Abstract
In this study, we show that conditioned media (CM) generated from bone marrow (BM)-derived mesenchymal stromal cells lead to BCR-ABL independent STAT3 activation. Activation of STAT3 is important not only for survival of CML cells but also for its protection against Nilotinib (NI), within the BM microenvironment. Reducing the expression of both JAK2 and TYK2 or utilizing a pan-JAK inhibitor blocked CM-mediated STAT3 activation and sensitized CML cells to NI-mediated cell death. Finally, we demonstrate that in patient-derived primitive leukemic cells, co-cultured with BM stromal cells, inhibition of BCR-ABL and JAK activity was a successful strategy to potentiate their elimination.
Keywords: STAT3, Conditioned media, Bone marrow microenvironment, Nilotinib, JAK, INC424
Introduction
Chronic Myeloid Leukemia (CML) is characterized by the presence of the BCR-ABL oncoprotein that has constitutive tyrosine kinase activity [1, 2]. This oncoprotein is distributed throughout the cytoplasm, where it interacts and activates multiple proteins leading to transformation to CML in patients [3]. Currently, the prototype BCR-ABL inhibitor imatinib (IM) as well as the new drugs nilotinib (NI) and dasatinib (DA) has proven to be very effective in inducing hematologic and cytogenic remission in CML patients [4–8]. However, complete eradication of the disease has not been possible due to the presence of minimal residual disease (MRD), as measured by quantitative real-time polymerase chain reaction (RT-PCR) [9, 10].
MRD has a very important prognostic value, since the presence of MRD has shown to result in the patient relapse after discontinuation of treatment with a BCR-ABL kinase inhibitor [11, 12]. One of the reasons for MRD is the development of BCR-ABL independent drug resistance in sanctuary sites such as the bone marrow (BM). For example, experimental evidence indicates that BCR-ABL inhibitors fail to kill the leukemic stem cell in the BM, in observation that did not correlate with failure to inhibit BCR-ABL kinase activity [13]. Also, CML cells when adhered to fibronectin, a component of the BM microenvironment, demonstrated significant resistance to BCR-ABL inhibitors via the phenomenon referred to as cell adhesion mediated drug resistance (CAM-DR) [14]. In addition to the physical components, the BM microenvironment also contains a milieu of cytokines and growth factors that contribute to drug resistance in CML [15, 16]. Therefore, in addition to BCR-ABL inhibition, overcoming BM microenvironment-mediated drug resistance, caused by direct physical contact and by soluble factors, is an essential strategy towards a disease-free clinical outcome in CML.
In a previous study, we showed that soluble factors secreted by immortalized HS-5 BM stromal cells activated STAT3 in CML cell lines and was sufficient to cause resistance to IM-mediated cell death [16]. Also, more recent studies have shown the importance of the presence of IM-resistant leukemic stem cells within the BM in ensuring maintenance of MRD [13]. Thus, it is attractive to speculate that the failure to eradicate the disease is due in part to the ability of the BM niche to activate survival pathways in a BCR-ABL independent fashion. In light of this, the present study was carried out to validate the BM-mediated STAT3-driven drug resistance phenotype in primary patient specimens and to delineate the most effective therapeutic strategies for inhibiting STAT3 activation in CML progenitor stem cells within the context of BM microenvironment. Our current results provide strong preclinical evidence for bypassing strategies that consider neutralizing antibodies and JAK specific inhibitors in favor of the use of a more promiscuous JAK inhibitor as a rationally designed strategy for increasing the efficacy of BCR-ABL inhibitors for eradicating MRD.
Materials and Methods
Cell Cultures
Human blastic phase CML derived K562 and KU812 cell lines and the human stromal cell line HS-5 (obtained from ATCC) were cultured in RPMI 1640 supplemented with 10% FBS, 1% penicillin/streptomycin at 37°C in 5% CO2 in a humidified incubator. For stable transfection, K562 cells engineered to express luciferase were transfected with a 5 μg of human pSM2 retroviral containing STAT3 shRNA (Open Biosystems, Huntsville, AL; Clone ID: V2HS_88502) or pSM2 retroviral empty vector using Amaxa Nucleofector methodology (Amaxa). Cells were incubated for 48 hr after transfection and subsequently selected with 5 μg/mL of puromycin. Clones were isolated and screened for STAT3 expression by Western blotting after clonal expansion.
Isolation of progenitor cells
For BM aspirate and peripheral blood, all patients were consented via the Total Cancer Care initiative at the Moffitt Cancer Center. Samples were de-identified before distribution to the laboratory. Normal BM aspirate was purchased from Lonza, Inc (Allentown, NJ). Mononucelar cells from the peripheral blood and BM aspirates were isolated by centrifugation through a Ficoll gradient. Peripheral blood mononuclear cells (PBMC) and BM mononuclear cells (BM-MNC) from healthy donors or CML patients were used to isolate CD34+ hematopoeitic progenitor cells with the aid of a CD34 MicroBead kit (Miltenyi Biotec Inc., Auburn, CA). BM-MNC were utilized for isolating Lin−CD34+ hematopoeitic progenitor cells by first depletion of Lin+ cells followed by positive selection of CD34+ cells with the aid of a Diamond CD34 Isolation kit (Miltenyi Biotec Inc, Auburn, CA).
Mesenchymal stromal cell (MSC) cell culture
The MSCs were isolated from the BM-MNC by plate adherence [17]. Briefly, BM-MNC were re-suspended in 5 mL of MSC growth medium (MGM) (MEMα/GlutaMAX™ supplemented with 10% fetal bovine serum and 1% 100× penicillin-streptomycin-glutamine (Invitrogen)), transferred to a T25 flask and incubated overnight. On the next day, after the medium was removed, the adhered cells were washed three times with PBS and grown further in MGM. MSCs were passaged after reaching 80% confluency by trypsinization and characterized by flow cytometry (see below) followed by culturing in MGM at 37°C in a humidified atmosphere containing 5% CO2.
Generation of CM
CM from HS-5 stromal cells was generated as described before [16]. For MSC CM, the cells were grown to 80–90% confluency, after which they were washed twice with PBS and the medium was replaced with RPMI 1640 supplemented with 10% FBS, 1% penicillin/streptomycin. After at least 16 hr, the medium was changed to fresh growth media. Twenty four hr later, the medium was collected, aliquoted and stored at −80°C.
Western blotting
Cells were plated in a 6-well plate at a concentration of 200,000 cells/mL. Cells were exposed to CM for 3 hrs followed by NI treatment for 24 hr. For the JAK inhibitor studies the cells were pretreated for 1 hr before exposure to CM. After treatment, cells were placed on ice and lysed in 30 μL of RIPA lysis buffer. Total protein (20 μg) was subjected to an 8% SDS-PAGE gel and indicated proteins were detected by Western blot analysis.
Semi-quantitative reverse-transcriptase-polymerase chain reaction (RT-PCR)
Cells were exposed to RM or CM for 24 hr followed by total RNA extraction using TRIzol as per the manufacturer's recommendation (Invitrogen). RNA (2 μg) was reverse transcribed using SuperScript III First-Strand synthesis superMix (Invitrogen) and 2 μL of the products were used as the template for PCR. The following primers were used: 5'CGG TAA TCG GAC TCA ACC TC3' and 5'CCT CCT TCT CCG TAG CCA A3' to amplify Mcl-1 and 5'CAT CTC TTG CTC GAA GTC CA3' and 5'ATC ATG TTT GAG ACC TTC AAC A3' to amplify β-actin;
Real-time reverse-transcriptase-polymerase chain reaction (qRT-PCR)
KU812 cells were pre-treated with 1000 nM JKI for one hour followed by exposure to RM or CM for 8 hr. Following treatment regimen, the total RNA extraction and cDNA preparation was carried out as described above. Real time PCR primers for Mcl-1 and GAPDH were purchased from Applied Biosystems (Carlsbad, CA). The qPCR reactions were performed using ABI 7900 sequence detection system (Applied Biosystems) and the Mcl-1 gene expression levels was normalized to the control GAPDH levels.
SCID-Hu model
The SCID-Hu model was performed as previously described [18]. After bone engraftment, 50,000 cells of the control K562 luciferase expressing cells or K562 luciferase expressing cells with reduced STAT3 were injected directly into the engrafted bone. The cells were allowed to engraft for 24 days and the tumor burden was measured utilizing Xenogen imaging system.
In vitro co-culture model
HS-5 BM stromal cells stably expressing GFP (HS-5-GFP) were plated at 200,000 cells/mL. After overnight incubation, KU812 cells (100,000 cells/mL) & HS-5-GFP cells where, individually, pretreated for 1 hr with indicated doses of INC424. The KU812 cells were then cultured over the HS-5-GFP cells for 3 hr followed by treatment with 25 nM of NI (INC424 treatment, were indicated, was continued throughout the experiment). After 24 hr, GFP-negative KU812 cells were subjected to cell death assay using Annexin V-APC.
For the primitive leukemic stem cell co-culture assay, HS-5-GFP cells were plated as above. After overnight incubation, freshly isolated Lin-CD34+ cells (100,000 cells/mL) from CML patients were cultured over the HS-5-GFP and incubated overnight. The next day (day 1) the media was changed and the co-culture was treated with either DMSO as control, 1000 nM INC424 and 100 nM NI or a combination of 1000 nM INC424 and 100 nM NI. On day 3, the media was changed and day 1 treatment regimen was repeated. At the end of day 5, GFP-negative cells were analyzed for cell death using Annexin V-APC.
Statistical Analysis
Experiments were independently performed three times, unless indicated otherwise, and the data were plotted as mean ± SD. The data were analyzed for statistical significance between treatment groups with the use of two-way ANOVA followed by post hoc Bonferroni's test. For the SCID-Hu mice model experiment the data was analyzed using a one-tailed unpaired t-test. A p<0.05 was accepted as statistically significant.
Results
CM from mesenchymal stromal cells (MSC) phosphorylates STAT3 and confers resistance to NI-mediated cell death
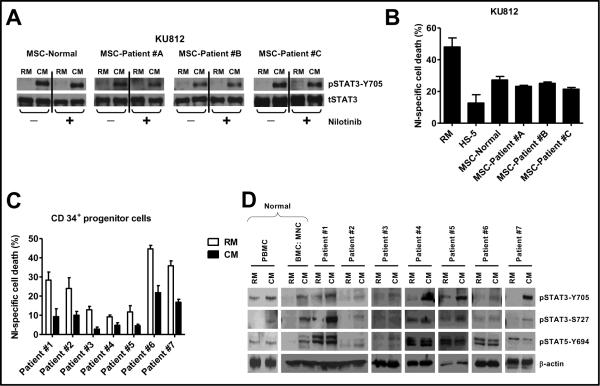
We first established that in CML cells, the HS-5 CM-induced phosphorylation of STAT1-Y701, STAT3-S727 and STAT5-Y694 but not of STAT3-Y705, was abolished in the presence of NI, indicating that phosphorylated STAT1-Y701, STAT3-S727, and STAT5-Y694 were not important players in mediating CM-induced NI resistance (Supplementary data S1). Next, we sought to determine whether a similar elevated phospho-STAT3-Y705 phenotype could be conferred by patient-derived components of the stem cell niche, specifically the MSC. MSC were obtained from a normal individual and three patients diagnosed with CML. MSC were confirmed by CD105, Stro-1, CD29 and CD73 positivity and CD34, CD33 and CD45 negativity (supplementary data S2). As shown in figure 1A, CM from MSC of 1 healthy and 3 CML patients all increased STAT3-Y705 phosphorylation in KU812 cells. The level of phosphorylation of STAT3 was sustained even in the presence of NI, indicating a BCR-ABL independent mechanism of activation. More importantly, the increased STAT3-Y705 phosphorylation correlated with protection against NI-induced cell death in KU812 cells exposed to the MSC-CM and was comparable to protection afforded by HS-5 stromal cell CM (figure 1B). Together these data indicate that CM derived from both normal and patient derived MSC can activate STAT3 in CML cells providing rationale for targeting STAT3 activation in combination with BCR-ABL inhibitors for the treatment of CML.
Figure 1. CM from BM stromal cells phosphorylates STAT3 and confers resistance against NI.
(A) (B) CM was collected from mesenchymal stromal cells (MSC) of 1 normal and 3 CML patient samples. KU812 cells (200,000 cells/mL) were pre-exposed to regular media (RM) and CM from MSC for 3 hr followed by treatment with vehicle (−) or 30nM NI (+). After 24 hr, cells were lysed and subjected to western blotting with phospho-STAT3-Y705 and total STAT3 antibody. (B) KU812 cells (200,000 cells/mL) were pre-exposed to RM, CM from MSC of 1 normal and 3 CML patient samples and CM from HS-5 for 3 hr followed by treatment with 30 nM NI. After 24 hr, cells were analyzed for cell death using Annexin V-FITC and FACS analysis with the help of CellQuest Pro software version 4.0.2 (BD Bioscience). (C) CD34+ cells isolated from PBMC of CML patients (200,000 cells/mL) where exposed to RM or CM for 3 hours followed by treatment with 100 nM NI for 48 hr. Apoptotic cells were detected by AnnexinV-FITC staining and FACS analysis with the help of CellQuest Pro software version 4.0.2 (BD Bioscience). (D) CD34+ cells were isolated from normal PBMC, normal BM aspirate and from PBMC of CML patients. CD34+ cells (200,000 cells/mL) were exposed to RM or CM for 24 hr and then lysed and subjected to SDS-PAGE with indicated antibodies. β-actin was used as loading control. The % NI-specific cell death was calculated by subtracting the background cell death in vehicle-treated control cells from NI-treated cell death.
CM mediates drug resistance in CD34+ progenitor cells in a STAT3-dependent mechanism
In order to further, pre-clinically validate the cell-line data we tested seven primary CML patient samples for STAT3 activation and levels of sensitivity to NI when CD34+ hematopoietic progenitor cells were cultured in HS-5 derived CM. As shown in figure 1C, seven out of seven CML patient specimens tested were resistant to NI-induced cell death when pre-exposed to HS-5 CM for three hours. In the seven primary patients tested, there was a mean 2.68 fold decrease in cell death when patient-derived CD34 positive cells were cultured in CM compared to RM. Interestingly, even though different CD34+ CML patient cells had different basal sensitivity towards NI, in all the cells tested, prior exposure to CM was sufficient to confer resistant in every patient specimen tested. Moreover, as shown in figure 1D, exposure to HS-5 CM resulted in consistent activation of STAT3-Y705 phosphorylation in all the samples tested, when compared to cells in RM. STAT3-S727 phosphorylation was up-regulated in CM only in a majority of the patient samples and, unlike STAT3 phosphorylation, some of the patient samples had very high basal levels of STAT5 phosphorylation and was not consistently up-regulated in CM (figures 1D). Together, these data continue to support the need to develop novel strategies for neutralizing the STAT3-Y705 activation within the BM niche for increasing the efficacy of BCR-ABL inhibitors and reducing MRD associated with CML.
STAT3 phosphorylation leads to increased STAT3 dependent transcription of Mcl-1
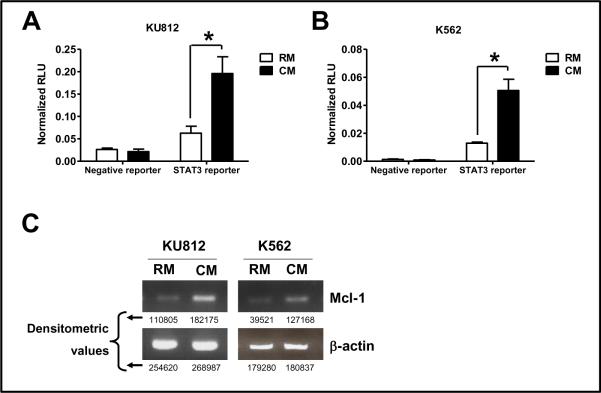
To determine whether increased levels of STAT3-Y705 corresponded to increased STAT3 mediated transcription, a STAT3-regulated luciferase reporter assay was performed. Cells that were incubated with CM showed increased STAT3 regulated promoter activity as measured by a mean 3.59 ± 0.9 fold increase in luciferase activity in K562 cells and a mean 3.31 ± 1.32 fold increase in luciferase activity in KU812 cells compared with cells incubated with RM (figure 2A and 2B). Increased transcriptional activity was indeed due to increase in binding of activated STAT3 to it response element was confirmed by EMSA (Supplementary data S3). We next examined the levels of a well-known STAT3 driven gene, Mcl-1. Increased expression of these genes has been causally linked to cell survival in CML cells [19]. As shown in figure 2C, in KU812 and K562 cells, transcription of Mcl-1 was increased in cells incubated in CM when compared to RM. Together, these data indicate that exposing CML cells to CM results in increased STAT3 dependent DNA binding and transcriptional activation of STAT3 and increased expression of Mcl-1, which are likely to contribute to drug resistance and persistence of disease in the confines of the BM microenvironment.
Figure 2. CM activates STAT3 and leads to increased transcription of STAT3-regulated Mcl-1 gene.
(A) KU812 or (B) K562 cells were transfected, using Amaxa Nucleofector per manufacture instructions, with 1 μg of either a STAT3 reporter or a negative reporter, both driving the firefly luciferase expression along with an internal control plasmid of β-actin driving renilla luciferase expression (SA BioSciences). Forty eight hr post-transfection, cells were exposed to RM or CM for another 24 hr. The cells were lysed and luciferase activity was determined as per manufacturer's recommendation (Promega). Data was normalized for differences in transfection efficiency. (C) Total RNA was obtained from cells exposed to RM or CM for 24 hrs. Products were amplified as described in materials and methods. The RT-PCR blots are representative of three individual experiments. Densitometric values of the amplified transcripts are provided below the blots. * denotes significant difference (p<0.05) (n=3).
STAT3 levels are a determinant of growth in vitro and in the SCID-Hu in vivo model
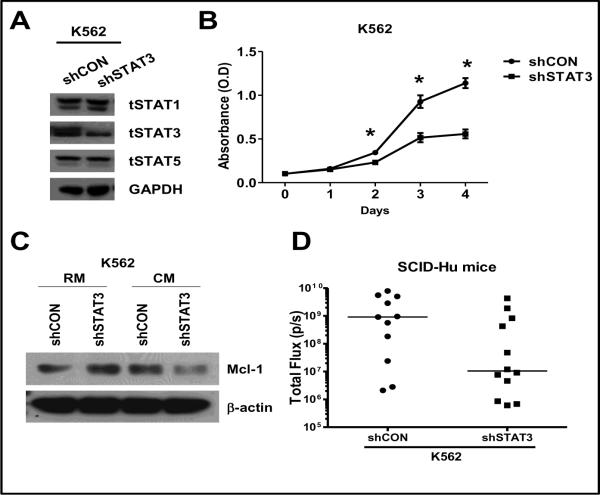
To further validate the role of STAT3, K562 cells expressing luciferase, were stably transfected with either a lentiviral shRNA construct targeting STAT3 or a lentiviral empty vector. As shown in figure 3A, we were able to establish a stable clone showing decreased STAT3 expression (shSTAT3) as compared to the control cell line (shCON). In the shSTAT3-K562 cells, the levels of STAT1 and STAT5 remained unchanged and similar to the shCON-K562 cells (figure 3A). In order to characterize the two stable cell lines, we compared the proliferation rate of the two stable cell lines. As shown in figure 3B, the proliferation rate of shSTAT3-K562 cells was significantly lower than the shCON-K562 cells. Also, to our surprise, shSTAT3-K562 cells when incubated in CM derived from HS-5 showed a significant decrease in Mcl-1 levels as compared to shCON-K562 cells incubated in the similar media (figure 3C).
Figure 3. STAT3 is required for growth of K562 cells in vitro and in vivo.
(A) K562 cells with stable luciferase expression were transfected with a shSTAT3 expression construct (shSTAT3) or the empty vector construct (shCON) and puromycin-resistant clones were expanded and analyzed for the expression of the indicated proteins by Western blotting. (B) Stable clones (shSTAT3 and shCON) were seeded at equal concentration (200,000 cells/mL) and proliferation was measured utilizing a CellTiter-blue cell viability assay kit as per the manufacturers instruction (Promega, Madison, WI). * denotes significant difference (p<0.05) (n=3). (C) Stable clones (shSTAT3 and shCON) were seeded at 200,000 cells/ml and then exposed to RM or CM for 24 hrs. The cells were lysed and subjected to western blot analysis with indicated antibodies. β-actin was used as loading control. (D) Stable clones (shSTAT3 and shCON) were injected into the BM cavity of the engrafted fetal tissue. Tumor burden was measured by bioluminescent imaging following 24 days of tumor engraftment.
Next we tested these cell lines in a SCID-Hu mouse model. As seen in figure 3D, shCON-K562 cells had an increased tumor burden compared to the shSTAT3-K562 cells 24 days after injection of cells into the fetal human BM. Interestingly, while all the mice injected with the shCON-K562 cells grew to form a tumor by the end of 24 days, 3 out of the 12 mice injected with shSTAT3-K562 cells, failed to engraft within the BM. Taken together, these data indicate that STAT3 is required for not only survival, but also for cell proliferation within the BM microenvironment.
CM phosphorylates STAT3 through the JAK family of proteins
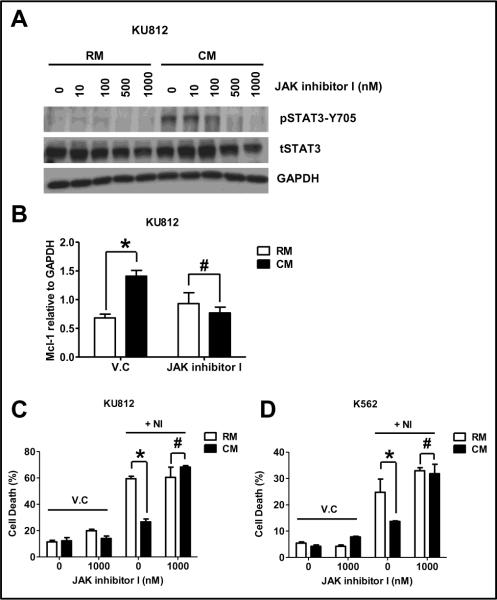
HS-5 CM contains a variety of cytokines and growth factors (GF) that are predictive for increased STAT3-Y705 levels via the JAK-STAT pathway [20]. We utilized a pan-JAK inhibitor to determine whether the inhibition of JAKs was sufficient to inhibit pSTAT3-Y705, decrease the levels of Mcl-1 and importantly reverse resistance associated with culturing CML cells in CM. As shown in figure 4A, the commercially available pan JAK inhibitor referred to as JAK inhibitor I, completely abolished pSTAT3-Y705 levels at a concentration of 1000 nM. We utilized real time RT-PCR to quantify the levels of Mcl-1 and as shown in figure 4B in the presence of the pan JAK inhibitor, the levels of Mcl-1 were similar irrespective of growth conditions. Furthermore, these data were consistent in both KU812 and K562 as confirmed by semi-quantitative RT-PCR (data not shown). As shown in figure 4C–D, the JAK inhibitor alone had no appreciable effects on cell death when cells were grown in RM or CM. Despite no activity as a single agent, the addition of the pan-JAK inhibitor completely reversed resistance to NI, when either KU812 or K562 cells were cultured in CM.
Figure 4. A pan-JAK inhibitor reverses CM-induced NI resistance by inhibiting the increased expression of Mcl-1.
(A) KU812 cells (200,000 cells/mL) where treated for 1 hr with indicated doses of JKI and then exposed to either RM or CM for 24 hr (JKI treatment was continued throughout the experiment). After 24 hr, cells were lysed and subjected to western blotting with indicated antibodies. GAPDH was used as the loading control. (B) KU812 cells (200,000 cells/mL) where treated for 1 hr with 1000 nM of JKI or vehicle control (V.C) and then exposed to either RM or CM for 8 hr. RNA was extracted and subjected to qRT-PCR as described in the methods. Mcl-1 expression was normalized by dividing each test condition with GAPDH expression. KU812 cells (C) or K562 cells (D) (200,000 cells/mL) where treated for 1 hr with 1000 nM of JKI or vehicle control (V.C) and then exposed to either RM or CM for 3 hr followed by treatment with 25 nM of NI (KU812) or 50 nM of NI (K562). JKI treatment, were indicated, was continued throughout the experiment. After 24 hr, cells were analyzed for cell death using Annexin V-FITC and FACS analysis with the help of CellQuest Pro software version 4.0.2 (BD Bioscience). # denotes no significant difference (p>0.05) and * indicates significant difference (P<0.05) (n=3).
CM phosphorylates STAT3 through a JAK2 and TYK2 pathway in K562 cells
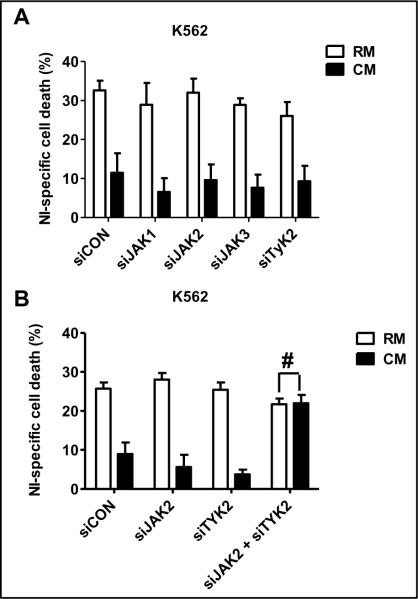
The currently clinically available JAK inhibitors have varying degrees of specificity for individual members of JAKs. We next sought to determine whether targeting a specific JAK member would be an appropriate strategy for reversing resistance in the context of the BM microenvironment. To delineate which of the JAK family of protein was involved in the CM-mediated STAT3 phosphorylation we used siRNA technology to individually knockdown JAK1, JAK2, JAK3 or TYK2 in K562 cells. For these experiments, K562 cells were chosen due to their high rate of transfection efficiency. As seen in figure 5A, reducing the expression of individual JAK family members was insufficient to reverse the resistance associated with culturing K562 cells in CM. The reduction in the individual members of JAK family following siRNA transfection was confirmed by Western blotting (Supplementary data S4–A). Since, western blot analysis demonstrates that KU812 cells cultured in CM showed prominent increases in the phosphorylation levels of JAK2 and TYK2 (data not shown) compared to cell cultured in RM, we simultaneously reduced the expression of JAK2 and TYK2 using siRNA strategies. Our data indicate that reducing the expression of both JAK2 and TYK2, was sufficient to reverse the resistance associated with culturing K562 cells in the BM milieu (figure 5B and Supplementary data S4–B). Taken together, these results indicate that CML cells exposed to CM derived from HS-5 stromal cells activates the JAK2-TYK2-STAT3 pathway.
Figure 5. JAK2 and TYK2 are necessary for CM-induced NI resistance in K562 cells.
(A) K562 cells where transfected with siCON (non-silencing siRNA), siJAK1 (siRNA towards JAK1), siJAK2 (siRNA towards JAK2), siJAK3 (siRNA towards JAK3) or siTYK2 (siRNA towards TYK2) independently. After 48 hr, cells were exposed to RM or CM for 3 hr followed by 48-hr treatment with 50 nM NI. Apoptotic cells were detected by Annexin V-FITC staining and FACS analysis with the help of CellQuest Pro software version 4.0.2 (BD Bioscience). Data is the representation of experiment performed twice. (B) K562 cells where transfected with siCON (non-silencing siRNA) or with siJAK2 (siRNA towards JAK2) and siTYK2 (siRNA towards TYK2), either independently or in combination. After 48 hr, cells were exposed to RM or CM for 3 hr followed by 48-hr treatment with 50 nM NI. Apoptotic cells were detected using Annexin V-FITC staining and FACS analysis. The data was performed three independent times and shown is a representative figure performed in triplicate. # no significant difference (p>0.05).
INC424 potentiate cell death in primitive leukemic cells when used in combination with NI
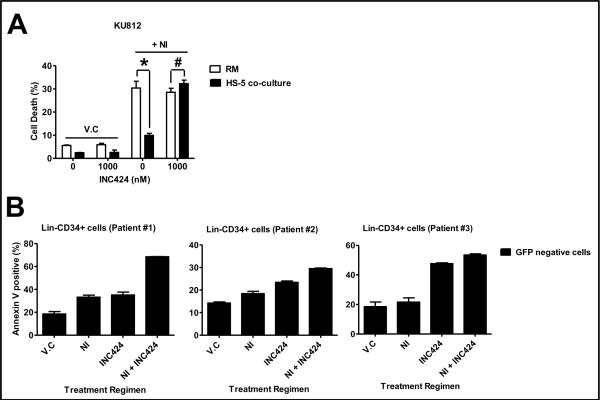
CML cells within the BM are protected against BCR-ABL inhibitors by not only the milieu of cytokines and GF present in the BM but also via its association with the BM stroma though the process of cell adhesion mediated drug resistance. Our earlier results showed that CM-mediated JAK-STAT3 activation is important for protection against NI-mediated cell death. To investigate whether JAK inhibition can also sensitize CML cells to NI within the BM microenvironment, we first tested KU812 cells co-cultured with HS-5-GFP cells and pretreated with INC424, a JAK inhibitor under clinical investigation [21, 22]. As seen in Figure 6A, KU812 cells in co-culture are very resistant to Ni-mediated cell death (30.37 ± 2.95 % in RM as compared to 9.93 ± 0.89 % in CM). However when this cells were pretreated with 1000 nM INC424 the cell death in RM and CM was comparable, demonstrating that the cells were sensitized to NI with inhibition JAK activity (28.55 ± 1.71 % in RM as compared to 32.28 ± 1.51 % in CM). Next, since CML is a leukemic stem cell disease, we tested our combination therapy in Lin−CD34+ leukemic progenitor cells isolated from CML patients. Figure 6B shows the cell death in Lin−CD34+ leukemic stem cells isolated from three CML patients. In all the patient leukemic progenitor cells, INC424 and NI-induced cell death that was above the background cell death. Moreover, combining both the drugs resulted in sensitization resulting in increased cell death in the progenitor cells in all the patient samples (68.54 ± 0.18% in patient #1, 29.51 ± 0.26% in patient #2 and 53.46 ± 0.87% in patient #3). Our result gives a strong evidence for clinical testing of INC424 with NI with the aim of eradicating leukemic stem cells from the BM and in the process treating CML.
Figure 6. INC424 sensitizes leukemic progenitor cells co-cultured with HS-5 GFP stromal cells to NI-mediated cell death.
HS-5-GFP cells were plated at 200,000 cells/mL and incubated overnight. (A) KU812 cells and overnight incubated HS-5 GFP cells were treated with INC424 for 1 hr separately. KU812 cells were then plated on top of HS-F GFP cells. Three hr, after incubation, the cells were treated with vehicle control or NI (25 nM). After 24 hr, cell death in GFP negative cell population was determined using Annexin V-APC staining and FACS analysis. (B) Lin−CD34+ cells from CML patients were isolated as described in the method section. The cells were then plated on the overnight incubated HS-5 cells. After overnight incubation the media was changes and the cells were treated with 1000 nM INC424 or 100 nM NI either individually or in combination. On the third day after treatment, the media was changed and cells were re-treated with INC424 and NI. At the end of five days, cell death in the GFP negative cell population was determined using Annexin V-APC staining and FACS analysis.
Discussion
In this study, we show that STAT3 is an important BCR-ABL-independent target that is only appreciated when studying CML cells in the context of the BM microenvironment. In doing so we identified a pharmacological strategy for inhibiting STAT3 activation in CML cells grown in the context of the microenvironment. Specifically, we show that utilizing a pan-JAK inhibitor was sufficient to block STAT3 activation, inhibit the up-regulation of Mcl-1, and sensitize CML cells to NI-mediated cell death in BM microenvironment. Finally, as proof of principle, we showed that inhibition of BCR-ABL and JAK activity increased elimination of patient-derived primitive leukemic cell in co-culture with BM stromal cells.
In our study, we have utilized soluble factors synthesized de novo in HS-5 stromal cells or normal and CML patient MSC to show STAT3 activation in both CML cell lines and in patient-derived CD34+ progenitor cells. Moreover, this STAT3 activity was sustained even in the presence of NI and resulted in increased gene expression of Mcl-1 and cell survival. The requirement of Mcl-1 in mediating survival in the BM compartment is supported by Opferman et al., who showed that ablation of Mcl-1, a gene tightly regulated by STAT3, led to cell death in the BM compartment [23]. Also, Aichberger et al. showed that IM together with a Mcl-1 antisense resulted in a cooperative anti-leukemic effect in CML [19]. In our study, K562 cells with reduced STAT3 showed decreased expression of Mcl-1 when exposed to CM. Accordingly, these cells showed a reduction in frequency of engraftment of tumor within BM microenvironment in the SCID-Hu mouse model. Taken together, our data indicate that CML cells are less dependent on BCR-ABL signaling for survival when protected by the BM and that inhibition of common downstream signaling pathway initiated by BM microenvironment and ending with STAT3 activation might be essential to circumvent the de novo resistance associated with BM niche.
There is a consensus expounded within published reports that exposure to exogenous cytokines and GF leads to development of resistance against BCR-ABL inhibitors in CML cells [15, 24–26]. In our study, use of CM from HS-5 stromal cells provides a more clinically relevant model to simulate the cytokine milieu found in the BM microenvironment than exogenous addition of a cocktail of cytokines and GF. The list of soluble factors present in HS-5 CM has been well documented and data mining the list showed up several interesting candidates like IL-6, G-CSF and VEGF that could potentially phosphorylate STAT3 via activation of multiple JAK proteins [20]. Indeed, we observed that multiple members of the JAK family of proteins are capable of activating STAT3 in the BM microenvironment. In our study, inhibition of STAT3 in CM exposed cells could only be achieved by using a pan-JAK inhibitor or reducing the expression of both JAK2 and TYK2 expression. This showed that STAT3 was activated within the BM microenvironment through the canonical JAK-STAT pathway. More importantly, our result indicates that there is a redundancy within the system such that independently knocking down individual JAK family of proteins was not sufficient for reversing CM-mediated drug resistance.
Experimental evidence indicates that strategies directed at curing CML must eliminate primitive leukemic progenitor stem cells from the BM. Within the BM, the leukemic stem cells are protected against BCR-ABL inhibitors by not only cell adhesion mediated drug resistance but also via the soluble factors mediated drug resistance. To reproduce these conditions, we have isolated Lin−CD34+ progenitor cells from CML patients and co-cultured them with HS-5 BM stromal cells. In this model we were successful in demonstrating that even though NI was inefficient in inducing cell death in primitive progenitor cell population, addition of INC424, a JAK kinase inhibitor, potentiated the NI-mediated leukemic progenitor cell death. Similar to our observation, Corbin et al showed that survival of CML stem and progenitor cells was BCR-ABL independent and so these cells were not sensitive to BCR-ABL kinase inhibitor-induced cell death [13]. However, the study did not look into the survival signals that are necessary for CML stem cell survival within the BM microenvironment. On the other hand, the inability of NI to induce cell death in primitive progenitor cells within the BM stromal cells is consistent with previous reports showing that cytokine support was sufficient to inhibit IM-mediated cell death in leukemic stem cells [13, 26, 27]. Interestingly, inhibition of JAK activity in primitive progenitor cells is sufficient to sensitize the cells to NI-mediated cell death, demonstrating the importance of the JAK-STAT pathway in the survival of the leukemic progenitor cells in the BM microenvironment.
In conclusion, we have demonstrated the importance of STAT3 in the survival of CML cells in the BM microenvironment, and, in doing so; we have opened up an avenue for future rationally designed combination therapy consisting of a pan-JAK inhibitor in combination with a BCR-ABL inhibitor to test whether this is a viable strategy for eradicating MRD, typically found in BM compartment in CML.
Supplementary Material
KU812 cells (200,000 cells/mL) where exposed to regular media (RM) or conditioned media (CM) for 3 hr followed by treatment with indicated doses of NI. After 24 hr, cells were lysed and subjected to Western blotting with indicated antibodies. GAPDH was used as the loading control.
BM-MNC cells were isolated from BM aspirates of one healthy individual and three CML patients. MSC were cultured as described in Materials and Methods. MSC were characterized by FACS analysis for cell surface markers using the indicated antibodies. Corresponding isotype IgG antibody was used by negative control.
Nuclear extracts (10 ug) from K562 cells exposed to RM or CM for 24 hrs were subjected to electrophoretic mobility shift assay using oligonucleotide for STAT3 binding (5' AGCTTCATTTCCCGTAAATCCCTA 3'). The specificity of the shifted band was determined by adding anti-STAT3 or IgG antibody (1 ug) to the nuclear extract.
(A) K562 cells were transfected with siCON (non-silencing siRNA), siJAK1 (siRNA towards JAK1), siJAK2 (siRNA towards JAK2), siJAK3 (siRNA towards JAK3) and siTYK2 (siRNA towards TYK2) independently. After 48 hrs, cells were lysed and the reduced expression of JAK1, JAK2, JAK3 and TYK2 was confirmed by Western blot analysis. (B) K562 cells were trasnfected with siCON (non-silencing siRNA) or with siJAK2 (siRNA towards JAK2) and siTYK2 (siRNA towards TYK2), either independently or in combination. After 48 hrs the cells were lysed and confirmation of knockdown of JAK2 and TYK2 was performed by Western blot analysis.
Acknowledgements
We are grateful to Novartis Pharma who supplied us with Nilotinib (AMN107) and to Incyte Corporation for providing us with INC424 (INCB018424/Ruxolitinib). We are grateful to Andrew Choi (Brown University) for his technical assistance in performing RT-PCR and the reporter assay. We would also like to thank Rasa G Hamilton (H Lee Moffitt Cancer Center) for her help with the preparation of the manuscript.
Funding Source This work was supported in part by the Leukemia and Lymphoma Society (LAH) and the by the National Cancer Institute R01CA122065 (LAH). Also, supported in part by Flow Cytometry core and Mouse Model core facilities at the H Lee Moffitt Cancer Center and Research Institute, Tampa, FL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authors' Contributions LH and RN designed the experiments; LZ and JP provided intellectual input with respect to specimens obtained; RN, JT and RA performed the laboratory work for this study; RN and LH interpreted the data and wrote the manuscript.
Conflict of Interest The authors report no potential conflicts of interest.
References
- [1].Lugo TG, Pendergast AM, Muller AJ, Witte ON. Science. Vol. 247. New York, NY: 1990. Tyrosine kinase activity and transformation potency of bcr-abl oncogene products; pp. 1079–82. [DOI] [PubMed] [Google Scholar]
- [2].Gishizky ML, Johnson-White J, Witte ON. Evaluating the effect of P210 BCR/ABL on growth of hematopoietic progenitor cells and its role in the pathogenesis of human chronic myelogenous leukemia. Seminars in hematology. 1993;30:6–8. [PubMed] [Google Scholar]
- [3].Johnson KJ, Griswold IJ, O'Hare T, Corbin AS, Loriaux M, Deininger MW, et al. A BCR-ABL mutant lacking direct binding sites for the GRB2, CBL and CRKL adapter proteins fails to induce leukemia in mice. PloS one. 2009;4:e7439. doi: 10.1371/journal.pone.0007439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hochhaus A, O'Brien SG, Guilhot F, Druker BJ, Branford S, Foroni L, et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23:1054–61. doi: 10.1038/leu.2009.38. [DOI] [PubMed] [Google Scholar]
- [5].Kantarjian H, Pasquini R, Levy V, Jootar S, Holowiecki J, Hamerschlak N, et al. Dasatinib or high-dose imatinib for chronic-phase chronic myeloid leukemia resistant to imatinib at a dose of 400 to 600 milligrams daily: two-year follow-up of a randomized phase 2 study (START-R) Cancer. 2009;115:4136–47. doi: 10.1002/cncr.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kantarjian H, Giles F, Wunderle L, Bhalla K, O'Brien S, Wassmann B, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. The New England journal of medicine. 2006;354:2542–51. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- [7].Kantarjian H, Shah NP, Hochhaus A, Cortes J, Shah S, Ayala M, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. The New England journal of medicine. 2010;362:2260–70. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- [8].Kantarjian HM, Hochhaus A, Saglio G, Souza CD, Flinn IW, Stenke L, et al. Nilotinib versus imatinib for the treatment of patients with newly diagnosed chronic phase, Philadelphia chromosome-positive, chronic myeloid leukaemia: 24-month minimum follow-up of the phase 3 randomised ENESTnd trial. The lancet oncology. 2011;12:841–51. doi: 10.1016/S1470-2045(11)70201-7. [DOI] [PubMed] [Google Scholar]
- [9].Ross DM, Branford S, Seymour JF, Schwarer AP, Arthur C, Bartley PA, et al. Patients with chronic myeloid leukemia who maintain a complete molecular response after stopping imatinib treatment have evidence of persistent leukemia by DNA PCR. Leukemia. 2010;24:1719–24. doi: 10.1038/leu.2010.185. [DOI] [PubMed] [Google Scholar]
- [10].Bhatia R, Holtz M, Niu N, Gray R, Snyder DS, Sawyers CL, et al. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 2003;101:4701–7. doi: 10.1182/blood-2002-09-2780. [DOI] [PubMed] [Google Scholar]
- [11].Cortes J, O'Brien S, Kantarjian H. Discontinuation of imatinib therapy after achieving a molecular response. Blood. 2004;104:2204–5. doi: 10.1182/blood-2004-04-1335. [DOI] [PubMed] [Google Scholar]
- [12].Rousselot P, Huguet F, Rea D, Legros L, Cayuela JM, Maarek O, et al. Imatinib mesylate discontinuation in patients with chronic myelogenous leukemia in complete molecular remission for more than 2 years. Blood. 2007;109:58–60. doi: 10.1182/blood-2006-03-011239. [DOI] [PubMed] [Google Scholar]
- [13].Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. The Journal of clinical investigation. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Damiano JS, Hazlehurst LA, Dalton WS. Cell adhesion-mediated drug resistance (CAM-DR) protects the K562 chronic myelogenous leukemia cell line from apoptosis induced by BCR/ABL inhibition, cytotoxic drugs, and gamma-irradiation. Leukemia. 2001;15:1232–9. doi: 10.1038/sj.leu.2402179. [DOI] [PubMed] [Google Scholar]
- [15].Hiwase DK, White DL, Powell JA, Saunders VA, Zrim SA, Frede AK, et al. Blocking cytokine signaling along with intense Bcr-Abl kinase inhibition induces apoptosis in primary CML progenitors. Leukemia. 2010;24:771–8. doi: 10.1038/leu.2009.299. [DOI] [PubMed] [Google Scholar]
- [16].Bewry NN, Nair RR, Emmons MF, Boulware D, Pinilla-Ibarz J, Hazlehurst LA. Stat3 contributes to resistance toward BCR-ABL inhibitors in a bone marrow microenvironment model of drug resistance. Molecular cancer therapeutics. 2008;7:3169–75. doi: 10.1158/1535-7163.MCT-08-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Quirici N, Soligo D, Bossolasco P, Servida F, Lumini C, Deliliers GL. Isolation of bone marrow mesenchymal stem cells by anti-nerve growth factor receptor antibodies. Experimental hematology. 2002;30:783–91. doi: 10.1016/s0301-472x(02)00812-3. [DOI] [PubMed] [Google Scholar]
- [18].Nair RR, Emmons MF, Cress AE, Argilagos RF, Lam K, Kerr WT, et al. HYD1-induced increase in reactive oxygen species leads to autophagy and necrotic cell death in multiple myeloma cells. Molecular cancer therapeutics. 2009;8:2441–51. doi: 10.1158/1535-7163.MCT-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Aichberger KJ, Mayerhofer M, Krauth MT, Skvara H, Florian S, Sonneck K, et al. Identification of mcl-1 as a BCR/ABL-dependent target in chronic myeloid leukemia (CML): evidence for cooperative antileukemic effects of imatinib and mcl-1 antisense oligonucleotides. Blood. 2005;105:3303–11. doi: 10.1182/blood-2004-02-0749. [DOI] [PubMed] [Google Scholar]
- [20].Graf L, Iwata M, Torok-Storb B. Gene expression profiling of the functionally distinct human bone marrow stromal cell lines HS-5 and HS-27a. Blood. 2002;100:1509–11. doi: 10.1182/blood-2002-03-0844. [DOI] [PubMed] [Google Scholar]
- [21].Shi JG, Chen X, McGee RF, Landman RR, Emm T, Lo Y, et al. The Pharmacokinetics, Pharmacodynamics, and Safety of Orally Dosed INCB018424 Phosphate in Healthy Volunteers. Journal of clinical pharmacology. 2011 doi: 10.1177/0091270010389469. [DOI] [PubMed] [Google Scholar]
- [22].Verstovsek S, Kantarjian H, Mesa RA, Pardanani AD, Cortes-Franco J, Thomas DA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. The New England journal of medicine. 2010;363:1117–27. doi: 10.1056/NEJMoa1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Opferman JT, Letai A, Beard C, Sorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–6. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- [24].Belloc F, Airiau K, Jeanneteau M, Garcia M, Guerin E, Lippert E, et al. The stem cell factor-c-KIT pathway must be inhibited to enable apoptosis induced by BCR ABL inhibitors in chronic myelogenous leukemia cells. Leukemia. 2009;23:679–85. doi: 10.1038/leu.2008.364. [DOI] [PubMed] [Google Scholar]
- [25].Chu S, Holtz M, Gupta M, Bhatia R. BCR/ABL kinase inhibition by imatinib mesylate enhances MAP kinase activity in chronic myelogenous leukemia CD34+ cells. Blood. 2004;103:3167–74. doi: 10.1182/blood-2003-04-1271. [DOI] [PubMed] [Google Scholar]
- [26].Konig H, Holtz M, Modi H, Manley P, Holyoake TL, Forman SJ, et al. Enhanced BCR-ABL kinase inhibition does not result in increased inhibition of downstream signaling pathways or increased growth suppression in CML progenitors. Leukemia. 2008;22:748–55. doi: 10.1038/sj.leu.2405086. [DOI] [PubMed] [Google Scholar]
- [27].Holtz MS, Slovak ML, Zhang F, Sawyers CL, Forman SJ, Bhatia R. Imatinib mesylate (STI571) inhibits growth of primitive malignant progenitors in chronic myelogenous leukemia through reversal of abnormally increased proliferation. Blood. 2002;99:3792–800. doi: 10.1182/blood.v99.10.3792. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
KU812 cells (200,000 cells/mL) where exposed to regular media (RM) or conditioned media (CM) for 3 hr followed by treatment with indicated doses of NI. After 24 hr, cells were lysed and subjected to Western blotting with indicated antibodies. GAPDH was used as the loading control.
BM-MNC cells were isolated from BM aspirates of one healthy individual and three CML patients. MSC were cultured as described in Materials and Methods. MSC were characterized by FACS analysis for cell surface markers using the indicated antibodies. Corresponding isotype IgG antibody was used by negative control.
Nuclear extracts (10 ug) from K562 cells exposed to RM or CM for 24 hrs were subjected to electrophoretic mobility shift assay using oligonucleotide for STAT3 binding (5' AGCTTCATTTCCCGTAAATCCCTA 3'). The specificity of the shifted band was determined by adding anti-STAT3 or IgG antibody (1 ug) to the nuclear extract.
(A) K562 cells were transfected with siCON (non-silencing siRNA), siJAK1 (siRNA towards JAK1), siJAK2 (siRNA towards JAK2), siJAK3 (siRNA towards JAK3) and siTYK2 (siRNA towards TYK2) independently. After 48 hrs, cells were lysed and the reduced expression of JAK1, JAK2, JAK3 and TYK2 was confirmed by Western blot analysis. (B) K562 cells were trasnfected with siCON (non-silencing siRNA) or with siJAK2 (siRNA towards JAK2) and siTYK2 (siRNA towards TYK2), either independently or in combination. After 48 hrs the cells were lysed and confirmation of knockdown of JAK2 and TYK2 was performed by Western blot analysis.