Abstract
The plant hormone auxin regulates virtually every aspect of plant growth and development. Auxin acts by binding to the F-box protein TIR1 and promotes the degradation of the Aux/IAA transcriptional repressors. Here, we show that efficient auxin binding requires assembly of an auxin co-receptor complex consisting of TIR1 and an Aux/IAA protein. Heterologous experiments in yeast and quantitative IAA binding assays using purified proteins showed that different combinations of TIR1 and Aux/IAA proteins form co-receptor complexes with a wide range of auxin-binding affinities. Auxin affinity appears to be largely determined by the Aux/IAA. As there are 6 TIR1/AFBs and 29 Aux/IAA proteins in Arabidopsis thaliana, combinatorial interactions may result in many co-receptors with distinct auxin sensing properties. We also demonstrate that the AFB5-Aux/IAA co-receptor selectively binds the auxinic herbicide picloram. This co-receptor system broadens the effective concentration range of the hormone and may contribute to the complexity of auxin response.
INTRODUCTION
Indole-3-acetic acid (IAA, 1) is a small tryptophan-derived phytohormone that regulates many plant growth and developmental processes1 including embryogenesis2,3, tropic growth4, leaf formation5, stem elongation6, root elongation7, and fruit development8. A number of synthetic auxinic compounds have also been identified, most notably 1-naphthaleneacetic acid (1-NAA, 2) and the important herbicides 2,4-dichlorophenoxyacetic acid (2,4-D, 3) and picolinate derivatives instanced by 4-amino-3,5,6-trichloro-2-pyridinecarboxylic acid (picloram, 4)9. Recent studies have produced a coherent model for auxin perception and transcriptional regulation. At low auxin levels, Aux/IAA transcriptional repressors10–12 together with co-repressor proteins including TOPLESS (TPL)13,14, repress genes targeted by Auxin Response Factor (ARF)15,16 transcriptional activators16,17. When auxin levels rise, the Aux/IAA proteins are degraded by the 26S proteasome18. This results in de-repression of ARFs and activation of transcriptional responses19. In Arabidopsis, 29 Aux/IAA proteins have been identified, most of which share a similar domain structure. Domain I (DI) binds to TPL and is required for transcriptional repression. Domain II (DII) contains the degron motif, a sequence of 13 amino acids that is required for the characteristic instability of Aux/IAA proteins. Domains III (DIII) and IV (DIV) mediate homo and heterodimerization, including interactions with ARF proteins11. Since synthesis of many Aux/IAA proteins themselves is rapidly auxin-induced, auxin signaling undergoes cycles of negative feedback regulation20,21.
Auxin-dependent degradation of the Aux/IAAs occurs through the action of a family of E3 ligases called SCFTIR1/AFB1–5. The F-box protein TIR1 (TRANSPORT INHIBITOR RESPONSE1) and related proteins AFB1–5 (AUXIN SIGNALING F-BOX PROTEIN1, 2, 3, 4, 5) are the substrate specificity determinants, or substrate receptors for the SCF22–24. Surprisingly, substrate recognition requires direct binding of auxin to the F-box protein24–26. Along with identifying a long-sought mechanism of auxin perception, this was the first demonstration of an SCF-substrate interaction that is regulated by the direct binding of a small ligand. The structure of TIR1 was determined in the presence of auxin and a 13 amino acid degron peptide from DII of Aux/IAA protein IAA727,28. The resulting model reveals key aspects of TIR1-auxin binding, as well as the TIR1-Aux/IAA interaction. First, the model shows that the mushroom-like fold of the TIR1-ASK1 complex, formed by the 18 Leucine-Rich-Repeats (LRRs) in the C-terminus of TIR1, is essential for Aux/IAA and auxin binding. Second, it confirms that post-translational modifications are not required for auxin or Aux/IAA binding to TIR1. Third, it reveals that auxin binding to TIR1 does not result in changes in TIR1 conformation. Rather, auxin appears to function by extending the protein interaction interface and increasing the affinity of TIR1 for the Aux/IAA protein. This is supported by the fact that auxin occupies the binding pocket in TIR1, just underneath the Aux/IAA binding site. Last, it reveals that an Inositol (1,2,3,4,5,6) hexakisphosphate (InsP6) cofactor is bound at the core of TIR1.
Pull-down experiments indicate that all six members of the TIR1/AFB family function as auxin receptors. However, a number of studies have shown that individual TIR1/AFB proteins have distinct biochemical properties and biological functions. For example, TIR1 and AFB2 exhibit a much stronger interaction with Aux/IAA proteins than AFB1 and AFB3 do24,29. AFB3 has been shown to have a unique role in the nitrate response of roots30. In addition, genetic experiments indicate that AFB4 negatively regulates auxin response, unlike other members of the family29.
Although important advances have been made in our understanding of auxin perception, a number of key questions remain including the nature of the auxin receptor itself. For example, it is not clear if TIR1/AFB receptors can bind auxin alone, or whether the Aux/IAA protein also contributes to auxin binding. Similarly, the binding properties of individual TIR1/AFB proteins for different auxins and Aux/IAA proteins have not been systematically determined. In this study, we use a variety of biochemical approaches to obtain answers to these questions. Our results demonstrate both the stunning complexity and diversity of the auxin perception mechanism.
Results
TIR1 and Aux/IAA proteins act as auxin coreceptors
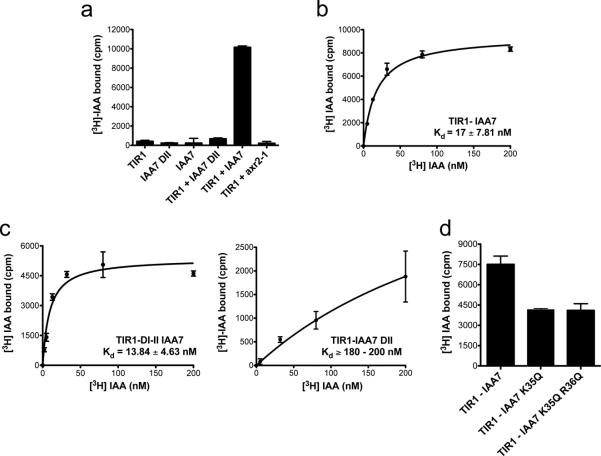
To evaluate the molecular requirements of auxin binding we carried out in vitro auxin binding assays using [3H]-IAA. For these experiments, we co-expressed a tagged version of TIR1 with ASK1 in Hi5 insect cells28. TIR1-ASK1 was incubated with labeled IAA and either a synthetic peptide derived from domain II of IAA7 (IAA7 DII)22 or full-length IAA7. We found that TIR1, the IAA7 DII, and IAA7 all lacked appreciable binding to IAA, while the combination of TIR1 together with a molar excess of IAA7 DII peptide exhibited relatively low binding to auxin (Fig. 1a). In strong contrast, TIR1 with full-length IAA7 bound auxin with high affinity (KD = 17.81 ± 7.81 nM) (Fig. 1b). We also evaluated auxin binding to TIR1 in combination with a mutated version of IAA7 that carries a Pro87Ser substitution in DII (IAA7axr2-1). This mutation corresponds to a dominant mutation that stabilizes IAA7 protein in vivo (called axr2-1) and abolishes the TIR1-Aux/IAA interaction31,32. The TIR1-IAA7axr2-1 combination did not bind auxin, confirming that both proteins are required for significant binding (Fig. 1a).
Figure 1. The Auxin Receptor Is a Co-Receptor System.
a.In vitro binding of 200 nM [3H] IAA to recombinantly expressed TIR1 and/or IAA7 full-length or a peptide corresponding to the DII, degron motif. Together, the TIR1-IAA7 pair constitutes an auxin co-receptor. A mutation that mimics a gain of function allele in the degron of IAA7 (IAA7axr2-1) abolishes auxin binding. B. and c. Saturation binding experiments of [3H] IAA to b. TIR1-IAA7 and c. TIR1-DI-DII (left) and TIR1-IAA7 DII co-receptor complexes (right). b. TIR1-IAA7 constitutes a high-affinity auxin co-receptor with a KD in the low nanomolar range. c. TIR1-DI-DII but not TIR1-IAA7 DII binds auxin with high affinity. d. [3H]-IAA binding at 200 nM to TIR1-IAA7 compared to TIR1-IAA7 K35Q and TIR1-IAA7 K35Q R36Q. Values are determined as mean ± SEM of either two or three independent experiments carried out in triplicate.
To further investigate the contribution of the Aux/IAA protein to auxin binding, we carried out binding experiments using TIR1 together with truncated versions of IAA7 containing the DI and DII domains or DII only. Saturation binding assays showed that the TIR1-DI-DII complex binds auxin with an affinity similar to that of TIR1-IAA7 (KD = 13.84 ± 4.63 nM) (Fig. 1c). On the other hand, the TIR1-IAA7 DII combination exhibited a binding affinity one order of magnitude lower (KD = 218.40 ± 25.80 nM) than the TIR1- IAA7 combination. This is in agreement with the results of surface plasmon resonance (SPR) experiments done with TIR1 and an immobilized DII peptide, in which we obtained a binding affinity for IAA of 111 nM (Supplementary Results, Supplementary Fig. 2). These results indicate that DII of the Aux/IAAs is essential for interaction with TIR1 but also indicate that other sequences within IAA7 contribute to complex formation. Previous studies show that a conserved Lys Arg (KR) motif between DI and DII is required for basal proteolysis of Aux/IAA proteins20,33. To determine the role of the KR motif in auxin binding, we mutated Lys35 and Arg36 to glutamine and tested IAA binding. In both Lys35Gln and Lys35Gln Arg36Gln mutants, auxin binding was diminished around 50% indicating that the KR motif also contributes to assembly of the complex (Fig. 1d). Taken together, our in vitro auxin binding assays demonstrate that TIR1 and the Aux/IAA are both necessary and sufficient for auxin binding and act as auxin co-receptors (Fig. 1a and Supplementary Fig. 1a).
Coreceptor pairs assemble at different auxin levels
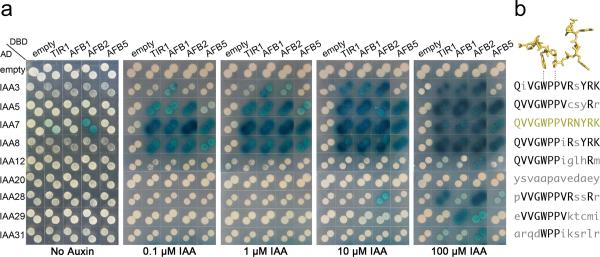
Previously, we showed that TIR1 and AFB1, 2, and 3 have similar but distinct roles in auxin signaling and speculated that these differences might relate to differential interactions with the Aux/IAA proteins24. To investigate this possibility, we analyzed a number of TIR1/AFB-Aux/IAA pairs in a yeast two-hybrid assay (Y2H) (Fig. 2). Nine Aux/IAA proteins representing distinct subclades34 were chosen for this analysis. Seven of these contained the canonical GWPPV degron motif, one (IAA31) contained a degenerate form of this motif, and one (IAA20) completely lacks DII (Fig. 2b). TIR1/AFB and Aux/IAA expression level in yeast was assessed by immunoblot analysis (Supplementary Fig. 7). This analysis showed that the TIR1, AFB1, AFB2 and AFB5 fusion proteins were similarly expressed. The Aux/IAA proteins alsoaccumulated to a roughly similar level, allowing a qualitative assessment of their relative ability to form co-receptors complexes. Each co-receptor combination was evaluated on media supplemented with increasing concentrations of auxin. Strikingly, we observed different dose-response relationships for different pairs of proteins. Among the Aux/IAAs tested, only IAA7 interacts with TIR1/AFBs in the absence of auxin. IAA5, IAA7, and IAA8 interact with all the TIR1/AFBs at 0.1 μM IAA. IAA3 also bound TIR1, AFB1, and AFB2 at this concentration but was a poor substrate for AFB5. In contrast IAA12, IAA28, and IAA29 required much higher concentrations of IAA to interact with the F-box proteins. IAA12 interacted specifically with TIR1 and AFB2 at 100 μM IAA, suggesting that at least in the yeast system, higher IAA levels are required to form stable TIR1 or AFB2-IAA12 complexes. The interaction between IAA28 and AFB2 and TIR1 was particularly strong at concentrations over 10 μM, whereas IAA29 interacted only with AFB1 and AFB2 at high auxin levels (Fig. 2a). Since all of these proteins include the GWPPv degron motif, our results suggest that additional amino acids, either within DII, or elsewhere in the protein, contribute to the interaction with TIR1/AFBs (Fig. 2b). Additionally, the evolutionarily divergent IAA31 protein interacted weakly with the TIR1/AFBs. Finally, IAA20 did not interact with any of the TIR1/AFB proteins even at high concentrations. This suggests that these Aux/IAAs are not substrates for SCFTIR1/AFB or that a different ligand is required to promote the interaction. Overall, the results of our Y2H experiments suggest that there are substrate preferences among the TIR1/AFB proteins. Certain Aux/IAA proteins, such as IAA3, IAA5, IAA7 and IAA8, are generally better substrates for TIR1/AFBs than IAA12, IAA28 and IAA31. Our assays also indicate that the degron motif is necessary for co-receptor assembly but that other sequences probably contribute to complex formation.
Figure 2. Differences in Auxin Dependent TIR1/AFB-Aux/IAA Interaction Are Not Exclusively Determined by the Degron Domain.
a. Yeast-two hybrid interaction experiments of TIR1, AFB1, AFB2 and AFB5 with IAA3, IAA5, IAA7, IAA8, IAA12, IAA20, IAA28, IAA29, IAA31, which represent the different subclades of Arabidopsis Aux/IAAs. Diploids containing LexA DBD-TIR1/AFBs and ADAux/IAAs were generated and spotted in selective media (Gal/Raff -Ura-His-Trp + X-Gal) containing increasing concentrations of IAA. β-galactosidase reporter expression evidenced IAA-induced protein-protein interactions 4 days after spotting. b. Aux/IAA proteins with a very similar DII domain interact differentially with TIR1/AFBs suggesting that regions outside of DII contribute to binding. IAA7 DII depicted as stick (yellow), with conserved tryptophan and second proline residues, which interact with the surrounding hydrophobic wall in the TIR1 pocket and stack against the auxin molecule lying underneath. DII sequences of the selected Aux/IAAs are shown for comparison (right).
Aux/IAA proteins determine coreceptor affinity for auxin
One of the most interesting questions in auxin biology is how the hormone controls so many different aspects of plant growth and development. Based on the results of our Y2H analysis, it is possible that different TIR1/AFB-Aux/IAA co-receptor pairs exhibit biochemical differences that enable specialized functions. For example, co-receptor pairs may have unique affinities for auxin and therefore respond to different auxin concentrations. To address this question, we carried out either saturation or homologous competitive IAA binding assays in which different TIR1-Aux/IAA combinations were incubated with a fixed concentration of radiolabeled IAA and increasing amounts of cold IAA (Fig. 1 and 3, Supplementary Fig. 1b–c). These experiments show that the binding affinity of IAA for the co-receptor complexes ranges from 10 nM to >1 μM. IAA14 is the closest IAA7 paralog and the two proteins have an identical GWPPVR amino acid sequence in DII. Indeed, the TIR1-IAA14 co-receptor pair binds auxin with very high affinity, KD ~10 nM, similar to TIR1-IAA7 co-receptor pair (Fig. 3a and Table 1). Interestingly, TIR1-IAA17 has a slightly lower affinity for IAA (KD ~30 nM) even though IAA17 is closely related to IAA7 and has a nearly identical DII (Table 1, Supplementary Fig. 1b). The TIR1-IAA1 and TIR1-IAA3 co-receptor pairs have affinities between 17 – 45 nM for IAA, while TIR1-IAA28 binds auxin with a KD of approximately 75 nM (Fig. 3b–c, Supplementary Fig. 1b, Table 1). Consistently, when we used stabilized DII mutant forms of the various Aux/IAA proteins in our binding assays, we observed that independent of the binding properties of the native TIR1-Aux/IAA pair, auxin binding was abolished (Fig. 3e).
Figure 3. Aux/IAA Proteins Determine the Affinity of the Co-receptor Complex for Auxin and, Together with TIR1, Form a Series of Co-Receptor Complexes With a Range of Auxin Sensing Properties.
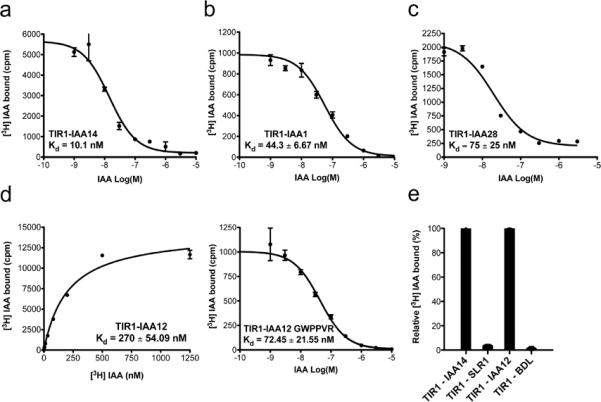
Homologous competitive binding experiments for a. TIR1-IAA14, b. TIR1-IAA1 and c. TIR1-IAA28 co-receptor complexes. Specific binding of a constant concentration of [3H] IAA by the different co-receptors in the presence of various concentrations of unlabeled IAA was measured. IC50 values were determined using appropriate concentrations (5, 10, 25 or 100 nM) of radiolabeled auxin, so that the concentration of hot IAA was less than half the IC50. KD was then calculated as the IC50- [[3H]-IAA] fitted to a built-in equation of one-site competition. d. Saturation binding of the TIR1-IAA12 co-receptor complex, which exhibits low auxin binding affinity in the high nanomolar range (left). The binding affinity of the TIR1-IAA12 co-receptor for IAA increases when the canonical degron motif GWPPVR is incorporated in IAA12 (right). e. Mutated versions of Aux/IAA proteins that mimic the stabilized versions of the proteins (solitary root (slr-1) and bodenlos (bdl), abolish specific auxin binding by the different TIR1-Aux/IAA co-receptor complexes. Binding of [3H] IAA (200 nM) to recombinant TIR1-ASK1 and IAA14, IAA12, slr and bdl, where error bars correspond to SEM of four replicates.
Table 1.
Auxin Binding Activity of Various Auxin Co-Receptor Complexes
| Auxin Co-Receptor | DI and Degron Domain (DII) of AUX/IAA | Auxin | Kd or Ki* (Mean ± SEM) (nM) |
n |
|---|---|---|---|---|
| TIR1-IAA7 | KRGFSETVDLMLNLQSNKEGSVDLKNVSAVPKEKTTLKDPSKPPAKAQVVGWPPVRNYRKNMMTQQKTSS | IAA | 17 ± 7.81 | 3 |
| 1-NAA | 113.50 ± 3.50 | 2 | ||
| 2,4-D | 248 – 1000 | 4 | ||
| Picloram | 3900 ± 910 | 2 | ||
|
| ||||
| AFB5-IAA7 | KRGFSETVDLMLNLQSNKEGSVDLKNVSAVPKEKTTLKDPSKPPAKAQVVGWPPVRNYRKNMMTQQKTSS | IAA | 51.32 ± 12.65 | 3 |
| Picloram | 54.70 ± 3.84 | 3 | ||
|
| ||||
| TIR1-IAA14 | KRGFSETVDLKLNLQSNKQGHVDLNTNGAPKEKTFLKDPSKPPAKAQVVGWPPVRNYRKNVMANQKSGE | IAA | 10.10 | 2 |
|
| ||||
| TIR1-IAA3 | KRVLSTDTEKEIESSSRKTETSPPRKAQIVGWPPVRSYRKNNIQSKKNES | IAA | 16.97 ± 3.43 | 3 |
|
| ||||
| TIR1-IAA17 | KRGFSETVDLKLNLNNEPANKEGSTTHDVVTFDSKEKSACPKDPAKPPAKAQVVGWPPVRSYRKNVMVSCQKSS | IAA | 33 ± 3.00 | 2 |
|
| ||||
| TIR1-IAA1 | KRKNNDSTEESAPPPAKTQIVGWPPVRSNRKNNNN | IAA | 44.33 ± 6.67 | 2 |
|
| ||||
| TIR1-IAA28 | KRLELRLAPPCHQFTSNNNINGSKQKSSTKETSFLSNNRVEVAPVVGWPPVRSSRRNLTAQLKE | IAA | 75 ± 25 | 2 |
|
| ||||
| TIR1-IAA12 | KRSAESSSHQGASPPRSSQVVGWPPIGLHRMNSLVNNQA | IAA | 270 ± 54.09 | 3 |
| TIR1-IAA12 GWPPVR | KRSAESSSHQGASPPRSSQVVGWPPVRLHRMNSLVNNQA | IAA | 72.45 ± 21.55 | 3 |
|
| ||||
| TIR1-IAA31 | MEVSNSCSSFSSSSVDSTKPSPSESSVNLSLSLTFPSTSPQREARQDWPPIKSRLRDTLKGRRL | IAA | > 1000 (NF) | 3 |
|
| ||||
| TIR1-DI-DII | MIGQLMNLKATELCLGLPGGAEAVESPAKSAVGSKRGFSETVDLMLNLQS NKEGSVDLKNVSAVPKEKTTLKDPSKPPAKAQVVGWPPVRNYRKNMMTQQKTSS | IAA | 13.84 ± 4.63 | 2 |
| TIR1-biotinylated peptide DII | AKAQVVGWPPVRNYRKN | IAA | 218 ± 25.80 | 3 |
Kd and Ki values were determined by nonlinear regression from saturation binding experiments and/or homologous competition binding assays, respectively, n, number of independent auxin binding assays performed in triplicates. NF, no fit. The highlighted residues match the KR motif and the canonical GWPPVR in DII of IAA7.
We next explored the auxin binding capabilities of the TIR1-IAA12 pair, since this complex assembles only at high auxin concentrations in Y2H assays (Fig. 2a). In accordance with the yeast results, the KD for auxin of the TIR1-IAA12 co-receptor was 270.25 ± 54.09 nM, suggesting that this co-receptor has comparatively low affinity for auxin (Fig. 3d).
To specifically determine the contribution of the canonical degron motif to the auxin binding affinity of the co-receptors, we mimicked the degron domain of IAA7 in IAA12 (IAA12 GWPPVR) and tested it in the auxin-binding assay. Although TIR1-IAA12 GWPPVR showed an increased affinity for IAA (KD = 72.45 ± 21.55 nM), we were not able to obtain KD values equal to those observed for the TIR1-IAA7 co-receptor pair (Fig. 1b and 3d). This is striking additional evidence that regions outside of DII of Aux/IAA proteins also contribute to interaction with TIR1 and to auxin binding. Constitutively expressed fusions of DIIs of IAA8, 9 and IAA28 to VENUS have been recently used as auxin sensors in vivo35, but our results indicate that DI-DII of Aux/IAAs are required for the co-receptor to have full auxin sensing properties. More importantly, our data show that Aux/IAA proteins determine the auxin binding properties of the TIR1-Aux/IAA co-receptor complex, which might allow the formation of a spectrum of auxin sensors.
TIR1 mutations affect coreceptor complex assembly
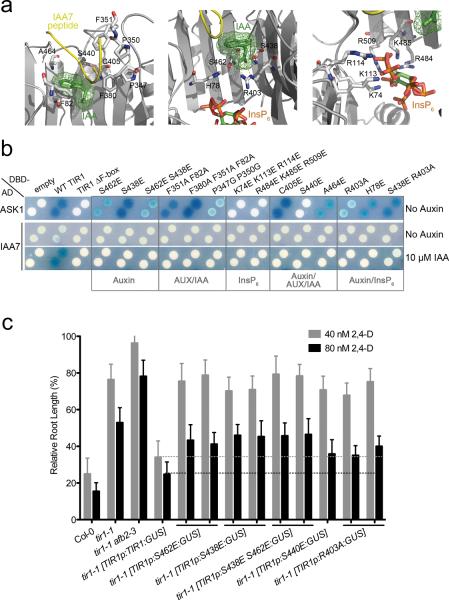
Structural studies identified a number of key residues that participate in co-receptor assembly and auxin binding28 (Fig. 4a). To explore the function of these residues, we generated mutant TIR1 proteins and evaluated their interaction with either ASK1 or IAA7 in Y2H assays (Fig. 4b). TIR1 and the AFBs interacted with ASK1 independently of auxin, and a mutant form of TIR1 that lacks the F-box motif was unable to interact with ASK1 (Supplementary Fig. 3a).
Figure 4. Mutations at Auxin, Aux/IAA and InsP6 Binding Sites Impair Auxin-Dependent TIR1-Aux/IAA Interaction And Compromise TIR1 Function In Vivo.
a. Side view of auxin (green) and IAA7 peptide (yellow) (left), auxin and InsP6 (rainbow) (middle), and InsP6 (right) binding sites in TIR1 (grey). Selected ligand-interacting residues in TIR1 are shown in mostly white stick representation. b. Yeast-two hybrid ASK1, and auxin-induced IAA7 interactions with wild type (WT) TIR1 or TIR1 carrying mutations on ligand-binding sites. c. Five-day-old seedlings grown on MS media were transferred to media containing either 40 or 80 nM 2,4-D. Root elongation was measured after additional 4 days and expressed as a proportion of growth in the absence of auxin (scale bars represent STDEV). Auxin inhibits root growth elongation in wild type plants (Col-0) but tir1-1, as well as the tir1-1afb2–3 auxin-receptor mutants are resistant to auxin treatment. Auxin resistance in tir1-1 mutants is reverted by introducing TIR1 wild type fused to the β-glucuronidase (GUS) reporter gene, under the TIR1 promoter. Interrupted lines indicate that unlike TIR1p:TIR1:GUS, versions of TIR1 that carry mutations in ligand binding sites are unable to restore auxin sensitivity in tir1-1.
Except for Ser440, substitution of residues implicated in auxin and/or Aux/IAA binding did not affect interaction with ASK1. However, our studies showed that these residues are all required for the auxin-dependent interaction of TIR1 with IAA7. Thus, substitution of residues within the auxin binding groove (Ser462 and Ser438), residues that contribute to Aux/IAA interaction (Phe82, Phe351, Phe380, Pro347, Pro350), residues that form the canopy of the auxin-binding groove (Cys405, Ser440, Ala464), and residues that are implicated in both auxin and InsP6 binding (His78, Arg403, Ser438) all abolish interaction with IAA7 (Fig. 4b).
We also addressed the role of InsP6 on TIR1 function (Fig. 4a–b). The InsP6 binding pocket is surrounded by positively charged residues including Lys74, Lys113, Arg114, Arg484, Lys485 and Arg509. These residues are thought to support the floor of the auxin-binding pocket. In addition, InsP6 contacts Arg403, the residue that interacts with the carboxyl group of auxin28. Two mutant proteins were generated, each of which replaced three of these residues with glutamate. Both mutant proteins failed to interact with IAA7 in the presence of auxin in yeast and in pull-down experiments using tagged-recombinant Aux/IAA protein (Fig. 4b and Supplementary Fig. 3d). This is consistent with an important role for InsP6 in TIR1 function. Further, neither mutant version of TIR1 was able to interact with ASK1 in yeast, suggesting that InsP6 has a broader role in determining TIR1 architecture. Finally, we characterized mutant proteins previously identified in forward genetic screens using the Y2H assay36,37. The tir1–1 and tir1–2 mutations completely abolished the interaction between the mutant protein and either ASK1 or IAA7, suggesting that these substitutions result in major changes in protein structure (Supplementary Fig. 3b). On the other hand, the tir1–6, tir1–7, and wei1 mutations have a modest effect on IAA7 interaction (Supplementary Fig. 3c). This correlates with the weaker phenotypes of these mutants.
To determine if these mutant proteins function in plants, we introduced mutant versions of TIR1 fused to GUS under control of the TIR1 promoter into wild-type and tir1–1 plants (Fig. 4c). We previously showed that expression of a TIR1 cDNA from the TIR1 promoter rescues the auxin resistant phenotype of tir1–1 plants in root elongation assays24. We introduced single and double mutants affecting putative auxin and Aux/IAA binding sites, including Ser438Glu, Ser462Glu, Ser440Glu or Arg403Ala. At least 4 transgenic lines were identified for each mutation and were characterized with respect to GUS expression and auxin response. None of the transgenes was able to restore auxin sensitivity to tir1–1 plants. This suggests that the TIR1 mutant proteins are not functional and indicates that Ser438, Ser462, Ser440 and Arg403 are essential for TIR1 function in planta (Fig. 4c). In summary, mutagenesis of TIR1 provides strong support for the published structural model. Our data indicate that TIR1-Aux/IAA co-receptor formation, and interaction with ASK1 are essential for auxin sensing in vivo.
Auxin agonists differentially stabilize TIR1-Aux/IAA
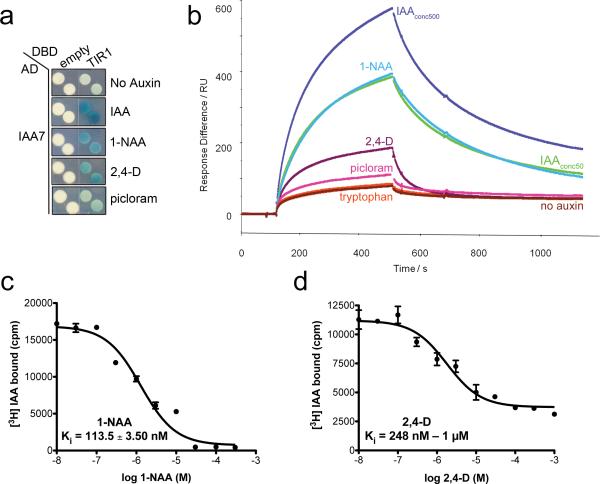
The auxin binding groove of TIR1 consists of a three walled hydrophobic cavity, with an open roof and two selective polar residues that anchor the auxins to the base of the pocket28. The pocket can accommodate various auxin analogues including 1-NAA and 2,4-D. Moreover, other studies have shown that various chemically diverse compounds have auxin agonist as well as antagonist activity9,38,39. The synthetic auxins 1-NAA, 2,4-D, and picloram exhibit auxin activity in a wide range of physiological responses and promote TIR1/AFB-Aux/IAA interactions in vitro39. We asked if the synthetic auxins 1-NAA, 2,4-D and picloram promote TIR1-Aux/IAA interaction in yeast. We observed that all three auxins promoted TIR1-IAA7 interaction, but not to the same extent as IAA (Fig. 5A). We next tested the contribution of the different auxins to the TIR1-Aux/IAA dynamic interaction using SPR analyses. In this experiment, we evaluated the kinetics of co-receptor complex formation using an immobilized biotinylated peptide corresponding to the degron motif of IAA7 (DII) and purified TIR1 protein, in the absence or presence of different auxins. A small amount of basal binding occurred between TIR1 and the IAA7 DII peptide in the absence of auxin. In contrast, co-injection of TIR1 with IAA, NAA, 2,4-D, and picloram enhanced association with the peptide. Strikingly, we also observed that the dissociation rate of the complexes differed, depending on the auxin in the system (Fig. 5b). Comparing the sensorgrams at 50 μM ligand concentration, little difference is observed between IAA and NAA on TIR1-DII association, while 2,4-D addition led to a much more rapid dissociation rate. This accounts for the apparent reduction in Rmax by working against association of the complex. TIR1-IAA7 DII complex formation with picloram was weaker and exhibited a rapid dissociation. We carried out heterologous competitive binding experiments for comparison and determined that the TIR1-IAA7 co-receptor complex has a higher binding affinity for 1-NAA than for 2,4-D (KD = 113.50 ± 3.50 nM and between 248 nM and 1 μM, respectively) (Fig. 5c, d, Table 1). Taken together, our data show that distinct auxins differentially stabilize the TIR1-IAA7 co-receptor complex and that an important determinant of affinity is the dissociation rate. Based on published structural studies, IAA, 1-NAA, and 2,4-D are anchored in the binding pocket through interactions between their carboxylate group and the TIR1 residues Arg403 and Ser438. However, only IAA establishes an additional connection to the walls of the pocket through an interaction between the -NH group of the indole ring of IAA and Lys439 residue of TIR1. Due to its double aromatic rings, 1-NAA provides a larger hydrophobic platform for association with the auxin binding pocket of TIR1 than 2,4-D28. Computational comparative analysis of hydrogen bond energy formation of TIR1-IAA7 and various auxins has also suggested that IAA forms stronger bond interactions with the co-receptor40. This is consistent with the slower dissociation rate observed for the TIR1-IAA-IAA7 DII complex.
Figure 5. Auxin Agonists Differentially Stabilize TIR1-Aux/IAA Complexes.
a. IAA, 1-NAA, and 2,4-D enhance TIR1-IAA7 interaction in yeast. Diploids carrying LexA DBD-TIR1 and AD-IAA7 were spotted on selective media containing 50 μM auxins. β-galactosidase expression is a measure of TIR1-IAA7 interaction. b. Surface Plasmon Resonance analysis of auxin-dependent TIR1-IAA7 DII interaction. Sensorgram shows the effect of no auxin (brown) and various auxinic compounds (500 μM IAA (dark blue), 50 μM IAA (light green), 50 μM 1-NAA (light blue), 50 μM 2,4-D (purple), 50 μM picloram (pink), 50 μM tryptophan (orange)) on TIR1-DII peptide association and dissociation. Auxins were mixed with TIR1 in solution prior to injection over DII peptide. c and d. Competitive [3H] IAA binding assays using 1-NAA and 2,4-D cold competitors. TIR1-IAA7 co-receptor complex binds 1-NAA and 2,4-D with a Ki (inhibition constant) of 113.50 ± 3.51 nM and > 1 μM, respectively.
The AFB5-IAA7 Co-Receptor Complex Binds Picloram
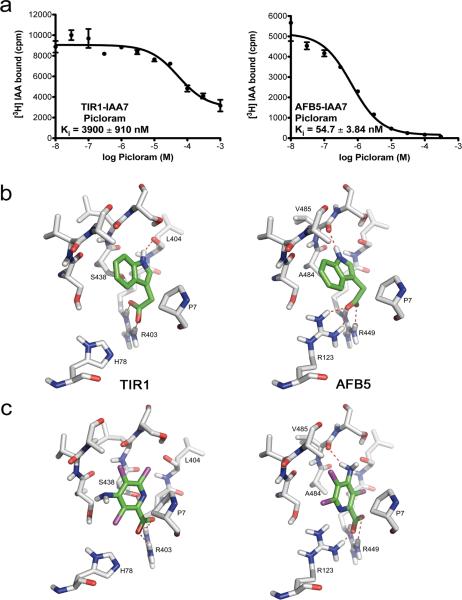
The AFB4 and AFB5 have diverged significantly from the other members of the TIR1/AFB family24. Previous studies indicate that the afb5 mutant plants are resistant to the synthetic auxin picloram while tir1 plants are not, suggesting that AFB5 exhibits agonist selectivity41. Recently, we presented evidence that both afb4 and afb5 are resistant to picloram and that the AFB4 clade may be the major targets for this class of herbicides29. To investigate the biochemical basis for this selectivity, we compared the auxin binding affinity of AFB5-IAA7 to TIR1-IAA7. In competition binding assays, we evaluated the binding of radiolabeled IAA to either TIR1-IAA7 or AFB5-IAA7 in the presence of increasing concentrations of unlabeled picloram (Fig. 6a). We found that while the AFB5-IAA7 co-receptor complex exhibited a relatively high affinity for picloram (Ki = 54.90 ± 3.84 nM), picloram was not able to displace IAA efficiently from TIR1-IAA7 (Ki = 3900 ± 910 nM). These results provide evidence for AFB5 selectivity and an explanation for the picloram resistance of afb4 and afb5 mutant plants. Further, we evaluated the ability of the AFB5-IAA7 co-receptor to bind IAA. Based on saturation binding curves, AFB5-IAA7 binds IAA with a KD of 51.32 ± 12.65 nM, slightly similar to that of TIR1-IAA7 for IAA (KD ~10 nM) or AFB5-IAA7 for picloram (Supplementary Fig. 4b, Table 1). Next, we compared the kinetics of association and dissociation of the AFB5-IAA7 and TIR1-IAA7 complexes in SPR experiments in the presence of various auxins, including picloram. We found that picloram promotes rapid assembly of the AFB5-IAA7 complex, less so the TIR1-IAA7 complex, although as for AFB5-IAA7, dissociation dominates the extent of complex assembly. Indeed, we found that dissociation of the AFB5-IAA7 complex was more rapid than that of TIR1-IAA7, irrespective of the auxin (Supplementary Fig. 4c).
Figure 6. TIR1-IAA7 And AFB5-IAA7 Co-Receptor Complexes Exhibit Differential Auxin Binding Affinities.
a. Heterologous competition binding experiments of TIR1-IAA7 and AFB5-IAA7 co-receptor complexes. Picloram displaces efficiently [3H] IAA binding by AFB5-IAA7, but not by the TIR1-IAA7 complex. b–c. 3D-structures of TIR1 and AFB5 show an almost identical fold with regard to secondary structure arrangements. b. Docking arrangements of IAA to TIR1 (left) and AFB5 (right) and c. of picloram to TIR1 (left) and AFB5 (right). IAA and picloram are stabilized in the TIR1 auxin binding pocket by hydrogen bonds to Arg403 (b and c left). IAA and picloram form strong hydrogen bonds (salt bridges) to Arg123 and Arg449 and to the backbone carbonyl group of Val485 in AFB5 (b and c (right)). b. Picloram lacks the hydrogen bond to the backbone carbonyl group of Leu404 in TIR1. The replacement of Arg123 (in AFB5) by His78 (in TIR1) is likely responsible for the reduced affinity of picloram to TIR1 compared to AFB5. c.
To address differences in the TIR1-IAA7 and AFB5-IAA7 co-receptor complexes, we built a structural model of the AFB5 protein by homology modeling using the Arabidopsis TIR1 structure as a template (Fig. 6b–c, Supplementary Fig. 5). AFB5 differs from TIR1 at two positions in the auxin binding pocket; His78 and Ser438 in TIR1 are replaced by Arg123 and Ala484 in AFB5, respectively. We carried out docking calculations of IAA in AFB5 and picloram in TIR1 and AFB5 binding pockets (Fig. 6b–c). The simulations indicate that the His-Arg exchange has a strong influence on the docking arrangements and affinities of TIR1 and AFB5 for IAA and picloram. Despite differences in their ring structure, picloram, IAA, and 2,4-D oriented in the cavity in a remarkably similarly way (Fig. 6b–c). However, electrostatic interactions differed. In TIR1, Arg403 tethers the carboxylate of picloram (and other auxins) through a salt bridge to the bottom of the auxin-binding pocket. By contrast, AFB5 Arg123 as well as Arg449, (TIR1 Arg403 equivalent), establish strong salt bridges with the carboxylate group of auxin. Also in AFB5, the imino group of IAA and the amino group of picloram form a hydrogen bond to the backbone carbonyl group of Val485. Hydrophobic interactions of the aromatic ring systems with Pro7 of IAA7 DII, further stabilize the docking arrangements for auxins in AFB5 (Fig. 6b, c). These simulations largely support our binding experiments and show that the affinity of AFB5 for picloram is nearly as high as TIR1 for IAA (Fig. 6b–c, Supplementary Figs. 4). Taken together, these data clearly show that AFB5 acts as a high affinity sensor for IAA and exhibits a stronger binding affinity for picloram. Further detailed studies will directly establish further the basis for this selectivity.
DISCUSSION
Recent studies have produced an exciting new view of auxin perception. Biochemical and structural experiments indicate that auxin acts directly to promote an interaction between the F-box protein TIR1 and the Aux/IAA transcriptional repressors. However, many details of the mechanism of auxin perception remain to be elucidated. Here we used a variety of approaches to perform a detailed characterization of the auxin receptor.
The structural model of TIR1 implicated a number of residues in auxin, Aux/IAA, and InsP6 binding. When we investigated the function of these residues in genetic studies, we found that these amino acids are essential for TIR1 structure and/or function in both yeast and Arabidopsis.
Based on our initial studies, we proposed that auxin may act by binding to both TIR1 and the Aux/IAA protein, thus stabilizing the SCFTIR1-Aux/IAA complex25,28. To test this possibility we performed auxin-binding assays with purified proteins. Our results show that high-affinity auxin binding requires the assembly of an auxin co-receptor complex consisting of TIR1 and an Aux/IAA protein. Further, we found that auxin binding with the combination of TIR1 and the IAA7 DII was one order of magnitude weaker compared to TIR1-full length Aux/IAA protein indicating that sequences outside DII contribute to complex formation.
The auxin co-receptor model has important implications. Since there are 29 Aux/IAA proteins in Arabidopsis, it is possible that a number of qualitatively different co-receptor pairs may exist. Our Y2H results strongly suggest that this is the case. We observed variation in the auxin dose-response for a number of the TIR1/AFBAux/IAA receptor pairs. Interestingly, the strength of TIR1-Aux/IAA interaction inversely correlates with Aux/IAA stability. For example, IAA7 exhibits a very strong interaction with TIR1/AFBs in yeast, and has a half-life of ~10 min in Arabidopsis upon auxin treatment23. On the other hand, IAA31, a relatively stable Aux/IAA, interacts poorly with the TIR1/AFBs. Although the yeast experiments provide information on the relative affinity of different co-receptor pairs for auxin, they are not quantitative. Since IAA is transported out of yeast cells, and is probably metabolized as well, the intracellular IAA concentration in is uncertain42. In addition Nishimura et al43 have shown that Aux/IAA proteins are degraded in yeast cells expressing TIR1 in the presence of auxin.
Our in vitro studies provide direct evidence that different co-receptor pairs have different auxin-binding affinities. The KD for IAA among the co-receptor pairs ranges from ~ 10 nM in the case of TIR1-IAA7, to ~ 300 nM for TIR1-IAA12, and > 1 μM for TIR1-IAA31 using full-length Aux/IAAs. To determine if changes in domain II alone are responsible for differences in affinity, we mutated this domain in IAA12 to match the sequence of IAA7. This change substantially increased the IAA affinity of the TIR1-IAA12 pair, but not to the level of TIR1-IAA7. Further we found that mutation of the conserved KR motif, located between DI and DII, significantly reduces IAA affinity.
The variation in binding affinity among the co-receptor pairs dramatically increases the dynamic range of auxin sensitivity. Thus, we would expect TIR1-IAA7 to respond to very low IAA concentration, whereas IAA12 would only be degraded at IAA levels in the high nanomolar range. Interestingly, axr2 mutants expressing a stable version of IAA7 exhibit an altered growth response to an extremely low concentration of auxin44 suggesting that IAA7 mediates responses to low hormone levels. On the other hand, IAA12 protein (also known as BDL) has an important function during embryogenesis when localized surges of auxin regulate development of the apical-basal axis and formation of the root and shoot meristems3,45. We would expect TIR1-IAA12 to be responsive to the high IAA levels that result from these surges. In this regard however, it is also important to note that we have not excluded the possibility that IAA12 has a high affinity for one of the other AFB proteins.
Based on our yeast data, the IAA affinity of the co-receptor seems determined primarily by the Aux/IAA protein and not by the F-box protein. There are a few exceptions to this rule. For example, IAA28 paired with AFB2 has a higher apparent affinity than TIR1, AFB1 or AFB5, while IAA29 produces the strongest response with AFB1. More striking is the behaviour of AFB5. Our results support earlier genetic studies indicating that AFB5 exhibits a specific interaction with the synthetic auxin picloram. Our experiments revealed that the AFB5-IAA7 complex has a much higher affinity than TIR1-IAA7 for picloram. Homology modeling suggests that this difference may be related to substitution of His78 in TIR1 with Arg123 in AFB5.
In summary, we have shown that auxin is perceived by a co-receptor complex consisting of a TIR1/AFB protein and of an Aux/IAA protein. We also determined that both protein partners contribute to ligand selectivity. Additionally, we showed that different co-receptor complexes exhibit very different affinities for auxin, dramatically increasing the dynamic range of the hormone. This likely contributes to the versatility of auxin as a signaling molecule throughout plant growth and development.
METHODS
Plant materials and growth assays
Wild-type Columbia (Col-0) and tir1-1 transgenic Arabidopsis plants expressing TIR1p:TIR1-GUS were described previously24. Various mutated versions of TIR1 were introduced by site-directed mutagenesis into the TIR1 cDNA cassette in the TIR1p:TIR1-GUS construct, and wild-type, as well as tir1-1 plants were transformed and selected on hygromycin. At least 4 T2 lines were used for root elongation assays (see Supplementary Methods).
LexA yeast two hybrid assays
Yeast two hybrid assays were carried out as in34. DBD-AFB1–5 and AD Aux/IAA constructs were generated and tested as described in Supplementary Methods.
[3H]-labeled Auxin Binding Assay
Radioligand binding assays were performed using purified ASK1-TIR1 and ASK1-AFB5 protein complexes recombinantly expressed in insect cells as in28. GST-tagged Aux/IAAs or their GST-Aux/IAA dominant mutated versions were expressed in E.coli. In brief, in a typical auxin binding assay, 0.44 μg of ASK1-TIR1 or ASK1-AFB5 complex and 1:10 molar radio of Aux/IAA proteins were incubated in 100 μL binding buffer (50 mM Tris-HCl pH 8.0, 200 mM NaCl, 10% glycerol, 0.1% Tween 20, protease inhibitors) with 200 nM [3H]-IAA for over an hour at 4°C. [3H]-IAA with a specific activity of 20 Ci/mmol from American Radiolabeled Chemicals, Inc. was used. The samples were immobilized in polyethelenemide pre-treated fiberglass filters and washed three times with binding buffer. Upon over-night incubation, auxin binding was quantified by scintillation counting. Nonspecific binding was determined using 1000x excess of cold IAA with respect to [3H]-IAA. Specific binding was determined as the average of at least three measurements of total binding – nonspecific binding. For saturation binding assays, samples were prepared as above and incubated with at least six IAA concentrations on either side of the KD of a given co-receptor pair. For homologous and non-homologous competition binding assays, various concentrations of cold IAA, 1-NAA, 2,4-D and picloram were added to the samples containing a fixed concentration of [3H]-IAA. Depending on the estimated IC50 of the competitive compound 5, 10, 25 or 100 nM of [3H]-IAA was used. For competitive binding assays, KD and Ki values were obtained by fitting the normalized specific binding affinity versus the logarithm of the unlabeled auxin concentration (Log[IAA], Molar) using the one-site competition model. Nonlinear regression in Prism 5, GraphPad Software, Inc. was used for all data analysis.
Surface Plasmon Resonance
SPR measurements were performed using Biacore 2000 and 3000 systems (Biacore GE Healthcare Biosciences). Biotinylated DII IAA7 peptide (Biotin-AKAQVVGWPPVRNYRKN) or respective Aux/IAA DII peptides were immobilized onto a streptavidin-coated Biacore SA sensor chip in sample cell. Reference cell was blocked with either a mutated version of DII IAA7 (Biotin-AKAQVVEWSSGRNYRKN) as negative control, or coated with biocytin. Purified ASK1-TIR1 or ASK1-AFB5 proteins were injected over the chip at a flow rate of 25 μl/min in HBSEP buffer (20 mM Hepes [pH 7.4], 150 mM NaCl, 3.4 mM ethylenediaminetetraacetic acid [EDTA], and 0.005% P20) ± various concentrations of either IAA, 1-NAA, 2,4-D, picloram or tryptophan. Data analysis was performed with BIAevaluation software (Biacore GE Healthcare Biosciences).
Aux/IAA Peptides
For radioligand binding assays, biotinylated peptides corresponding to the wild-type and mutated degron domain of IAA7 were purchased from Midwest Bio-Tech, Fischers, IN, USA. 5 mg of Biotin (LC) AKAQVVGWPPVRNYRKN and Biotin (LC) AKAQVVGWSPVRNYRN peptides were synthesized to a purity of 90–95% by HPLC, according to manufacturer. For SPR analyses, biotinyl -(NH)-AKA QVV GWP PVR NYR KN-(COOH) and mutant peptide biotinyl -(NH)-AKAQVVEWSSGRNYRKN-(COOH) were synthesized by Thermo Fisher Scientific, Ulm, Germany to a >80% purity, generally 94–95% pure.
Homology Modeling of AFB5
AFB5 was modeled using the Prime module of the Schrodinger 2010 software package46, using the Arabidopsis TIR1 structure as a template (PDB code 2P1Q:chain B). After pair wise sequence alignment (Supplementary Fig. 6a) a model structure was built, followed by side-chain optimization and energy minimization. The quality of the model was first analyzed with PROCHECK47. The Ramachandran Plot showed 88.3% of all backbone dihedral angles in most favored areas, 10.5% in additional allowed ones, 3 amino acid residues in generally allowed ones and one outlier (Ser298) appeared (Supplementary Fig. 5a). Since for the template X-ray structure of TIR1 (2P1Q_B) the corresponding values are slightly better (90.5%, 9.3% with no outlier), the model was refined with the md-refinement tool of YASARA using the YASARA2 force field. The resulting structure was slightly improved with regard to the Ramachandran plot analysis. No longer an outlier was detected and 89.1% of all backbone dihedral angles were in the most favored area, 10.5% in additionally allowed ones. All other criteria such as planarity of peptide bonds, bond length and angles for a good stereochemical quality obtained by the PROCHECK analysis were fulfilled. To analyze the model structure for a putative native fold, an analysis with PROSA II was performed48 (Supplementary Fig. 5b). The energy graph is almost identical to the X-ray structure template. The resulting combined z score value for the model of AFB5 with 566 amino acids is −13.85 which is close to the one of the X-ray structure with 571 amino acids and a z score of - 14.30. This result is a strong indication for a native like folded model structure. Both 3D-structures show an almost identical fold with regard to secondary structure arrangements.
Molecular Modeling of Interactions between Picloram and TIR1 and AFB5
Automated docking simulations were performed with the algorithm-based Induced FIT Docking protocol of the Schrödinger suite. Prior to the calculation, each of the protein and ligand structures was prepared with hydrogens added according to the expected protonation at pH 7, using the protein preparation and ligand preparation wizards available within Maestro. The best docking poses were selected by conformational clustering analysis and the binding score. The refined AFB5 structure and the TIR1 structure (after 3D-protonated with MOE49) were taken for docking of IAA and picloram as ligands using PLANTS with the ChemPLP scoring function and the Lennard Jones potential for intra ligand energy calculations50. For each ligand and protein, 30 docking poses were calculated from which the most favored (lowest docking scores) were compared (Supplementary Fig. 6b).
Supplementary Material
Acknowledgments
We thank Elisabeth J. Chapman for helpful discussions and comments to the manuscript; and Ray Shao and Ina Kim for technical assistance. We also thank Andrew McCammon for hosting part of the computational analyses. We gratefully acknowledge financial support from the National Institutes of Health (R01 CA107134 to N.Z. and T32 GM07270 to L.B.S.), Howard Hughes Medical Institute (M.E. and N.Z.), BBSRC (BB/F013981/1 to S.K. and BB/F014651/1 to R.N.). The McCammon group, including A.I. and C.D.O. is supported by NIH, NSF, and HHMI.
We dedicate this work to the memory of Laura B. Sheard, a talented young scientist who made key contributions at the early stage of this work. Her life was tragically cut short while this manuscript was under review.
Footnotes
Author Contributions. L.I.A.C.V. and M.E. prepared the manuscript. L.I.A.C.V. designed, performed the experiments and analyzed the data. X.T. and H.M. purified TIR1-ASK1 complex. L.I.A.C.V. expressed and purified AFB5-ASK1 complex. G.P. contributed to the generation of Y2H clones, C.D.O., A.I., and W.B. carried out homology modeling of AFB5 and docking experiments of picloram and IAA. S.L., L.A.R.N. and S.K. expressed TIR1-ASK1 and AFB5-ASK1 constructs. S.L., S.K. and R.N. designed and performed SPR experiments. L.B.S. and N. Z. helped the expression and purification of TIR1-ASK1, AFB5-ASK1 complexes and the initial radioligand binding experiments. S.K., R.N., L.S. and N.Z. provided input to the manuscript. M.E. oversaw the project and approved the intellectual content.
Competing Financial interests The authors declare no competing financial interest.
References
- 1.Auxin Signaling-From Synthesis to Systems Biology. Cold Spring Harbour Laboratory Press; New York: 2011. [Google Scholar]
- 2.Hamann T, Benkova E, Baurle I, Kientz M, Jurgens G. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev. 2002;16:1610–5. doi: 10.1101/gad.229402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weijers D, et al. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell. 2006;10:265–70. doi: 10.1016/j.devcel.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Holland JJ, Roberts D, Liscum E. Understanding phototropism: from Darwin to today. J Exp Bot. 2009;60:1969–78. doi: 10.1093/jxb/erp113. [DOI] [PubMed] [Google Scholar]
- 5.Scarpella E, Barkoulas M, Tsiantis M. Control of leaf and vein development by auxin. Cold Spring Harb Perspect Biol. 2010;2:137–153. doi: 10.1101/cshperspect.a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vernoux T, Besnard F, Traas J. Auxin at the shoot apical meristem. Cold Spring Harb Perspect Biol. 2010;2:107–120. doi: 10.1101/cshperspect.a001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Overvoorde P, Fukaki H, Beeckman T. Auxin control of root development. Cold Spring Harb Perspect Biol. 2010;2:121–136. doi: 10.1101/cshperspect.a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundberg E, Ostergaard L. Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb Perspect Biol. 2009;1:155–168. doi: 10.1101/cshperspect.a001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodward AW, Bartel B. Auxin: regulation, action, and interaction. Ann Bot. 2005;95:707–35. doi: 10.1093/aob/mci083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abel S, Theologis A. Early genes and auxin action. Plant Physiol. 1996;111:9–17. doi: 10.1104/pp.111.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overvoorde PJ, et al. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell. 2005;17:3282–300. doi: 10.1105/tpc.105.036723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ. AUX/IAA proteins are active repressors and their stability and activity are modulated by auxin. The Plant Cell. 2001;13:2809–2822. doi: 10.1105/tpc.010289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long JA, Ohno C, Smith ZR, Meyerowitz EM. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312:1520–3. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- 14.Szemenyei H, Hannon M, Long JA. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science. 2008;319:1384–6. doi: 10.1126/science.1151461. [DOI] [PubMed] [Google Scholar]
- 15.Okushima Y, et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell. 2005;17:444–63. doi: 10.1105/tpc.104.028316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weijers D, et al. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005;24:1874–85. doi: 10.1038/sj.emboj.7600659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell. 1997;9:1963–71. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell. 2001;13:2349–60. doi: 10.1105/tpc.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–85. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- 20.Dreher KA, Brown J, Saw RE, Callis J. The Arabidopsis Aux/IAA protein family has diversified in degradation and auxin responsiveness. The Plant Cell. 2006;18:699–714. doi: 10.1105/tpc.105.039172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Worley CK, et al. Degradation of Aux/IAA proteins is essential for normal auxin signalling. Plant J. 2000;21:553–62. doi: 10.1046/j.1365-313x.2000.00703.x. [DOI] [PubMed] [Google Scholar]
- 22.Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9:109–19. doi: 10.1016/j.devcel.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 24.Parry G, et al. Complex regulation of the TIR1/AFB family of auxin receptors. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–5. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 26.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435:446–51. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 27.Calderón-Villalobos LI, Tan X, Zheng N, Estelle M. Auxin perception-structural insights. Cold Spring Harb Perspect Biol. 2010;2:47–62. doi: 10.1101/cshperspect.a005546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan X, et al. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–5. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 29.Greenham K, et al. The AFB4 Auxin Receptor Is a Negative Regulator of Auxin Signaling in Seedlings. Curr Biol. 2011;21:520–5. doi: 10.1016/j.cub.2011.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Vidal EA, et al. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2010;107:4477–82. doi: 10.1073/pnas.0909571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dharmasiri N, Dharmasiri S, Jones AM, Estelle M. Auxin action in a cell-free system. Curr Biol. 2003;13:1418–22. doi: 10.1016/s0960-9822(03)00536-0. [DOI] [PubMed] [Google Scholar]
- 32.Timpte C, Wilson AK, Estelle M. The axr2-1 mutation of Arabidopsis thaliana is a gain-of-function mutation that disrupts an early step in auxin response. Genetics. 1994;138:1239–49. doi: 10.1093/genetics/138.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abel S, Oeller PW, Theologis A. Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci U S A. 1994;91:326–30. doi: 10.1073/pnas.91.1.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prigge MJ, Lavy M, Ashton NW, Estelle M. Physcomitrella patens Auxin-Resistant Mutants Affect Conserved Elements of an Auxin-Signaling Pathway. Curr Biol. 2010;20:1907–12. doi: 10.1016/j.cub.2010.08.050. [DOI] [PubMed] [Google Scholar]
- 35.Vernoux T, et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol Syst Biol. 2011;7:508. doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alonso JM, et al. Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc Natl Acad Sci U S A. 2003;100:2992–7. doi: 10.1073/pnas.0438070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruegger M, et al. The TIR protein of Arabidopsis function in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev. 1998;12:198–207. doi: 10.1101/gad.12.2.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi K, et al. Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc Natl Acad Sci U S A. 2008;105:5632–7. doi: 10.1073/pnas.0711146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Hasenstein KH. Physiological interactions of antiauxins with auxin in roots. J Plant Physiol. 2010;167:879–84. doi: 10.1016/j.jplph.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 40.Hao GF, Yang GF. The role of Phe82 and Phe351 in auxin-induced substrate perception by TIR1 ubiquitin ligase: a novel insight from molecular dynamics simulations. PLoS One. 2010;5:e10742. doi: 10.1371/journal.pone.0010742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walsh TA, et al. Mutations in an auxin receptor homolog AFB5 and in SGT1b confer resistance to synthetic picolinate auxins and not to 2,4-dichlorophenoxyacetic acid or indole-3-acetic acid in Arabidopsis. Plant Physiol. 2006;142:542–52. doi: 10.1104/pp.106.085969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prusty R, Grisafi P, Fink GR. The plant hormone indoleacetic acid induces invasive growth in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4153–7. doi: 10.1073/pnas.0400659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature methods. 2009;6:917–22. doi: 10.1038/nmeth.1401. [DOI] [PubMed] [Google Scholar]
- 44.Evans ML, Ishikawa H, Estelle M. Responses of Arabidopsis roots to auxin studied with high temporal resolution-comparison of wild-type and auxin-response mutants. Planta. 1994;194:215–222. [Google Scholar]
- 45.Jenik PD, Jurkuta RE, Barton MK. Interactions between the cell cycle and embryonic patterning in Arabidopsis uncovered by a mutation in DNA polymerase epsilon. The Plant cell. 2005;17:3362–77. doi: 10.1105/tpc.105.036889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prime. V 2.1. Schoedinger, LLC; New York, NY: 2010. [Google Scholar]
- 47.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. Procheck-A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993:283–291. [Google Scholar]
- 48.Sippl MJ. Calculation of conformational ensembles from potentials of mean force. An approach to the knowledge-based prediction of local structures in globular proteins. J Mol Biol. 1990;213:859–83. doi: 10.1016/s0022-2836(05)80269-4. [DOI] [PubMed] [Google Scholar]
- 49.MOE, version 2008.10; Molecular Operating Environment. Chemical Computing Group Inc.; Montreal, Canada: 2009. [Google Scholar]
- 50.Korb O, Monecke P, Hessler G, Stutzle T, Exner TE. pharmACOphore: multiple flexible ligand alignment based on ant colony optimization. J Chem Inf Model. 2010;50:1669–81. doi: 10.1021/ci1000218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.