology samples, the analysis used here relies solely on the grouping of training images into their respective classes. Our previous work has shown that independence of user parameters allows this same software to be used in a variety of image processing tasks without modification or task-specific tuning. These include problems common in machine vision such as texture identification and face recognition (34), as well as biomedical applications such as classification of sub-cellular organelles (34), scoring of images in high-content screens (35), and determining physiological age in C.elegans (36,37) as well as medical imaging applications such as the diagnosis (38) and prediction (39) of osteoarthritis in human knee X-rays. The application of this software to the classification of melanoma in this study implies its potentially general utility in histopathology.
Materials and Methods
Tissue Samples
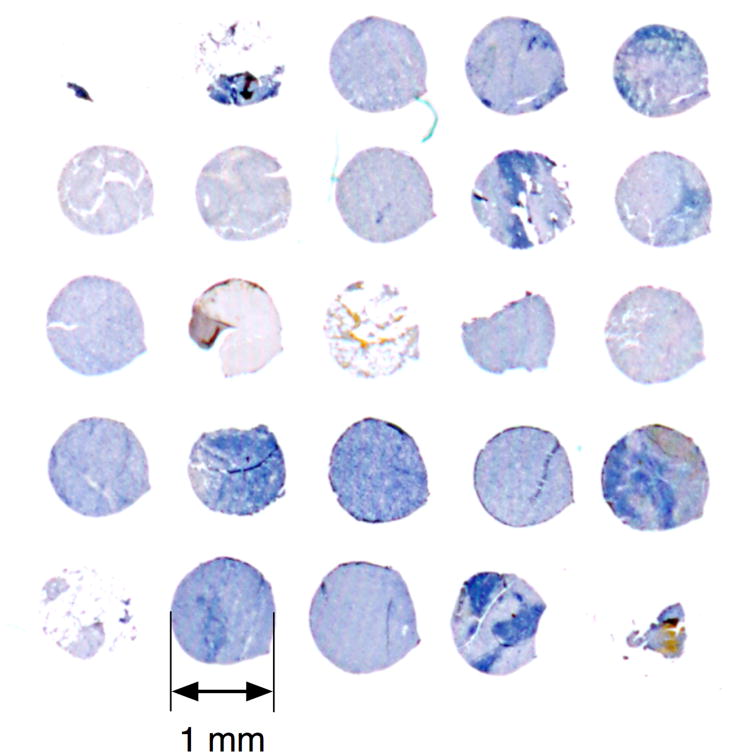
This study focused on a set of malignant tissues (40) corresponding to all four AJCC stages of melanoma. We imaged early melanocytic lesions having radial (RD) or vertical (VT) growth belonging to AJCC stage I. Secondary sites revealed at more advanced stages include subcutaneous type (SQ, stage II); lymph node metastatic (LN, stage III); and two distinct forms of stage IV: gastrointestinal (GI) and soft tissue (ST). Additionally, we included non-malignant tissue as a control (e.g., benign nevus tissue, NV). The melanoma progression TMA was constructed as described in Leotlela et al (41). The 1 mm diameter cores (5 um sections) were arrayed on four glass slides. The H&E staining procedure (42) was applied simultaneously to all samples, reducing variability between the samples. Fig. 1 shows a portion of one slide with the stained cores of the lymph node metastatic tissues. Depending on completeness of the cores (absence of tears, etc.), up to nine high-resolution (50×) non-overlapping fields of view could be taken from a 1 mm core. Magnified representative samples of all seven tissue types used in the study are shown in Fig. 2. The primary CMMs contained 52 cores of VT and eight cores of RD types; four secondary sites included 28 SQ cores, 8 ST type samples, 13 GI samples, and 31 LN samples. We had a total of 23 cores of the control NV tissue. Overall, we imaged 164 TMA cores. The melanoma progression TMA was constructed as described in Leotlela et al (41). Tissue microarrays were built at National Cancer Institute (NCI) using de-identified tissue obtained from the co-operative human tissue network (CHTN). They are part of the Tissue Array Research Program (TARP) at the NCI under NIA IRB protocol exemption number 2004-147.
Figure 1.
TMA cores arranged on a microscope slide (lymph node metastatic tissue – LN - stained with H&E). The core size is 1 mm.
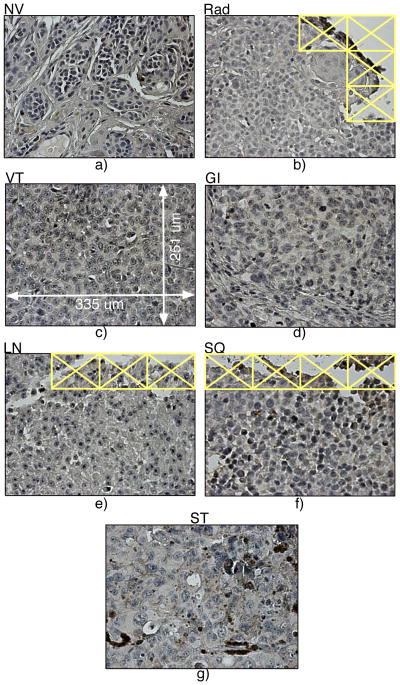
Figure 2.
Representative panels from primary and secondary tumor sites: a) nevus (healthy tissue, NV), b) primary site, radial (Rad) growth phase, c) primary site, vertical (VT) growth phase, d) gastro-intestinal (GI) tissue, e) lymph node (LN) metastatic tissue, f) subcutaneous (SQ) type, and g) soft tissues (ST). The panel size is 335 by 251 um. Images with little or no tissue (as in b, e and f) were excluded from the analysis.
Imaging
We used a light microscope (Zeiss Axioscope) with a 50× objective lens, a color RGB camera (AxioCam MR5) and halogen bright-field illumination. Imaging conditions and parameters of the instrument (objective lens, camera, light source, exposure time) remained unchanged over the course of acquisition, so no further normalization was performed. The frame size (i.e. a CCD panel or field of view) was set to 1292 × 968 pixels with 2×2 binning (0.2 um/pixel) and recorded as uncompressed RGB TIFFs at 12 bits/channel. The 12-bit pixel depth is higher than the more typical 8-bits, and may be important when converting range-sensitive color spaces, but the effect of channel depth on classification accuracy was not examined further.
Automated acquisition
The motorized microscope stage and the Zeiss software interface allow navigating across the slide using a master position list, as discussed below. The tissue deposition tool stamps the cores on the glass slide (as shown in Fig. 1). The cores form a grid-like pattern, but the core placement did not have sufficient precision to be imaged at high resolution directly. Instead, we used low-resolution imaging to find the core positions on each slide. A 4× objective lens and a 323 × 242 pixel frame (8×8 binning) were used to image the entire slide in grayscale. These low-resolution images, the position of one reference core, and the core-to-core pitch (in x- and y-) were used to reconstruct the core positions of the entire grid. Next, we generated a 3 × 3 template of stage positions for each core, resulting in a master position list for unattended high-resolution imaging (50× objective, 1292 × 968 frame size with 2×2 binning and 0.2 um/pixel resolution). Each of the resulting fields of view were subdivided into a 4 × 4 grid of contiguous 323 × 242 pixel images for further analysis. Tiling was done to both reduce the number of pixels to be processed during feature extraction, and to provide a statistical sample for scoring each field of view. We excluded from the analysis images containing little or no tissue as shown in Fig. 2.
Representing color
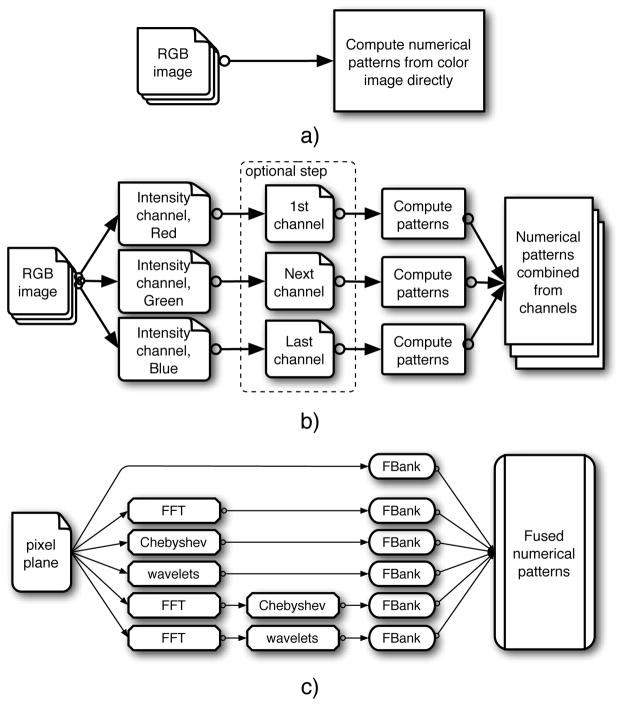
In histopathology imaging problems, color itself is irrelevant because it acts only as a contrast agent to reveal the underlying cellular morphology recognized by the chemical properties of the stain. Because these images are typically acquired with RGB color cameras, the relevant morphological signal must be extracted from the color representation imposed by the imaging technique. Fig. 3 illustrates several alternative approaches for color images commonly used in computer vision. Computed numerical patterns from RGB images can be used directly with some color features (Fig. 3a). The difficulty with this approach is that the color features are sensitive to absolute color values, and these may vary between stain preparations. A different approach is to compute features from each of the RGB channels independently (Fig. 3b) and combine the numerical patterns into a single feature space. This preserves all of the information captured by the RGB detector, but a potential limitation is the redundancy of the information in the separate RGB channels. A nonlinear scaling of RGB colors may address this redundancy by deriving a color space (Fig. 3b, the frame with a dashed border) different from the original instrument-specific RGB space, such as the CIE-L*a*b* space (subsequently referred to as LAB). In LAB, distances between colors are more closely correlated to color perception than they are in the RGB color space (43,44), making LAB a common choice for applications with color information (24,31). Lastly, the separate stains can be deconvolved from the RGB image using an algorithm proposed by Ruifrock and Johnston (45). In this technique, areas of isolated stain are manually selected in one or more images, which allows the algorithm to determine the color profiles of the two dyes, and subsequently reconstruct the stain concentrations into two separate channels (hematoxylin and eosin – HE) based on the Beer-Lambert law. In this work, areas of “pure” eosin and hematoxylin were selected from all four slides to get average color profiles, which were then used to reconstruct all of the RGB images into separate grayscale H and E channels.
Figure 3.
Three alternatives for processing color images: a) numerical patterns directly from the color image; b) separate numerical patterns either from RGB channels or from channels of a derived color space (marked with a dashed frame. Note that the number of derived channels might be different); c) the block ‘Compute patterns’ implements the computational framework as described in the corresponding Section (see Computational framework).
Computational Framework
Supervised learning
We used a supervised leaning approach that required ground truth annotation for the portion of the data used in training the classifier. This ground truth as provided by the authors (A.T.W. and S.M.H) directly correlates with the tissue origin, so it did not require manual reading or subjective assessment other than ensuring that each core was not a heterogeneous mixture of benign and malignant tissue.
Feature extraction
Visual patterns in the pixel plane P form visual cues that are specific to the imaging problem. These patterns can be analyzed quantitatively, through their mapping
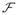 from the pixel plane to sets of numerical patterns (NPs)
, where P ⊂
from the pixel plane to sets of numerical patterns (NPs)
, where P ⊂
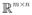 and
and
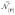 ∈
∈
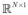 . We use general, low-level image features that describe the pixel plane with no reference to any particular imaging task. We then use a supervised learning approach to force this general set of descriptors into a context that is specific to the particular application. These global descriptors are implemented through the Feature Library (FL) as reported in (34). Briefly, this FL contains a collection of algorithms (feature families) each contributing to the overall feature vector representation of each image. The eleven families used were: features derived from coefficients of Chebyshev polynomials, features derived from coefficients of Chebyshev-Fourier polynomials, coefficients of Zernike polynomials, features derived from Gabor filters, features derived from the Radon transform, multi-scale histograms, comb filters for the first four moments, Haralick textures, Tamura textures, features based on object statistics, and features based on edge statistics.
. We use general, low-level image features that describe the pixel plane with no reference to any particular imaging task. We then use a supervised learning approach to force this general set of descriptors into a context that is specific to the particular application. These global descriptors are implemented through the Feature Library (FL) as reported in (34). Briefly, this FL contains a collection of algorithms (feature families) each contributing to the overall feature vector representation of each image. The eleven families used were: features derived from coefficients of Chebyshev polynomials, features derived from coefficients of Chebyshev-Fourier polynomials, coefficients of Zernike polynomials, features derived from Gabor filters, features derived from the Radon transform, multi-scale histograms, comb filters for the first four moments, Haralick textures, Tamura textures, features based on object statistics, and features based on edge statistics.
Image transforms as expansions of the pixel plane
In general, an image transform (such as Fourier transform) generates a spectral plane representation of the original pixel plane. Should an inverse for the given transform exist, the original image could be reconstructed from this spectral plane. Thus, the spectral plane is an alternative representation of the original image, and the numerical patterns in it (i.e. image features) can be computed from the spectral plane similarly to the original image plane. Super-spectral planes can also be computed, resulting from transforming spectral planes repeatedly, and may also be used for extracting additional numerical (super-spectral) patterns. Fig. 3c illustrates the use of transforms in computing NPs. This study used Fourier (FFTW (46)), wavelets (symlets5, level-1 details – implemented in MATLAB Wavelet Toolbox), and Chebyshev transforms, as well as compound transforms of Chebyshev of Fourier and wavelets of Fourier. These types of spectral and super-spectral features have not previously been used for the analysis of melanoma in the current literature. The CHARM-set of numerical image patterns proposed in (34) combines plain, spectral and super-spectral NPs and is shown in Fig. 3c, resulting in a total of 1025 image features. When multiple channels are used together (e.g. RGB and LAB), the feature vector is extended by computing the same features on each channel, resulting in 3075 features for RGB and LAB.
Classifiers
Three statistical classifiers were used in this work. The WND classifier works in a weighted feature space, and using Fisher scores (47) as weights for the feature values. Therefore, the distance ρc(t⃗) between a test sample and all training samples in a class c is a weighted distance: , while t⃗ is the test sample, and X⃗(j(c)) are the training samples of the class c. For a given test sample t⃗ the classifier calls cpred (the predicted class) by maximizing similarity sc(t⃗) between t⃗ and a class c, where the similarity is defined as , and the parameter p is fixed at 5 (34). We also used a classical kNN (48) classifier, where the test sample is classified by checking the labels of the k nearest neighbors of the same class in training set and then taking a vote comparing all classes.
Lastly, we used RBF (48) classifier. Although this classifier represents a network, it has only one hidden layer of neurons, so no topology optimization is necessary. Similarly to the two other classifiers, RBF performs mapping
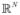 →
→
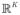 of the high-dimensional feature space
of the high-dimensional feature space
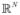 (as an input layer) to the low-dimensional
(as an input layer) to the low-dimensional
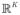 , where K is the number of classes. Specifically, the RBF output is given by y⃗ = Σiw⃗i · φ(X⃗, μ⃗i), where y⃗, w⃗ ∈
, where K is the number of classes. Specifically, the RBF output is given by y⃗ = Σiw⃗i · φ(X⃗, μ⃗i), where y⃗, w⃗ ∈
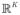 (0 ≤ yc ≤ 1 for the output components) and X⃗, μ⃗ ∈
(0 ≤ yc ≤ 1 for the output components) and X⃗, μ⃗ ∈
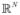 . The function centers μ⃗k of the Gaussian forms of the radial functions φ(X⃗, μ⃗k) were assumed to be the cluster centers for the k-mean algorithm. For both RBF and kNN the feature values were not further weighted by their Fisher scores.
. The function centers μ⃗k of the Gaussian forms of the radial functions φ(X⃗, μ⃗k) were assumed to be the cluster centers for the k-mean algorithm. For both RBF and kNN the feature values were not further weighted by their Fisher scores.
Feature selection
For feature selection (dimensionality reduction), features were added to a classifier until its performance stopped improving in random train/test splits. The same greedy hill climbing algorithm was used for all three classifiers used. For comparison, we also employed mRMR (49) and Pearson correlation (47) to select features instead of greedy hill climbing.
Marginal probability and classification accuracy
The marginal probability of the sample t⃗ belonging to a class cpred is defined as . The class assignment for the given sample t⃗ is given by . Marginal probability may be used for evaluating accuracy of calling an individual image, as well as groups of images by averaging. Specifically, when classifying a field of view (16 images), the marginal probabilities are averaged over all 16 constituent images, and the highest marginal probability in the average is used for the class assignment. Similarly, when classifying an entire core with several fields of view, the marginal probabilities are averaged over all of the core’s constituent images. The reported accuracy is the frequency of correct class assignments divided by all attempted assignments within the given test set or class.
Cross-Validation
When classifying fields of view, cross-validation is based on eight random splits of the data set into training and test portions. We used 224 randomly selected images (each 323 × 242 pixels in size) of each class for training the classifier, leaving the remaining images for testing. Because of differences in core size (due primarily to tears), this resulted in uneven numbers of test images: 3088, 64, 3488, 1200, 2896, 656 and 60 for NV, RD, VT, SQ, LN, GI, and ST respectively. The reported accuracy is the average of eight random test/train splits.
When testing accuracy of classifying entire cores, up to 144 images from one randomly selected core (for each class) contributed to the test set, while the remaining cores were used for training. Here the marginal probabilities of all constituent images were averaged before making the classification call for the core. The accuracy of classifying the fields of view available for each core (up to 9) could also be assessed independently from the accuracy reported for the whole core. As before, there was variation in the number of test images so that for example, the RD tissue type had 128, 96, 80, and 48 test images in the four CV splits. The number of training images in each class was fixed at 224 as above.
Results
Color Spaces and Classification
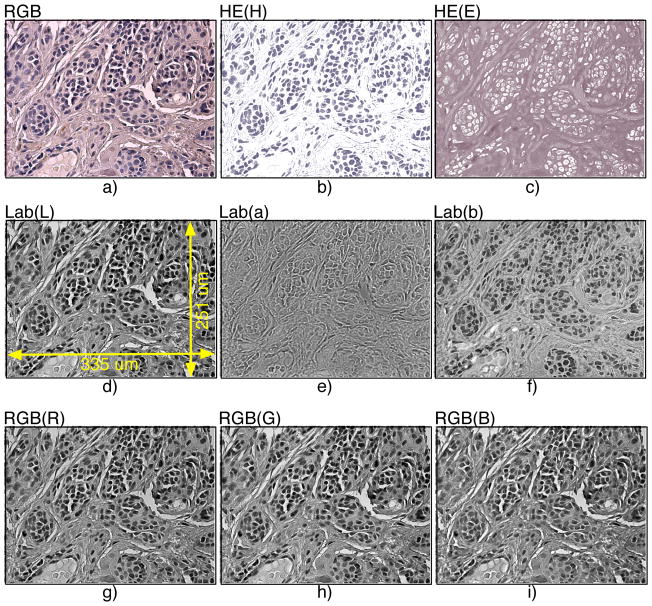
In our work we compared three color spaces for analyzing color images: RGB, LAB, and the deconvolved HE color space using the Ruifrock and Johnston algorithm (45). Example image panels in these three color schemes are shown in Fig. 4 using a portion of a TMA core with benign (nevus) tissue. The component channels of the same color image panels (4a) are displayed using HE (H and E in 4b and 4c, respectively), LAB (channels L, a*, b* in Fig. 4d–f, respectively), and RGB (R, G, and B in Fig. 4g–i, respectively). As one can easily see in Fig. 4, visual patterns in the three RGB channels (Fig. 4g–i) appear very similar to each other. This is not the case for H and E channels (Fig. 4b–c), and the component channels in LAB space (Fig. 4d–f) appear somewhat less redundant morphologically, but not as independent as the HE channels. The H&E’s stain concentrations carry the most relevant biological signal, corresponding to cell nuclei and cytoplasm. The size and shape of nuclei have been consistently considered important and indicative markers in cancer diagnosis (50,51), and thus we used the H channel for subsequent classification.
Figure 4.
Different color spaces used in tissue classification: a) original tricolor RGB, b) HE (H), c) HE (E), d) Lab (L), e) Lab (a), f) Lab (a), g) RGB (Red), h) RGB (Green), and i) RGB (Blue).
The classification scheme shown in Fig. 3b allows working with arbitrary kinds of multi-channel data because it relies on a global set of image features and does not rely on additional parameters or segmentation that may effect subsequent classification. This non-parametric approach allows for direct comparisons of the relative information content of different color schemes for classification purposes using one or more channels. Table 1 shows the average classification accuracy for the seven tissues using three classifiers (WND, RBF, and kNN) and three color spaces (RGB, LAB, and HE). Although the three classifiers perform similarly to each other in the three color spaces, there is a significant and consistent difference in classification accuracy between the three color spaces tested. The average accuracy over the three classifiers in LAB and RGB was modest (72% and 75%, respectively), but was markedly higher in the HE space (95%). We conclude that the HE space provides the best signal, allowing the single H-channel to outperform the three-channel spaces RGB and LAB.
Table 1.
Comparison of classifier accuracy (%) on RGB, LAB and HE data sets (the error is the standard deviation for eight different data splits). All three classifiers achieved their highest accuracy on HE set; average of all three accuracies is 94% -- noticeably above the results for RGB and LAB sets.
| Data Set | RGB | LAB | HE |
|---|---|---|---|
| WND | 82.1 ± 2.3 | 73.1 ± 2.8 | 95.7 ± 0.4 |
| RBF | 70.3 ± 4.1 | 73.2 ± 1.6 | 94.4 ± 1.6 |
| kNN | 72.2 ± 0.8 | 69.9 ± 3.0 | 93.6 ± 3.3 |
| Classifiers’ average | 74.9 | 72.1 | 94.6 |
All classifiers performed very similarly to each other within each color space, with the exception of RGB where WND outperformed kNN and RBF by 10% and 12%, respectively. We also performed a comparison of different feature selection and ranking methods for the WND classifier operating on the H channel of the HE color space. The accuracy of the WND classifier with Fisher ranking and weighting (96%) was similar to the result obtained using Pearson correlations (94%). We obtained similar results (96%) using the mRMR algorithm (49), which constructs a feature sub-space using a minimal redundancy and maximal relevance criterion. These observations led us to conclude that the choice of classifier and feature selection or ranking method makes little difference in the classification accuracy of this data set. In contrast, the findings in Table 1 show that the choice of color space is more important for accurate classification than the choice of classifier or feature selection algorithm, and that the reconstructed HE color space provides the best classification results because it is more representative of the underlying biologically significant morphological signal.
Cross-Validation Experiments
Table 2 reports average classification accuracies (as an average for the seven tissue types) using per-field of view and per-core cross-validation tests. Table 2 only shows results for the WND classifier on the H channel of the HE color space. When classifying fields of view (the first column), the panels are assigned randomly into training and test data without regard to the core they originate from. Therefore, the same TMA core could contribute data to both training and test sets. This type of cross-validation resulted in 95.7% classification accuracy (for comparison, it is also shown in Table I, as WND/HE result).
Table 2.
Comparison of classification accuracies for each class in two cross-validation scenarios: per-field and per-core. Per-field cross-validation results (eight random splits used) is shown in the first column. For per-core cross-validation, classification accuracies are reported per-field (second column) as well as per-core (third column) using “votes” from each field in the core. WND classifier was used on the H dataset in both cases. The error in the first column is the same as in Table I; the error in the second column is the standard deviation for 28 test cores used in the per-core test.
| Cross-validation | Per-Field | Per-Core | |
|---|---|---|---|
| Classification | Per-Field | Per-Field | Per-Core |
| Average | 95.7 ± 0.4 | 93.8 ± 11 | 100 |
Columns two and three in Table 2 show results of cross-validation on whole cores. The accuracy reported in column 2 is the average accuracy of correctly classifying a field of view within each core (93.8% for all seven tissue types). Note the full range of variation of the different cores is not fully represented in each experiment’s training set, potentially explaining the slight decline in the overall accuracy in this test compared to the per-field test that used all cores for training (95.7%; Table 2, column 1). The third column in Table 2 reports classification accuracies when the marginal probabilities of all of the constituent images of a core are averaged together before making the final classification call for the core. All 28 cores are classified with 100% accuracy when all constituent images “vote” on the final classification. The per-field accuracies in the per-core cross-validation provide a measure of reliability for this voting. It remains to be tested if this level of performance can be maintained through different stain preparations and laboratories, which would also be required in a “real world” diagnosis setting. The overall accuracy in diagnosing new patients is quite high even for single panels, indicating that with consistent sample preparation, the classifier is able to generalize to new cases of melanoma after training on previously known cases.
Sub-Pattern Discovery
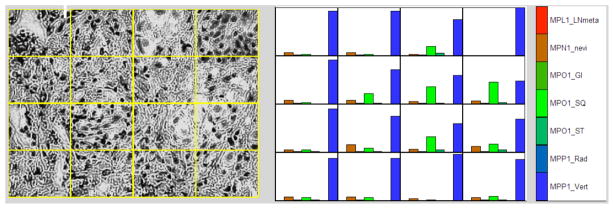
As discussed above, the WND classifier reports a set of marginal probabilities (one per class) for each field of view by averaging the set of marginal probabilities computed on each of its sixteen constituent images. For purposes of classification, the entire field is assigned to the class with the highest average marginal probability. Fig. 5 illustrates how the marginal probabilities are distributed in the individual images comprising a single field of a vertical growth primary tumor (VT). The left part of Fig. 5 shows a field of view in grayscale (H channel of HE) overlaid with the grid pattern of the constituent images. The right part shows the per-class marginal probabilities of each image as a bar graph. It can be seen that although the whole panel is classified correctly because in all but one of the images the highest marginal probability is VT, the individual images display considerable heterogeneity in their similarity to other classes in the dataset. One reason for that may be that VT (as a stage) marks the beginning of active metastasis into secondary sites.
Figure 5.
Assignment of marginal probabilities to images in a panel. The WND classifier was trained on ‘learning’ portion of TMA cores with all seven tissue types present and then tested on ‘test’ TMA cores different from the learning cores. On the left: a panel of the test core split into 16 images, – primary site, vertical growth stage (the scale is the same as in Figs 2 and 4: 335 um by 251 um). On the right: per-image marginal probabilities computed by the classifier.
Interestingly, the heterogeneity observed in this panel isn’t random. Although the classifier computes marginal probabilities independently for each image in the panel, when they are reassembled into the original field, we can see that a substantial subcutaneous (SQ) probability is present in a contiguous patch of images, and that this pattern is absent in a contiguous area surrounding this patch. The contiguity of this signal across several images indicates that this is not likely to be an artifact of the classifier, or random noise. Although this effect is not present in every panel, we were able to find several examples like this by manual inspection. This is an example where the subject specifics (actual observations) can go beyond the employed techniques (assigned ground truth): supervised learning assumes homogeneity of the groups, while in reality there can be considerable divergence which can be used to discover new patterns.
Discussion
Stain-Specific Color Space Provides the Best Classification Signal
Considering that the H-channel is an incomplete representation of the information contained in RGB and LAB, the higher accuracy obtained with it may seem surprising. In principle, one could attribute this to an artifact of the classifier or other internal machinery of the study, or systematic errors in non-HE color spaces. As discussed in the introduction, the feature bank we used in this study (CHARM) is not application-specific and was used in a wide variety of imaging studies. We also did not use segmentation, which is expected to be highly sensitive to image preprocessing such as the representation of color. These two properties allowed us to directly compare classification performance between different color representations of the same images. The performance we observed for the three color spaces was consistent between the three classifiers we tested, and has also been observed when classifying lymphoma sub-types (52). Taken together, these findings argue against systematic error, leading us to conclude that the best signal for classifying H&E stained tissue is a reconstructed HE color space regardless of the classifier employed, or even the specific tissue being studied.
Histochemical stains are commonly used as colored contrast agents to reveal subcellular morphology. The underlying biological processes which forms the basis for classification, is expressed through differential morphology, and is only tangentially related to the specific color response in the acquired RGB signal. The RGB channels contain a convolution of the underlying morphological signal because the color response of an RGB camera is not well matched to the color response of the contrast agents. The use of CIE-L*u*v* and L*a*b* is very common (24,31) and is well justified because the purpose of these spaces is to maximize channel independence, thus minimizing the redundancy of information presented to downstream classifiers. However, the LAB space is still dependent on the specific color response of the stains and their representation by RGB filters. In contrast, a color space based on reconstructed stain intensity is a better representation of the underlying morphology because it effectively factors out the largely irrelevant color responses of the intervening contrast agents and instrumentation.
Sub-Classification of Primary Tumors
In a previous study of adenocarcinomas (53), a gene expression signature was found that allowed differentiating primary tumors from secondary metastases. A subset of primary tumors was found to carry the gene expression signature associated with secondary metastases, and this subset was associated with increased metastasis and poorer clinical outcome than primary tumors that did not carry this signature. Similarly, using imaging, we observed that a subset of primary melanoma tumors carry a morphological signature associated with secondary sites. This marker allows us to further sub-classify primary tumors, but the clinical relevance of this sub-classification with regards to better prediction of disease outcome or prognosis remains to be definitively determined using a dataset with associated follow-up clinical outcomes.
Non-Parametric Pattern Recognition
There are obvious advantages to using multi-purpose features, like those contained in our feature library. Generality of the features means that a broad range of problems may be investigated with a single collection of algorithms. This point appears particularly important in analyzing cancer tissues: metastatic tissues present diverse cell morphologies, in varying patterns where often the underlying cellular mechanisms are not yet known. The generality ensures not only consistency for borderline cases but the potential for discovery of new sub-types or sub-grades, because the features are not constrained by specific a priori knowledge of the underlying biological process.
Despite its generality, this method demonstrated high accuracy in discriminating the stages of melanoma progression without making use of any specific features or parameters relevant exclusively to melanoma’s morphology. Thus, the techniques should be useful for other tissue types or imaging modalities. For example, multi-wavelength tissue scanning could be directly plugged into this framework in the same fashion as we used multiple color channels in this work. The promise of generalized, non-parametric image processing techniques is that clinicians and researchers do not have to engage in development or parameter tuning in domains where they lack expertise in order to address every particular imaging problem.
Conclusions
A major contribution of this study is the demonstrated capacity of a general classifier to automatically discriminate markers of melanoma progression without selecting algorithms or adjusting parameters other than assigning images to corresponding groups. The highest classification accuracy we achieved was 96% (per-field average for the seven tissue types analyzed) using the WND on the H channel of a reconstructed HE color space. When classifying whole cores of cases not previously seen by the classifier, we were able to achieve 100% accurate classification, indicating that accurate diagnostics can be performed in a clinically relevant setting where new cases must be accurately classified based on previous training data. We also demonstrate that appropriate, physiologically-relevant choice of color representation is much more important than the choice of classifier or feature-selection scheme.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging (Z01: AG000685-02).
Literature Cited
- 1.Clark JWH, From L, Bernardino EA, Mihm MC. The histogenesis and biologic behavior of primary human malignant melanomas of the skin. Cancer Research. 1969;29:705–726. [PubMed] [Google Scholar]
- 2.Slominski A, Ross J, Mihm MC. Cutaneous melanoma: pathology, relevant prognostic indicators and progression. British Medical Bulletin. 1995;51:548–569. doi: 10.1093/oxfordjournals.bmb.a072979. [DOI] [PubMed] [Google Scholar]
- 3.Huang C-L, Liao H-C, Chen M-C. Prediction model building and feature selection with support vector machines in breast cancer diagnosis. Expert Systems with Applications. 2008;34:578–587. [Google Scholar]
- 4.Yang L, Chen W, Meer P, Salaru G, Goodell LA, Berstis V, Foran DJ. Virtual microscopy and grid-enabled decision support for large-scale analysis of imaged pathology specimens. IEEE Tr on Information Technology in Biomedicine. 2009;13:636–644. doi: 10.1109/TITB.2009.2020159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biswas SK, Mukherjee DP. Recognizing architectural distortion in mammogram: a multiscale texture modeling approach with GMM. IEEE Tr on Biomedical Engineering. 2011;58:2023–2030. doi: 10.1109/TBME.2011.2128870. [DOI] [PubMed] [Google Scholar]
- 6.Dundar MM, Badve S, Bilgin G, Raykar V, Jain R, Sertel O, Gurcan MN. Computerized classification of intraductal breast lesions using histopathological images. IEEE Tr on Biomedical Engineering. 2011;58:1977–2030. doi: 10.1109/TBME.2011.2110648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wittke C, Mayer J, Schweiggert F. On the classification of prostate carcinoma with methods from spatial statistics. IEEE Tr on Information Technology in Biomedicine. 2007;11:406–414. doi: 10.1109/titb.2006.888703. [DOI] [PubMed] [Google Scholar]
- 8.Tabesh A, Teverovsky M, Pang HY, Kumar VP, Verbel D, Kotsianti A, Saidi O. Multifeature prostate cancer diagnostics and Gleason grading of histological images. IEEE Transactions on Medical Imaging. 2007;26(10):1366–1378. doi: 10.1109/TMI.2007.898536. [DOI] [PubMed] [Google Scholar]
- 9.Huang P-W, Lee C-H. Automatic classification for pathological prostate images based on fractal analysis. IEEE Tr on Medical Imaging. 2009;28:1037–1050. doi: 10.1109/TMI.2009.2012704. [DOI] [PubMed] [Google Scholar]
- 10.Tahir MA, Bouridane A. Novel round-robin tabu search algorithm for prostate cancer classification and diagnosis using multispectral imagery. IEEE Tr on Information Technology in Biomedicine. 2006;10:782–793. doi: 10.1109/titb.2006.879596. [DOI] [PubMed] [Google Scholar]
- 11.Belkacem-Boussaid K, Pennell M, Lozanski G, Shanaah A, Gurcan M. Computer-aided classification of centroblast cells in follicular lymphoma. Analytical and Quantitative Cytology and Histology. 2010;32:254–260. [PMC free article] [PubMed] [Google Scholar]
- 12.Basavanhally AN, Ganesan S, Agner S, Monaco JP, Feldman MD, Tomaszewski JE, Bhanot G, Madabhushi A. Computerized image-based detection and grading of lymphocytic infiltration in HER2+ breast cancer histopathology. IEEE Transactions on Biomedical Engineering. 2010;57(3):642–653. doi: 10.1109/TBME.2009.2035305. [DOI] [PubMed] [Google Scholar]
- 13.Fatakdawala HJX, Basavanhally A, Bhanot G, Ganesan S, Feldman MD, Tomaszewski JE, Madabhushi A. Contour With Overlap Resolution (EMaGACOR): Application to Lymphocyte Segmentation on Breast Cancer Histopathology. IEEE Tr on Biomedical Engineering. 2010;57:1676–1689. doi: 10.1109/TBME.2010.2041232. [DOI] [PubMed] [Google Scholar]
- 14.Doyle S, Feldman M, Tomaszewski J, Madabhushi A. IEEE Tr on Biomedical Engineering. 2010. A Boosted Bayesian Multi-Resolution Classifier for Prostate Cancer Detection from Digitized Needle Biopsies. [DOI] [PubMed] [Google Scholar]
- 15.Tuzel O, Yang L, Meer P, Foran D. Classification of hematologic malignancies using texton signatures. Pattern Analysis and Applications. 2007;10(4):277–290. doi: 10.1007/s10044-007-0066-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daskalakis A, Kostopoulos S, Spyridonos P, Glotsos D, Ravazoula P, Kardari M, Kalatzis I, Cavouras D, Nikiforidis G. Design of a multi-classifier system for discriminating benign from malignant thyroid nodules using routinely H&E-stained cytological images. Computers in Biology and Medicine. 2008;38(2):196–203. doi: 10.1016/j.compbiomed.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 17.McDonagh S, Fisher RB, Rees J. Using 3D information for classification of non-melanoma skin lesions. Univ. of Dundee; Scottland: 2008. Jul 2–3, [Google Scholar]
- 18.Ganster H, Pinz A, Rohrer R, Wildling E, Binder M, Kittler H. Automated melanoma recognition. IEEE Tr on Medical Imaging. 2001;20(3):233–239. doi: 10.1109/42.918473. [DOI] [PubMed] [Google Scholar]
- 19.Gerger A, Wiltgen M, Langsenlehner U, Richtig E, Horn M, Weger W, Ahlgrimm-Siess V, Hofmann-Wellenhof R, Samonigg H, Smolle J. Diagnostic image analysis of malignant melanoma in in vivo confocal laser-scanning microscopy: a preliminary study. Skin Research and Technology. 2008;14:359–363. doi: 10.1111/j.1600-0846.2008.00303.x. [DOI] [PubMed] [Google Scholar]
- 20.Handels H, Ross Th, Kreusch J, Wolff HH, Poppl SJ. Computer-supported diagnosis of melanoma in profilometry. Methods of Information in Medicine. 1999;38:43–49. [PubMed] [Google Scholar]
- 21.Cheng Y, Swamisai R, Umbaugh SE, Moss RH, Stoecker WV, Teegala S, Srinivasan SK. Skin lesion classification using relative color features. Skin Research and Technology. 2008;14:53–64. doi: 10.1111/j.1600-0846.2007.00261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilgten M, Gerger A, Smolle J. Tissue counter analysis of benign common nevi and malignant melanoma. International Journal of Medical Informatics. 2003;69:17–28. doi: 10.1016/s1386-5056(02)00049-7. [DOI] [PubMed] [Google Scholar]
- 23.Iyatomi H, Oka H, Celebi ME, Ogawa K, Argenziano H, Soyer HP, Koga H, Saida T, Ohara K, Tanaka M. Computer-based classification of dermoscopy images of malonocytic lesions on acral volar skin. Journal of Investigative Dermatology. 2008;128:2049–2054. doi: 10.1038/jid.2008.28. [DOI] [PubMed] [Google Scholar]
- 24.Comaniciu D, Meer P, Foran D. Image-guided decision support system for pathology. Machine Vision and Applications. 1999;11:213–224. [Google Scholar]
- 25.Yang L, Tuzel O, Chen W, Meer P, Salaru G, Goodell LA, Foran DJ. PathMiner: A web-based tool for computer-assisted diagnostics in pathology. IEEE Transactions on Information Technology in Biomedicine. 2009;13(3):291–299. doi: 10.1109/TITB.2008.2008801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masood K, Rajpoot N. Texture based classification of hyperspectral colon biopsy samples using CBLP. 2009:1011–1014. [Google Scholar]
- 27.Naik J, Doyle S, Basavanhally A, Ganesan S, Feldman M, Tomaszewski J, Madabhushi A. A boosted distance metric: application to content based Image retrieval and classification of digitized histopathology. Lake Buena Vista, FL, USA: SPIE; 2009. Feb 10, p. 72603F-12. [Google Scholar]
- 28.Nielsen B, Albregsten F, Danielsen H. Low dimensionality adaptive texture feature vectors from class distance and class difference matrices. IEEE Transactions on Medical Imaging. 2004;23:73–84. doi: 10.1109/TMI.2003.819923. [DOI] [PubMed] [Google Scholar]
- 29.Kong J, Sertel O, Shimada H, Boyer KL, Saltz JH, Gurcan MN. Computer-aided evaluation of neuroblastoma on whole-slide histology images: Classifying grade of neuroblastic differentiation. Pattern Recognition. 2009;42(6):1080–1092. doi: 10.1016/j.patcog.2008.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burroni M, Corona R, Dell’Eva G, Sera F, Bono R, Puddu P, Perotti R, Nobile F, Andreassi L, Rubegni P. Melanoma computer-aided diagnosis: reliability and feasibility study. Clinical Cancer Research. 2004;10:1881–1886. doi: 10.1158/1078-0432.ccr-03-0039. [DOI] [PubMed] [Google Scholar]
- 31.Sertel O, Kong J, Lozanski G, Shana’ah A, Catalyurek U, Saltz J, Gurcan M. Texture classification using nonlinear color quantization: Application to histopathological image analysis. Proceedings of IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP 2008); Las Vegas, NV. 2008. [Google Scholar]
- 32.Alonso SR, Tracey L, Ortiz P, Perez-Gomez B, Palacios J, Linares MP, Serrano S, Saez-Castillo AI, Sanchez L, Pajares R, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Research. 2007;67(7):3450–3460. doi: 10.1158/0008-5472.CAN-06-3481. [DOI] [PubMed] [Google Scholar]
- 33.Cooper L, Sertel O, Kong J, Lozanski G, Huang K, Gurcan M. Feature-based registration of histopathology images with different stains: An application for computerized follicular lymphoma prognosis. Computer Methods and Programs in Biomedicine. 2009;96(3):182–192. doi: 10.1016/j.cmpb.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orlov N, Shamir L, Macura T, Johnston J, Eckley DM, Goldberg IG. WND-CHARM: Multi-purpose image classification using compound image transforms. Pattern Recognition Letters. 2008;29:1684–1693. doi: 10.1016/j.patrec.2008.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orlov N, Johnston J, Macura T, Shamir L, Goldberg I. Computer Vision for Microscopy Applications. In: Obinata G, Dutta S, editors. Vision Systems: Segmentation and Pattern Recognition. Vienna, Austria: I-Tech Education and Publishing; 2007. pp. 221–242. [Google Scholar]
- 36.Orlov N, Johnston J, Macura T, Wolkow C, Goldberg I. Pattern recognition approaches to compute image similarities: application to age related morphological change. Arlington, VA: 2006. Apr 6–9, pp. 1152–1156. [Google Scholar]
- 37.Johnston J, Iser WB, Chow DK, Goldberg IG, Wolkow CA. Quantitative image analysis reveals distinct structural transitions during aging in Caenorhabditis elegans tissues. PLoS ONE. 2008;3(7):e2821. doi: 10.1371/journal.pone.0002821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shamir L, Ling SM, Scott W, Orlov N, Macura T, Eckley DM, Ferrucci L, Goldberg IG. Knee X-ray image analysis method for automated detection of Osteoarthritis. IEEE Transactions on Biomedical Engineering. 2009;56(2):407–415. doi: 10.1109/TBME.2008.2006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shamir L, Ling SM, Scott W, Hochberg M, Ferrucci L, Goldberg IG. Early Detection of Radiographic Knee Osteoarthritis Using Computer-aided Analysis. Osteoarthritis and Cartilage. 2009;17(10):1307–1312. doi: 10.1016/j.joca.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dissanayake SK, Olkhanud PB, O’Connell MP, Carter A, French AD, Camilli TC, Emeche CD, Hewitt KJ, Rosenthal DT, Leotlela PD, et al. Wnt5A regulates expression of tumor-associated antigens in melanoma via changes in signal transducers and activators of transfcription 3 phosphorylation. Cancer Research. 2008;68(24):10205–10213. doi: 10.1158/0008-5472.CAN-08-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leotlela PD, Wade MS, Duray PH, Rhode MJ, Brown HF, Rosenthal DT, Dissanayake SK, Earley R, Indig FE, Nickoloff BJ, et al. Claudin-1 overexpression in melanoma is regulated by PKC and contributes to melanoma cell motility. Oncogene. 2007;26:3846–3856. doi: 10.1038/sj.onc.1210155. [DOI] [PubMed] [Google Scholar]
- 42.Kiernan JA. Histological and histochemical methods: theory and practice. Bloham Mill: Scion Publishing; 2008. p. 606. [Google Scholar]
- 43.Wyszecki G, Stiles WS. Color science: concepts and methods, quantitative data and formulae. New York: John Wiley & Sons; 1982. [Google Scholar]
- 44.Acharya T, Ray KK. Image processing: principles and applications. Wiley Interscience; 2005. [Google Scholar]
- 45.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23(4):291–299. [PubMed] [Google Scholar]
- 46.Frigo M, Johnston SG. The design and implementation of FFTW3. Proceedings of the IEEE. 2005;93:216–231. [Google Scholar]
- 47.Fukunaga K. In: Introduction to statistical pattern recognition. Rheinboldt W, editor. San Diego: Academic Press; 1990. p. 591. [Google Scholar]
- 48.Bishop C. Neural networks for pattern recognition. Oxford University Press; 1996. p. 504. [Google Scholar]
- 49.Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Tr on Pattern Analysis and Machine Intelligence. 2005;27(8):1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 50.Harris NL, Jaffe ES, Diebold J, Flandrin G, Muller-Hermelink HK, Vardiman J, Lister TA, Bloomfield CD. The World Health Organization classification of neoplastic diseases of the hematopoietic and lymphoid tissues. Ann Oncol; Report of the Clinical Advisory Committee meeting; Airlie House, Virginia. November, 1997; 1999. pp. 1419–32. [DOI] [PubMed] [Google Scholar]
- 51.Herlyn M, Ferrone S, Ronai Z, Finerty J, Pelroy R, Mohla S. Melanoma biology and progression. Cancer Research. 2001;61:4642–4643. [PubMed] [Google Scholar]
- 52.Orlov NV, Chen W, Eckley DM, Macura T, Shamir L, Jaffe ES, Goldberg IG. Automatic classification of lymphoma images with transform-based global features. IEEE Tr on Information Technology in Biomedicine. 2010;14(4):1003–1013. doi: 10.1109/TITB.2010.2050695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nature Genetics. 2003;33(1):49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]