Summary
Arf GTPases are key regulators of both retrograde and anterograde traffic at the Golgi complex. The Golgi-localized Arf activators, Arf-GEFs (guanine exchange factor) of the BIG/GBF family, are poorly understood in terms of both their regulatory and localization mechanisms. We have performed a detailed kinetic characterization of a functional Golgi Arf-GEF, the trans-Golgi network (TGN)-localized Sec7 protein from yeast. We demonstrate that Sec7 is regulated by both autoinhibition and positive feedback. We show that positive feedback arises through the stable recruitment of Sec7 to membranes via its HDS1 domain by interaction with its product, activated Arf1. This interaction mediates localization of Sec7 to the TGN, as deletion of the HDS1 domain or mutation of the HDS1 domain in combination with deletion of Arf1 significantly increases cytoplasmic localization of Sec7. Our results lead us to propose a model in which Arf-GEF recruitment is linked to Golgi maturation via Arf1 activation.
Introduction
The Golgi complex is the primary membrane and protein sorting station in the secretory pathway of eukaryotic cells (De Matteis and Luini, 2008; Glick and Nakano, 2009). Virtually all protein traffic out of the Golgi is controlled by GTPases of the Arf family that act by recruiting effectors, including cargo adaptors and vesicle coats, to sort cargo and generate transport carriers (D’Souza-Schorey and Chavrier, 2006; Gillingham and Munro, 2007; Kahn, 2009; Donaldson and Jackson, 2011). Arf family GTPases are also thought to contribute some of the mechanical force required to deform membranes during transport carrier formation, because activated Arf proteins can tubulate membranes in vitro and in vivo (Aridor et al., 2001; Lee et al., 2005; Beck et al., 2008; Krauss et al., 2008).
The structural and biochemical mechanism of Arf activation via nucleotide exchange by GEF domains is well characterized (Beraud-Dufour et al., 1998; Goldberg, 1998), and important regulatory features of the peripherally localized Arf-GEFs ARNO/Cytohesin-1/Grp1 have recently been described (DiNitto et al., 2007; Stalder et al., 2011). However, despite the essential function of Arf GTPases in membrane trafficking at the Golgi, their activation at this organelle remains poorly understood. Arf-GEF proteins of the BIG and GBF families are responsible for Golgi-localized Arf activation (Morinaga et al., 1996; Peyroche et al., 1996; Casanova, 2007), but the BIG/GBF family proteins share no detectable sequence homology with the ARNO/Cytohesin-1/Grp1 family outside of the GEF domain. Moreover, the BIG/GBF Arf-GEFs are fundamental regulators of intra-Golgi and Golgi-derived traffic in all eukaryotes, whereas the ARNO/Cytohesin-1/Grp1 Arf-GEFs appear to have cell-type specific functions at the plasma membrane (Kolanus et al., 1996; Klarlund et al., 1997; Venkateswarlu et al., 1998). The importance of the BIG/GBF Arf-GEFs is underscored by the association of mutations in the BIG2/ARFGEF2 gene with neuronal disease (Sheen et al., 2004; de Wit et al., 2009).
Yeast possess a single member of the BIG subfamily, Sec7, which activates the Arf1 and Arf2 (Arf1/2) GTPases at the trans-Golgi network (TGN) (Franzusoff et al., 1991), and two members of the GBF family, Gea1 and Gea2, which activate Arf1/2 at early Golgi compartments (Peyroche et al., 1996; Spang et al., 2001). SEC7 was among the first genes identified to act in the secretory pathway, and temperature-sensitive sec7 mutants accumulate greatly exaggerated TGN membrane compartments (Novick et al., 1980; Rambourg et al., 1993), consistent with its role in regulating virtually all anterograde traffic out of the TGN.
The two major unresolved questions regarding the Golgi Arf-GEFs are how their activity is regulated and how they achieve their subcellular localization. We now demonstrate that Sec7 activates Arf1 through a positive feedback mechanism and is also subject to autoinhibitory regulation. Using in vitro assays, we show that positive feedback occurs through Arf1-GTP dependent recruitment of Sec7 to the membrane surface via a conserved domain, HDS1. Our data suggest that the HDS1 domain switches from an autoinhibitory state to an activating state upon binding to Arf1-GTP. To probe the physiological significance of our in vitro data, we determine that the HDS1 domain also mediates stable interaction between Sec7 constructs and Arf1 in vivo. Remarkably, we find that the HDS1 domain cooperates with Arf1 to mediate localization of Sec7 to the TGN in vivo. We further determine that the HDS2-4 domains exert an autoinhibitory role and provide additional TGN-directed targeting, possibly through coincidence detection. Our results lead us to propose a model for Arf-GEF recruitment to the Golgi that is intimately linked to Golgi cisternal maturation.
Results
Purification of a Sec7 protein that provides essential SEC7 function
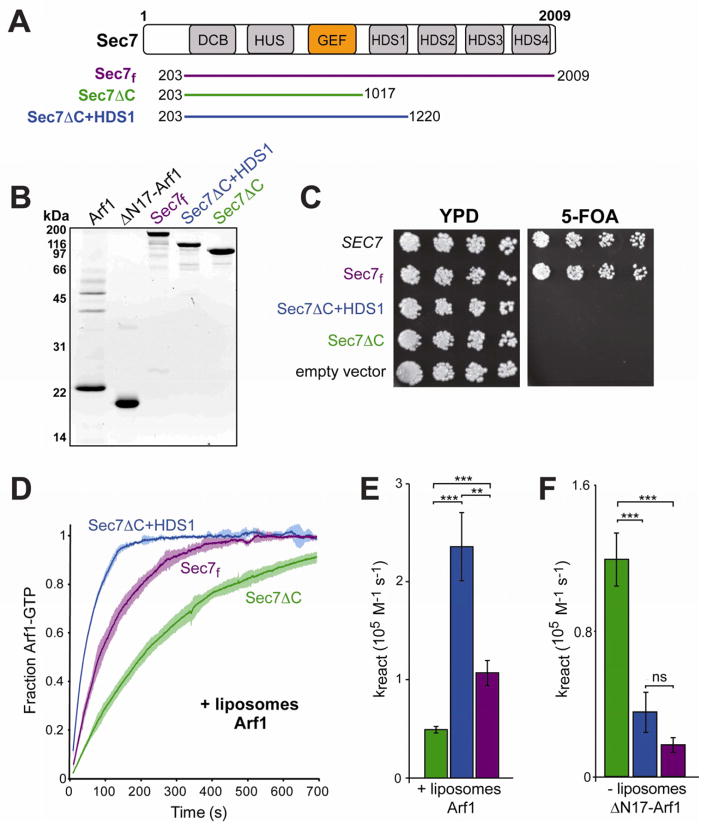
The prototypical member of the BIG and GBF families, Sec7 (for which the “Sec7” GEF domain is named), is 2009 amino acid residues in length, but the GEF domain itself comprises only ~200 amino acids. The remainder of Sec7 is highly conserved through humans. Sequence conservation was previously used to identify conserved regions within the BIG and GBF family members, and these regions have been ascribed domain names based on this conservation (Mouratou et al., 2005; Bui et al., 2009) (Figure 1A).
Figure 1. Purification of a functional Sec7 construct uncovers distinct regulatory domains in the C-terminus.
(A) Schematic diagram of the Sec7 truncated constructs used for this study, shown with the conserved domain structure of Sec7. The domain names are: DCB, dimerization and cyclophilin binding; HUS, homology upstream of Sec7 domain; GEF, guanine exchange factor (sometimes referred to as the Sec7 domain); HDS, homology downstream of Sec7 domain. Note that the HDS1, 2, 3, and 4 domains are not homologous with each other.
(B) 5 μg of each construct used in this study was run on a 15% SDS-PAGE gel and stained for total protein.
(C) Plasmid-borne Sec7 constructs (top to bottom: pCF1045, pCF1046, pCF1136, pCF1135, pRS415) with SEC7 promoters were tested for their ability to complement a sec7Δ mutation via 5-FOA counter-selection plasmid shuffling, using yeast strain CFY409.
(D) The nucleotide-bound state of Arf1 was monitored by increase in native tryptophan fluorescence. Dark lines represent the average of three normalized reactions; lighter surrounding areas represent the corresponding 95% confidence intervals for each time point.
(E) Quantification of reaction rate from curves in (D). Curves were fit to a single exponential and normalized for measured [Sec7] to obtain the overall reaction rate. Bars are colored as per construct coloring in (A). Error bars represent 95% confidence intervals; significance is measured by one-way ANOVA with post-processing to correct for multiple comparisons.
(F) Quantification of reaction rates in the absence of liposomes. A construct lacking the membrane insertion domain of Arf1 (ΔN17-Arf1) was used to permit exchange in the absence of liposomes. Error bars as in (E).
See also Figure S1.
Characterization of this family of ArfGEFs has been hindered by the difficulty in purifying protein constructs encoding the entirety of the functional gene products. To investigate the function of the non-GEF domains of Sec7, we sought to produce a purified fragment that retained the essential function of the full-length protein. We achieved robust expression of a well-behaved N-terminal deletion construct of Sec7 encoding residues 203–2009 (Figure 1A,B). The N-terminal 202 amino acids missing from this construct are poorly conserved and are predicted to lack secondary structural elements, so we expected this region to be dispensable for Sec7 function. As SEC7 is an essential gene in yeast, we tested the ability of the Sec7(203–2009) fragment to complement a sec7Δ null mutant, and found that the N-terminal 202 residues are indeed dispensable for growth in vivo (Figure 1C). Thus, the Sec7(203–2009) protein fragment that we have recombinantly produced and purified encodes the full essential function of the endogenous SEC7 gene product; for simplicity we hereafter refer to this purified protein as “Sec7f” to denote that this is a fully functional construct.
Two further recombinant Sec7 fragments were purified for this study: a construct comprising residues 203–1017 (“Sec7ΔC”), which contains the N-terminal region and the GEF domain, and a construct comprising residues 203–1220 (“Sec7ΔC+HDS1”), which contains the N-terminal region, the GEF domain, and the conserved HDS1 (homology downstream of Sec7) domain (Figure 1A,B). Neither construct complemented a sec7Δ mutant, indicating that the C-terminus is required for the essential function of Sec7 (Figure 1C).
Sec7 GEF activity is stimulated by region(s) outside of the GEF domain
We hypothesized that the non-GEF domains of Sec7 might regulate the activity of the GEF domain. To test this hypothesis, we used native tryptophan fluorescence to monitor Sec7-catalyzed Arf1 nucleotide exchange in real-time. This assay (Higashijima et al., 1987) has been used extensively to investigate the enzymatic kinetics of several different GEF proteins, including activators of Arf-family proteins (Beraud-Dufour et al., 1998; Futai et al., 2004; DiNitto et al., 2007). Our experiments were performed using approximately physiological concentrations of GEF (100 nM, based on ~3,700 Sec7 molecules per cell (Ghaemmaghami et al., 2003)) and GTPase (670 nM, based on ~19,000 Arf1 molecules per cell (Ghaemmaghami et al., 2003)).
When we measured the GEF activity of these Sec7 constructs towards Arf1 (N-terminally myristoylated form) in the presence of TGN-like synthetic liposomes (see Supplemental Experimental Procedures), we found that all were considerably more active than the isolated GEF domain (Figures 1D,E, and S1A,B). Furthermore, Sec7f was significantly more active than Sec7ΔC (Figures 1D,E and S1C). While constructs containing regions C-terminal to the GEF domain, but lacking the N-terminal region (residues 1–815) could not be tested due to poor behavior in solution (our unpublished results), the results thus far indicate that regions in both the N-terminus and C-terminus enhance the activity of the GEF domain.
Membranes modulate the autoinhibitory or activating potential of the Sec7 C-terminus
Although removing the C-terminus from the Sec7f construct (to generate Sec7ΔC) results in a loss of activity, reintroducing just the HDS1 domain to generate Sec7ΔC+HDS1 results in a construct with activity even higher than that of Sec7f (Figure 1D,E). Therefore, the HDS2-4 domains have an autoinhibitory function, whereas the HDS1 domain has an activating function, relative to the Sec7ΔC construct.
To determine the role of membranes in the autoregulatory behavior of the C-terminus, we performed the GEF activity assay using an Arf1 construct lacking the amphipathic N-terminal helix (ΔN17-Arf1) as a substrate. In contrast to myristoylated Arf1, ΔN17-Arf1 does not require the presence of biological membranes to become activated (Kahn et al., 1992; Antonny et al., 1997), permitting their removal from the assay. We note that it is not necessarily informative to compare the rates of a given Sec7 construct between reactions with and without liposomes: the different reactions involve different substrates (ΔN17-Arf1 versus Arf1) known to possess different intrinsic activation rates (Antonny et al., 1997). Therefore, we focus our analysis on the relative rates of the different Sec7 constructs for each substrate.
Surprisingly, the absence of membranes resulted in a markedly different activity profile of the constructs. In contrast to its activating role in reactions with Arf1 and liposomes, the HDS1 domain has an autoinhibitory effect in solution, as the activity of Sec7ΔC+HDS1 was less than that of Sec7ΔC under these conditions (Figures 1F, S1D). We found that Sec7ΔC+HDS1 exhibits a smaller hydrodynamic volume (Stokes radius) than Sec7ΔC, despite possessing a larger native molecular weight (Table S1). This implies that autoinhibition is associated with a more tightly closed conformation of the GEF in solution, in which the HDS1 domain may sequester the GEF domain against the N-terminus in order to prevent its access to substrate.
Taken together, these results suggest that the HDS1 domain acts as a switch, exerting either an inhibitory or an activating function, and switching between the two states is modulated by membranes.
Activated Arf1 stably recruits Sec7 to membranes through interaction with the conserved HDS1 domain
To further characterize the membrane-dependent switch, we sought to determine the membrane-bound status of the various Sec7 constructs under our reaction conditions. The peripherally-localized Arf-GEFs of the ARNO/Grp1/Cytohesin family contain a Sec7-GEF domain and a C-terminal PH domain that mediates its binding to membranes containing the signaling phospholipids PI(3,4,5)P3 or PI(4,5)P2 (Chardin et al., 1996; Klarlund et al., 2000). Structural elements proximal to the PH domain are autoinhibitory in solution (DiNitto et al., 2007), but the presence of the PH domain significantly increases the activity of the GEF domain in these proteins by enforcing membrane proximity (Chardin et al., 1996; Klarlund et al., 1997). More recently, the PH domain of these proteins has also been shown to interact with the activated, membrane-bound GTPases Arf6, Arf1, and Arl4, enabling it to modulate GEF activity and localization via GTPase cascades or positive feedback (Cohen et al., 2007; DiNitto et al., 2007; Hofmann et al., 2007; Li et al., 2007; Stalder et al., 2011).
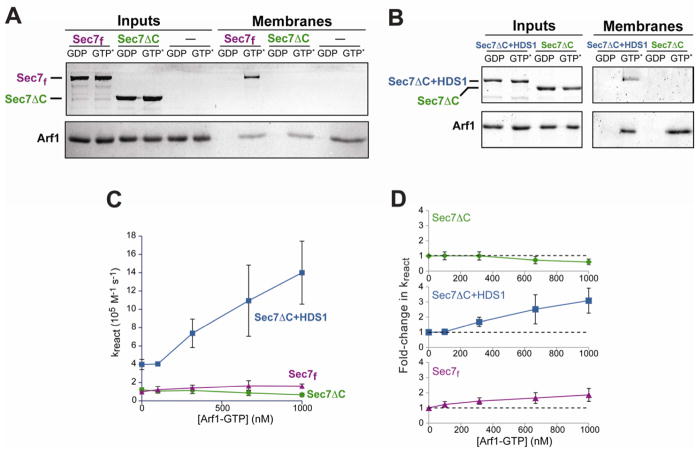
Sec7 has no PH domain or other obvious membrane binding motifs. To determine whether Sec7f can stably associate with membranes, we utilized an in vitro membrane-binding assay. Liposomes were incubated with purified proteins in the presence of guanine nucleotides, and membrane-bound proteins were isolated by floating the liposomes on a sucrose gradient. We tested for stable membrane binding of the three Sec7 constructs and found that none bound autonomously to membranes (Figures 2A,B “GDP” lanes). Surprisingly, Sec7f and Sec7ΔC+HDS1 instead were recruited to membranes in reactions that also contained activated Arf1 (Arf1 bound to the non-hydrolyzable GTP analog GMP-PNP) (Figures 2A,B, “GTP*” lanes). In contrast, the Sec7ΔC construct was not recruited to membranes under these conditions (Figures 2A,B). Similar results were obtained when GTP was used instead of GMP-PNP (data not shown). These results indicate that activated Arf1 stably recruits its activator Sec7 to the membrane surface and that this recruitment requires the HDS1 domain.
Figure 2. The HDS1 domain mediates positive feedback via stable recruitment of Sec7 to membranes.
(A), (B) Purified Sec7 constructs were added to liposomes pre-incubated with active (GMP-PNP-bound, denoted GTP*) or inactive (GDP-bound) purified Arf1, and lipid-bound proteins were separated from unbound proteins by flotation on a sucrose gradient. Input (left) and membrane-bound (right) protein content was determined by SDS-PAGE and total protein staining.
(C) A Sec7/liposome/GTP mixture was preincubated with varying amounts of Arf1-GTP as indicated. A constant amount of additional Arf1-GDP (670 nM) was then added and the rate of nucleotide exchange determined.
(D) Rates from (C) normalized to the rate following a mock (buffer only) preincubation. Error bars in (C) and (D) represent 95% confidence intervals.
See also Figure S2.
We observed the same Arf1-dependent phenomenon when testing binding of Sec7f to liposomes prepared from Folch lipids (which BIG1 and BIG2 were found to bind in a proteomic study (Tsujita et al., 2010)), or when the phosphoinositide PI(4)P was omitted from the synthetic TGN-like lipid mix (Figure S2A). This suggests the presence of activated Arf1 may be more important than lipid composition for stable association of Sec7 with membranes. Accordingly, we found that the reaction rates exhibited by the Sec7 constructs were not significantly different in the absence of PI(4)P (Figure S2B).
Sec7-GEF domains can stably bind their Arf GTPase substrates, but only in their nucleotide-free state, and these enzyme-substrate complexes can dynamically associate with membranes (Beraud-Dufour et al., 1999). However, such an interaction is unlikely to be responsible for the membrane recruitment we observe. First, the interaction we observe is GTP-dependent, inconsistent with GEF domain mediated binding. Second, the Sec7ΔC construct (which includes the GEF domain) was not recruited to membranes, providing further evidence that the membrane recruitment we observe is not due to an interaction between the active site and its substrate or product.
Despite numerous attempts, we were unable to produce well-behaved constructs comprising only the HDS1 domain. Therefore, we cannot say if the HDS1 domain alone is sufficient for membrane recruitment. However, the membrane-dependent activity of HDS1 in conjunction with the Arf1 dependence of membrane recruitment makes a direct interaction between HDS1 and Arf1 the most likely explanation for these effects.
Arf1 activation by Sec7 occurs through HDS1 domain-dependent positive feedback
The observation that Arf1-GTP, the product of Sec7 activity, stably recruits Sec7f to the membrane surface led us to hypothesize that activation of Arf1 by Sec7 may occur through a positive feedback loop, analogous to what has been observed with ARNO (Stalder et al., 2011) and other GEFs (Bose et al., 2001; Lippe et al., 2001; Butty et al., 2002; Margarit et al., 2003). To test this hypothesis, we performed a series of GEF assays in which we titrated Arf1-GTP (product) into the reaction starting conditions, while keeping the amount of Arf1-GDP (substrate) constant. We found that adding increasing amounts of Arf1-GTP to the reaction increased the rates of exchange catalyzed by both Sec7f and Sec7ΔC+HDS1 (although the effect on Sec7f is modest), but not Sec7ΔC (Figure 2C,D). This effect was seen whether the Arf1-GTP added to the reaction was itself activated by the GEF construct being investigated (as performed for the experiments shown in Figure 2C,D), or instead by EDTA-induced nucleotide exchange (data not shown). These results confirm that the product of Sec7 function, Arf1-GTP, stimulates Sec7 activity, indicative of positive feedback.
When we closely examined the earliest time-points of Arf1 activation in the absence of initial Arf1-GTP, we noted that the data consistently deviated from the expected single-exponential curve for both the Sec7f and the Sec7ΔC+HDS1 constructs (Figure S2C,D). The shape of the curve indicates that Arf1 activation by these two constructs initially proceeds slowly (a lag phase) before accelerating. Our interpretation of this phenomenon is that these two constructs are autoinhibited at the beginning of the time-course, until sufficient Arf1 is activated to trigger release of autoinhibition via positive feedback. The Sec7ΔC construct displays no such lag phase (Figure S2E), providing additional evidence for the central role of the HDS1 domain in switching the GEF from the autoinhibited state to the feedback-activated state. This observation raised the possibility that, instead of activating Sec7f and Sec7ΔC+HDS1, Arf1-GTP may simply relieve autoinhibition. However, the fact that Sec7f and Sec7ΔC+HDS1 display significantly higher reaction rates than Sec7ΔC (Figure 1E) indicates that Arf1-GTP exerts a stimulatory effect.
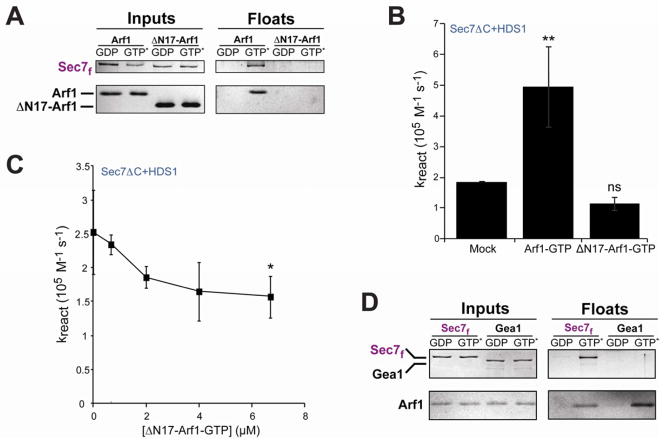
As expected, we found that ΔN17-Arf1-GTP did not recruit Sec7f to membranes, as ΔN17-Arf1-GTP itself is not membrane bound (Figure 3A). In contrast to the stimulatory effect of Arf1-GTP, we found that an equivalent concentration of ΔN17-Arf1-GTP (670 nM) did not significantly increase the activity of Sec7ΔC+HDS1 (Figure 3B). Addition of increasingly higher concentrations of ΔN17-Arf1-GTP resulted in a modest but significant decrease in activity of Sec7ΔC+HDS1 (Fig. 3C), most likely by competing with Arf1-GTP for binding to the HDS1 domain. These results suggest that Arf1-GTP must be bound to the membrane surface to exert its full activating effect on the Sec7 constructs. Taken together with the observation that only the two constructs containing the HDS1 domain exhibited positive feedback behavior, this result strongly suggests that positive feedback arises through stable recruitment of Sec7 to the membrane surface by direct interaction between Arf1-GTP and the HDS1 domain.
Figure 3. Membrane-bound activated Arf1 stably recruits Sec7f, but not Gea1, to membranes.
(A) Liposome flotation comparing Arf1 to ΔN17-Arf1 for recruitment of Sec7f to membranes.
(B) Sec7ΔC+HDS1 was preincubated with GTP, liposomes, and buffer (mock), Arf1-GTP (670 nM), or ΔN17-Arf1-GTP (670nM) before measuring rate of exchange on Arf1-GDP (670 nM). Error bars represent 95% confidence intervals.
(C) Sec7ΔC+HDS1 was preincubated with GTP, liposomes, and varying concentrations of ΔN17-Arf1-GTP before assaying exchange activity on Arf1-GDP (670 nM). Error bars represent 95% confidence intervals. The highest ΔN17-Arf1-GTP concentration shows statistically significant inhibition relative to mock incubation, and linear regression of all data points indicates a slope significantly different from 0 (P=.0017).
(D) Liposome flotation comparing membrane recruitment of purified Sec7f to purified full-length Gea1.
See also Figure S3.
The early-Golgi localized ArfGEFs (Gea1/2 in yeast, GBF1 in humans) share a similar domain architecture to the TGN-localized Sec7 and human BIG1/2, and also function on the same Arf GTPase substrates. However, they may be regulated differently given their distinct subcellular location. To assess whether the early-Golgi ArfGEFs are likely to exhibit positive feedback behavior, we assayed whether activated Arf1 could stably recruit purified yeast Gea1 to membranes using the liposome flotation assay. In contrast to Sec7f, we found that Gea1 was not recruited to liposomes by activated Arf1 (Figure 3D). Thus, although Gea1/2 and GBF1 possess an HDS1 domain, it may not serve the same function in these proteins as it does in Sec7. Indeed, the HDS1 domain is the least conserved of the recognized homology domains when comparing Gea1 to Sec7, yet is more strongly conserved between Sec7 and its human homolog BIG1 (Figure S3).
The HDS1 domain mediates interaction with Arf1 in vivo
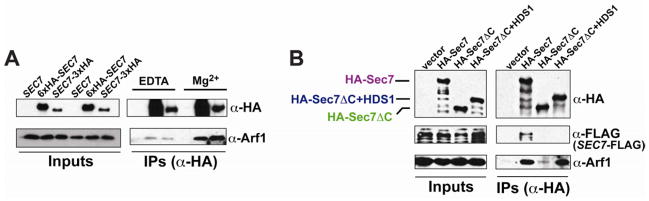
To establish the physiological relevance of our in vitro studies, we sought to detect a stable interaction between Arf1-GTP and Sec7 in cells. We reasoned that preparing cell extracts in the presence of Mg2+ would maintain the nucleotide-bound state of Arf1, whereas including EDTA instead of Mg2+ would destabilize the Arf1 binding to nucleotide, as is the case for all small GTPases; the EDTA condition would specifically disrupt HDS1-Arf1-GTP interactions while preserving GEF domain mediated interactions (Klebe et al., 1995), permitting distinction of the two modes of binding. We immunoprecipitated Sec7 using two different chromosomal HA-tags (SEC7-3xHA or 6xHA-SEC7) from detergent-solubilized cell extracts prepared in buffers containing either 5 mM EDTA or 5 mM Mg2+, and monitored the co-precipitation of Arf1 using anti-Arf1 antibody. We found that when precipitating Sec7 via either the N-terminal or C-terminal tag, significantly more Arf1 co-precipitated with Sec7 under conditions stabilizing the nucleotide-bound state of Arf1 (Figure 4A), indicating that the primary mode of the observed Sec7-Arf1 interaction is not mediated by the GEF domain.
Figure 4. Arf1 interacts with Sec7 in vivo, dependent upon the HDS1 domain.
(A) Arf1 co-immunoprecipitates with HA-tagged Sec7, and this interaction is enriched when Mg2+ is included in the buffer. Three strains were compared using the endogenous SEC7 locus: untagged (SEC7, CFY403), chromosomal 6xHA N-terminal tag (6xHA-SEC7, CFY743), and chromosomal 3xHA C-terminal tag (SEC7-3xHA, CFY512).
(B) The robust Arf1 interaction requires the HDS1 domain. Plasmid-borne 3xHA-tagged Sec7 constructs were introduced into a SEC7-FLAG strain (CFY362), and immunoprecipitations were performed using Mg2+-containing buffer. The plasmids tested were pRS415 (vector), pCF1101 (HA-Sec7), pCF1135 (HA-Sec7ΔC), and pCF1136 (HA-Sec7ΔC+HDS1). In both (A) and (B), different exposure times are shown for Inputs and IPs for clarity. See also Figure S4 for uniform exposures of these same experiments shown in order to gauge percent recovery.
To identify the domain mediating this interaction, we monitored the amount of Arf1 that co-precipitated with different HA-tagged Sec7 constructs. These constructs were introduced on plasmids into a strain containing C-terminally FLAG-tagged Sec7 (SEC7-FLAG). We found that both the HA-Sec7 and HA-Sec7ΔC+HDS1 constructs were able to co-precipitate significantly more Arf1 than did the HA-Sec7ΔC construct (Figure 4B). The low level of Arf1 co-precipitating with HA-Sec7ΔC is similar to that seen with HA-Sec7 in the absence of Mg2+ (Figure 4A, “EDTA”), suggesting that both represent background interaction with the active site in the GEF domain. Importantly, the observed dependence upon the HDS1 domain did not arise through interaction of the HA-tagged truncation constructs with the endogenous Sec7-FLAG protein, as neither HA-Sec7ΔC nor HA-Sec7ΔC+HDS1 co-precipitated Sec7-FLAG (Figure 4B). Therefore, the HDS1 domain and GTP-bound Arf1 were both required for co-precipitation of Arf1 under these conditions, matching the in vitro results.
The HDS1 domain mediates Sec7 localization to the trans-Golgi network
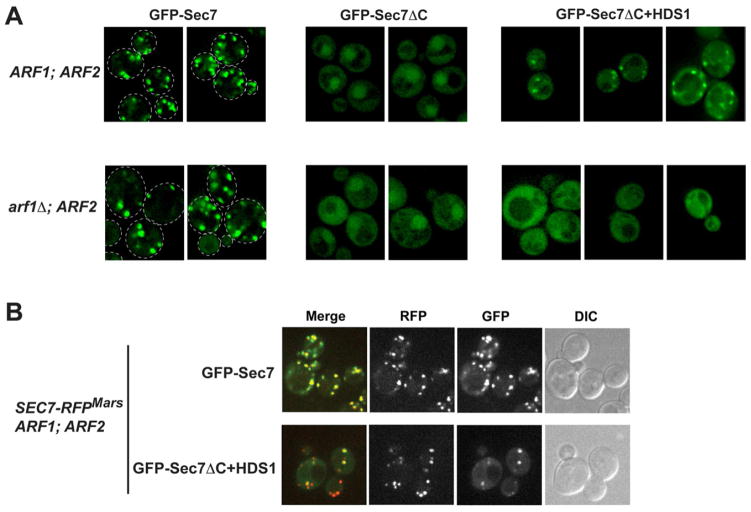
As Sec7 and Arf1-GTP interact in cell extracts, we hypothesized that this interaction might play a role in localizing Sec7 to the TGN. We examined GFP-Sec7, GFP-Sec7ΔC, and GFP-Sec7ΔC+HDS1 plasmid constructs in cells in which the endogenous SEC7 gene was intact. Whereas GFP-Sec7 decorated punctate structures known to correspond to the TGN (Franzusoff et al., 1991), and exhibited very faint cytoplasmic labeling, GFP-Sec7ΔC was exclusively cytoplasmic and nuclear, with no observable punctae (Figure 5A). Remarkably, GFP-Sec7ΔC+HDS1 restored partial localization to punctae, with the extent of localization varying among cells (Figure 5A). All GFP-Sec7ΔC+HDS1 expressing cells examined exhibited several observable GFP-positive punctae, whereas no GFP-positive punctae were observed in any cells expressing GFP-Sec7ΔC. The HDS1 domain is therefore required to localize the remaining N-terminal regions of Sec7 to punctae under these conditions. Furthermore, these results indicate that the HDS2-4 domains also play a role in localization to punctae, consistent with a report that a 48-amino acid deletion within the HDS4 domain resulted in partial mislocalization of Sec7 (Dehring et al., 2008).
Figure 5. The HDS1 domain cooperates with Arf1 to mediate localization of Sec7 to the TGN.
(A) Wild-type yeast cells (CFY103: ARF1; ARF2; SEC7) and arf1Δ yeast cells (CFY392: arf1Δ; ARF2; SEC7) expressing plasmid-borne GFP-tagged Sec7 constructs were imaged. Single deconvolved focal planes are shown at equivalent light levels. The plasmids used were pCF1084 (GFP-Sec7), pCF1140 (GFP-Sec7ΔC), and pCF1141 (GFP-Sec7ΔC+HDS1).
(B) SEC7-RFPMars yeast cells (CFY589) expressing pCF1084 or pCF1141 were imaged to examine the co-localization of GFP and RFP signals by confocal microscopy.
We considered the possibility that the partial delocalization of GFP-Sec7ΔC+HDS1 could be due to a difference in its multimeric state, as part of the HDS4 domain is reported to form a homotypic coiled-coil (Marino-Ramirez and Hu, 2002). We analyzed purified Sec7f and Sec7ΔC+HDS1 using combined sedimentation/Stokes analysis (Erickson, 2009) to determine that both constructs are dimers (Table S1), consistent with previous reports indicating that the N-terminus mediates homodimerization of BIG/GBF family GEFs (Grebe et al., 2000; Ramaen et al., 2007). Therefore, the partial delocalization of GFP-Sec7ΔC+HDS1 is not due to loss of dimerization of this construct.
To confirm that the GFP-Sec7ΔC+HDS1 punctae correspond to the TGN, we examined the colocalization of this construct with chromosomal SEC7-RFPMars (Figure 5B). Plasmid-borne GFP-Sec7 showed a near complete localization with endogenous Sec7-RFPMars. Similarly, the colocalization of plasmid-borne GFP-Sec7ΔC+HDS1 punctae with Sec7-RFPMars was significant. Whereas each GFP-Sec7ΔC+HDS1 puncta was also positive for Sec7-RFPMars, not every Sec7-RFPMars punctae was positive for GFP-Sec7ΔC+HDS1, most likely due to the relatively weaker labeling of punctae by this construct. These data indicate that the GFP-Sec7ΔC+HDS1 punctae do indeed correspond to the TGN, although perhaps not all TGN compartments within a cell have detectable levels of GFP-Sec7ΔC+HDS1. Taken together, these results demonstrate that both the HDS1 domain and the HDS2-4 domains play a role in localizing Sec7 to the TGN, as loss of HDS2-4 results in partial mislocalization to the cytoplasm and additional loss of HDS1 results in complete mislocalization.
To determine whether the HDS1 domain dependent localization of Sec7 required Arf1, we examined the localization of the GFP-tagged constructs in arf1Δ cells. As the ARF1 gene encodes approximately 90% of the total Arf1 and Arf2 protein in cells, arf1Δ strains express ~10% of Arf1/2 relative to ARF1 strains (Stearns et al., 1990) and represent the best steady-state alternative to the synthetically lethal arf1Δ arf2Δ double mutant. Strikingly, we found that the GFP-Sec7ΔC+HDS1 construct is completely mislocalized to the cytoplasm in arf1Δ cells, whereas GFP-Sec7 is only slightly mislocalized to the cytoplasm in arf1Δ cells, primarily appearing as toroids likely corresponding to enlarged TGN compartments known as Berkeley bodies (Figure 5A). Therefore, the HDS1-dependent TGN localization of the GFP-Sec7 constructs is mediated by Arf1.
Mutations in the HDS1 domain perturb Sec7 localization synergistically with arf1Δ
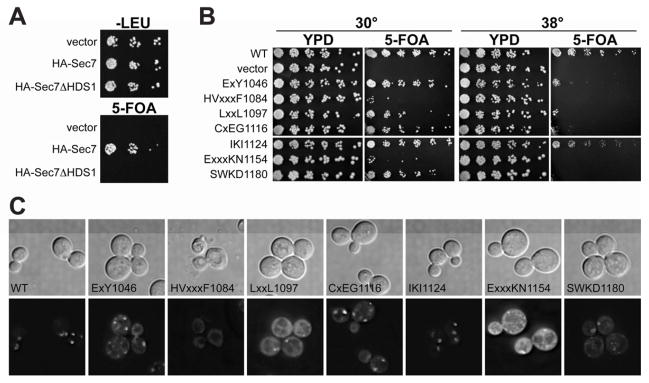
As expected from its effect on GEF activity and role in localization to the TGN, we found that the HDS1 domain is required for the essential function of Sec7, as an HA-Sec7ΔHDS1 construct is unable to complement a sec7Δ mutation (Figure 6A), despite being expressed at a level similar to that of HA-Sec7 (Figure S5A). We examined the localization of this construct (GFP-Sec7ΔHDS1), and found that it was partially mislocalized to the cytoplasm in both wild-type and arf1Δ cells (Figure S5B,C). We observed that Sec7ΔHDS1 forms stable complexes with endogenous Sec7 (Figure S5D), supporting a model in which partial localization of Sec7ΔHDS1 to the TGN is mediated by complex formation with endogenous Sec7, which remains properly localized in arf1Δ cells (Figure 5B). Thus, the partial mislocalization of GFP-Sec7ΔHDS1 to the cytoplasm is consistent with a critical role for HDS1 in Sec7 localization.
Figure 6. Missense mutations in HDS1 cause defects in growth and TGN localization synthetic with arf1Δ.
(A) Plasmids were introduced into a Sec7 shuffling strain (CFY409) and assayed for the ability to complement a sec7Δ null mutant, monitored by growth on media containing 5-FOA. Vector (pRS415), HA-Sec7 (pCF1101) and HA-Sec7ΔHDS1 (pCF1139) were tested.
(B) Plasmid-borne GFP-sec7 alleles with the indicated residues substituted with alanine were tested for their ability to complement sec7Δ arf1Δ (CFY863). Plates were imaged after 3 days of growth at the indicated temperature.
(C) GFP-tagged sec7 alleles with the indicated HDS1 alanine substitutions were imaged in log phase sec7Δ arf1Δ cells (CFY863). The GFP signal of the ExxKN1154 mutant was consistently brighter, suggesting cells require multiple copies of the plasmid for viability.
See also Figures S5, S6, and S7.
To specifically perturb the function of the HDS1 domain while circumventing complications arising from the presence of endogenous SEC7, we sought HDS1-domain mutants that perturbed Sec7 function yet remained viable. We used plasmid shuffling to replace the endogenous SEC7 gene with seven different plasmid-borne HDS1 mutant sec7 alleles created by making alanine substitutions in patches of conserved residues (Figure S6). When these sec7 alleles were shuffled into an arf1Δ mutant background, two alleles displayed severe synthetic growth defects, four alleles displayed varying degrees of temperature sensitivity, and one allele displayed little to no growth defect (Figure 6B). Each HDS1 mutant was expressed at a level similar to that of wild-type Sec7 when expression was assayed in a wild-type ARF1 background (Figure S5E). The viability of these alleles permitted the determination of their effects on Sec7 localization in cells. In ARF1 cells, all of the HDS1 mutants were correctly localized to the TGN (data not shown), consistent with normal growth in this background (Figure S5F). However, in arf1Δ cells, the degree of TGN-localization correlated with the severity of the growth phenotype: whereas wild-type Sec7 and alleles displaying no growth defect had no or low cytosolic signal, the alleles with the strongest growth defects exhibited brighter cytosolic fluorescence and a significantly lower TGN:cytosolic fluorescence ratio (Figures 6C, S7A,B,C,D). We note that the localization of the temperature-sensitive alleles was not significantly altered upon shift to the restrictive temperature (data not shown). We confirmed that two of the HDS1 mutants (one with a severe growth phenotype and one with a temperature-sensitive growth phenotype in arf1Δ cells) exhibited a reduced physical interaction with Arf1 when shuffled into wild-type ARF1 cells (Figure S7E,F), suggesting these mutants have a reduced affinity for Arf1/2-GTP. Taken together, these data indicate that the HDS1 domain serves a critical role, in cooperation with Arf1, to localize Sec7 to the TGN.
Discussion
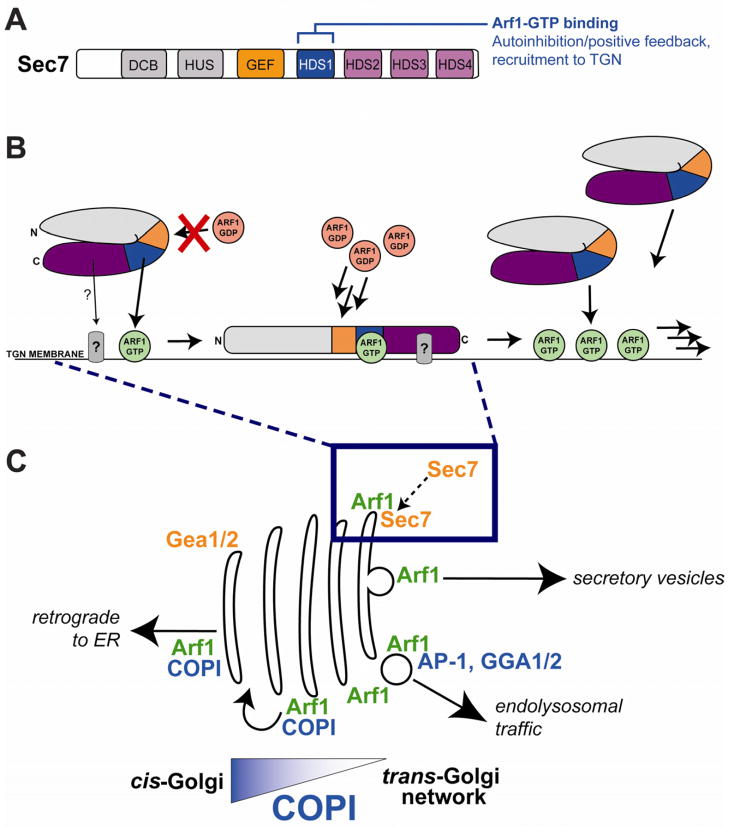
The Golgi is the primary cellular sorting station for protein and membrane secretory traffic. Most of the traffic within and out of the Golgi is controlled by Arf GTPases, yet the Arf-GEFs that activate these GTPases at the Golgi are poorly understood. In this study we have elucidated two key autoregulatory features of the TGN-localized Arf-GEF Sec7: autoinhibition and positive feedback. Both features require the function of a previously uncharacterized domain, the HDS1 domain. Positive feedback arises through interaction of the HDS1 domain with Arf1-GTP, the product of Sec7 activity, resulting in stabilization of Sec7 on the membrane surface (Figure 7A,B).
Figure 7. Feedback activation model for Sec7 recruitment to the TGN.
(A) Schematic representation of Sec7 homology domains with annotation of the function of the HDS1 domain.
(B) Model for autoinhibition and positive feedback regulation of Sec7. Although Sec7 is dimeric, for simplicity only one monomer is schematized. Sec7 autoinhibition in solution is represented by a putative intramolecular interaction. Release of autoinhibition is concomitant with recruitment to the TGN membrane by direct interaction of the HDS1 domain (blue) with Arf1-GTP (green circle). An additional factor (“?”) may also contribute to the recruitment of Sec7 to the TGN through interaction with the HDS2-4 domains. Positive feedback arises through the generation of more Arf1-GTP by Sec7, which leads to the recruitment of more Sec7, etc.
(C) Model for Sec7 recruitment to the TGN. Although Arf1-GTP is localized to the entire Golgi, Sec7 might only be recruited to the TGN because COPI may outcompete Sec7 for binding to Arf1-GTP at earlier Golgi compartments.
The rate enhancement that accompanies stable binding of Sec7 to the membrane can be explained by an increased frequency of productive encounters between Sec7, Arf1-GDP, and the membrane surface to form the tripartite enzyme-substrate-membrane complex that is a prerequisite for Arf1 activation. Alternatively, the observed rate enhancement may arise through relief of the autoinhibitory effects of the C-terminus via an allosteric conformational change triggered by binding to Arf1-GTP and concomitant association with the membrane. We favor a hybrid model in which Arf1-GTP recruitment of Sec7 to the membrane by Arf1-GTP results in both allosteric relief of autoinhibition and an increased frequency of productive encounters between Sec7, Arf1-GDP, and the membrane.
The Arf-GEF ARNO requires the presence of an activated Arf GTPase, either Arf6 or Arf1, for robust activity on membranes; when Arf1 is used as an activator and a substrate for ARNO, positive feedback is observed (Stalder et al., 2011). The positive feedback effect that we observed in our assays was less pronounced than that seen for ARNO. For both Sec7 and ARNO, positive feedback also involves relief of autoinhibition. It is possible that the larger positive feedback effect exhibited by ARNO is due to a greater degree of autoinhibition. Indeed, PH domain-proximal elements of ARNO inhibit the activity of its GEF domain by approximately 14-fold (Stalder et al., 2011), whereas we found that the C-terminus of Sec7 inhibited its GEF domain by approximately 7-fold when comparing the rates of Sec7f and Sec7ΔC in solution. In addition, we were unable to assay Sec7 at very low concentrations (< 50 nM) due to its instability under such conditions. We may have observed a more pronounced positive feedback effect if we had been able to perform the GEF assays at concentrations approaching those used for ARNO (7.5 nM). Furthermore, our data suggest that a key function of positive feedback for Sec7 is its role in localization of the GEF to the TGN, in addition to its role in rate enhancement. In contrast, positive feedback is not a likely localization mechanism for ARNO, as Arf1, the most likely substrate for ARNO, is primarily localized to the Golgi, but ARNO is primarily localized to the cell periphery. ARNO localization depends instead upon lipid interactions with its PH domain (Venkateswarlu et al., 1998), as well as GTPase cascades (Cohen et al., 2007; DiNitto et al., 2007; Hofmann et al., 2007; Li et al., 2007).
Our data indicate that Arf1-GTP recruits Sec7 to the TGN via interaction with the HDS1 domain. As Sec7 is a major source of Arf1-GTP, what is the origin of the initial Arf1-GTP that recruits Sec7 to the TGN? While residual activity of autoinhibited Sec7 suffices in vitro, the early-Golgi ArfGEFs (Gea1/2 in yeast) might represent a more robust source in vivo. As the Golgi matures, Gea1/2 provide the Arf1-GTP needed at the early compartments, most prominently to recruit coatomer to generate COPI vesicles for retrograde cargo sorting. The localization mechanism of Gea1/2 (as well as human GBF1) remains unresolved, although a transmembrane receptor has been proposed (Chantalat et al., 2003), and SNAREs have been shown to recruit Arf1-GDP to the membrane surface (Honda et al., 2005), which may in turn recruit Gea1/2 through a substrate-enzyme interaction. The localization mechanism of the early-Golgi Arf-GEFs is likely distinct from that of Sec7, as we found that Gea1 was not stably recruited to membranes by Arf1-GTP.
Given that Arf1-GTP is present throughout early-Golgi as well as late-Golgi compartments, how does Arf1-GTP specifically recruit Sec7 to the TGN? One possibility is coincidence detection involving an unknown factor binding to Sec7, either to the HDS1 domain or to the N-terminus: although Sec7ΔC+HDS1 is partially mislocalized to the cytoplasm, we found that the visible punctae correspond to the TGN. An alternative model is that Arf1-GTP is sufficient to provide TGN specificity because Sec7 must compete with other Arf1 effectors for binding to Arf1-GTP (Figure 7C). In this scenario, at early Golgi compartments, most of the Arf1-GTP is bound to the COPI coat, which is recruited to the cis- and medial-Golgi by interaction with cargo tails and Arf1-GTP. Therefore, at compartments where COPI cargo is present, the affinity of COPI for the membrane, and thus for Arf1-GTP, may be great enough to effectively outcompete Sec7 for binding to Arf1-GTP. In the maturing Golgi, COPI cargo is relatively absent from later compartments because it has been trafficked to earlier compartments (to the cis-Golgi and ER). Free from competition with COPI, Sec7 would be able to bind to Arf1-GTP only at the TGN. Under this speculative model, Arf1-GTP dependent Sec7 recruitment could serve as a checkpoint in Golgi maturation, preventing the premature recruitment of TGN-localized effectors until COPI sorting has completed. Of course, Sec7 would also compete for binding to Arf1-GTP at the TGN with TGN-localized Arf1 effectors, primarily the clathrin adaptors, but perhaps the affinity of these effectors for Arf1-GTP is less than that of coatomer. Future studies involving in-depth characterization of binding constants in the presence of membrane-bound cargo tails may be needed to test this hypothesis.
An intriguing possible consequence of Sec7 positive feedback at the TGN is that it could be used to generate TGN-derived vesicles that traffic to the PM, which have not been clearly demonstrated to require a vesicle coat. Vesiculation could occur through the rapid activation of a high local concentration of Arf1 via Sec7-mediated positive feedback, peaking in activity only after cargos destined for the endolysosomal system have been sorted away from the TGN by Arf1-dependent clathrin-coated vesicles. The reduced fraction of Arf1-GTP bound to clathrin cargo adaptors at the TGN would be free to recruit more Sec7, stimulating positive feedback. Arf1-GTP, at high concentrations, is sufficient to generate the membrane curvature needed for vesiculation (Beck et al., 2008; Krauss et al., 2008), and recently has been shown to be directly involved in vesicle fission (Beck et al., 2011). Thus, Sec7-mediated positive feedback activation of Arf1 could drive vesiculation of the TGN without the need for a vesicle coat.
Although Arf1 is the primary Arf at the Golgi in yeast cells (Arf2 is redundant and dispensible), there are four paralogous Arfs at the Golgi in human cells. Recent work has indicated that these human paralogs may exhibit some specificity for early versus late Golgi compartments (Ben-Tekaya et al., 2010; Manolea et al., 2010), although this analysis is complicated by robust functional redundancy among the paralogs (Volpicelli-Daley et al., 2005). Therefore in human cells it is possible that GBF1 activates one or two specific Arf paralogs that then serve to recruit BIG1/2 to activate distinct Arf paralogs at the TGN. Despite this important potential distinction, the overall mechanism of GEF recruitment by activated Arf GTPases is very likely conserved between yeast and humans, given the high degree of sequence conservation between Sec7 and BIG1/2, and between the yeast and human Golgi-localized Arfs.
Our results establish autoinhibition and positive feedback as important features of Sec7 regulation, and demonstrate a role for positive feedback in recruitment of Sec7 to the TGN. Our data suggest that the interplay between Arf1 activation and Arf1 effector recruitment must be intimately connected, and the dynamics of competition between GEF and effector for Arf1-GTP binding may play a role in Golgi maturation.
Experimental Procedures
Plasmids, Strains, Antibodies, and Proteins
Standard techniques were used for generating yeast strains by homologous recombination (Longtine et al., 1998; Gauss et al., 2005) and by mating. All yeast SEC7 plasmids encode Sec7 constructs driven by the native SEC7 promoter. Plasmids and yeast strains are presented in Supplemental Experimental Procedures.
The anti-Arf1 polyclonal antibody were a kind gift from the Schekman lab. The anti-FLAG monoclonal “M2” antibody and anti-G6PDH (yeast Zwf1) polyclonal antibody were purchased from Sigma. The anti-HA monoclonal “12CA5” antibody was purchased from Roche.
All Sec7 constructs generated for purification contain a C-terminal motif of the GEF domain, “loop>J”, recently shown to be important for GEF activity (Lowery et al., 2011).
Protein purifications are detailed in the Supplemental Experimental Procedures.
Preparation of Liposomes and flotation assay
Unilamellar liposomes were generated from a mixture of lipids (see Supplemental Experimental Procedures) approximating the endogenous TGN lipid composition determined in a published lipidomics study (Klemm et al., 2009), plus added DiR near-infrared dye (Avanti Polar Lipids) to aid in visualization and quantitation of lipids. Following vacuum drying, lipid films were hydrated in 20 mM HEPES pH 7.4, 150 mM KOAc, followed by extrusion through 100 nm filters (Whatman) to generate liposomes.
The liposome flotation assay was performed essentially as described (Matsuoka et al., 1998). Further details are provided in the Supplemental Experimental Procedures.
Arf1 nucleotide exchange kinetics assay
The nucleotide-bound state of Arf1 was monitored in real-time by native tryptophan fluorescence (297.5 nm excitation, 340 nm emission), similar to published procedures (Higashijima et al., 1987; Antonny et al., 1997). Liposomes, Sec7 constructs, Arf1 constructs, and 200 μM GTP were combined at 30°C, and the change in fluorescence was tracked for 10 to 40 minutes. Triplicate traces were fit to a single exponential curve as described in Supplemental Experimental Procedures to obtain the reaction rate. Due to documented variability in exchange rates for different batches of liposomes (Stalder et al., 2011), all reactions shown within any one figure panel were performed using a single batch of liposomes.
Microscopy
Cells were grown in synthetic dropout media and imaged in log phase (OD600 ~ 0.5) after spotting onto coverslips in growth media. Live cells were imaged at room temperature on two different microscopes. Details on the microscopes and image processing are described in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
The Golgi Arf-GEF Sec7 is regulated by autoinhibition and positive feedback
Positive feedback results from stable recruitment of Sec7 to the membrane surface
Positive feedback arises through interaction between the HDS1 domain and Arf1-GTP
The HDS1 domain and Arf1 mediate recruitment of Sec7 to the trans-Golgi network
Acknowledgments
We acknowledge the generosity of the Emr, Bretscher, and Schekman labs in sharing reagents, strains, plasmids, equipment, and advice. We are grateful to Y. Mao for assistance with insect cell culture. We thank J. MacGurn for making strains CFY403 and CFY589, and M. Rogals for making plasmid pMR1. We thank Scott Emr and Tony Bretscher for many fruitful discussions. We received helpful advice on microscopy from T. Bretscher, K. Donovan, D. Garbett, B. Judson, A. Manford, and C. Stefan. The authors are supported by Cornell University startup funding and by NIH/NIGMS grant R01GM098621.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonny B, Beraud-Dufour S, Chardin P, Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- Aridor M, Fish KN, Bannykh S, Weissman J, Roberts TH, Lippincott-Schwartz J, Balch WE. The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J Cell Biol. 2001;152:213–229. doi: 10.1083/jcb.152.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R, Prinz S, Diestelkotter-Bachert P, Rohling S, Adolf F, Hoehner K, Welsch S, Ronchi P, Brugger B, Briggs JA, et al. Coatomer and dimeric ADP ribosylation factor 1 promote distinct steps in membrane scission. J Cell Biol. 2011;194:765–777. doi: 10.1083/jcb.201011027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck R, Sun Z, Adolf F, Rutz C, Bassler J, Wild K, Sinning I, Hurt E, Brugger B, Bethune J, et al. Membrane curvature induced by Arf1-GTP is essential for vesicle formation. Proc Natl Acad Sci USA. 2008;105:11731–11736. doi: 10.1073/pnas.0805182105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tekaya H, Kahn RA, Hauri HP. ADP ribosylation factors 1 and 4 and group VIA phospholipase A regulate morphology and intraorganellar traffic in the endoplasmic reticulum-Golgi intermediate compartment. Mol Biol Cell. 2010;21:4130–4140. doi: 10.1091/mbc.E10-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraud-Dufour S, Paris S, Chabre M, Antonny B. Dual interaction of ADP ribosylation factor 1 with Sec7 domain and with lipid membranes during catalysis of guanine nucleotide exchange. J Biol Chem. 1999;274:37629–37636. doi: 10.1074/jbc.274.53.37629. [DOI] [PubMed] [Google Scholar]
- Beraud-Dufour S, Robineau S, Chardin P, Paris S, Chabre M, Cherfils J, Antonny B. A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the beta-phosphate to destabilize GDP on ARF1. EMBO J. 1998;17:3651–3659. doi: 10.1093/emboj/17.13.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose I, Irazoqui JE, Moskow JJ, Bardes ES, Zyla TR, Lew DJ. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J Biol Chem. 2001;276:7176–7186. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- Bui QT, Golinelli-Cohen MP, Jackson CL. Large Arf1 guanine nucleotide exchange factors: evolution, domain structure, and roles in membrane trafficking and human disease. Mol Genet Genomics. 2009 doi: 10.1007/s00438-009-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butty AC, Perrinjaquet N, Petit A, Jaquenoud M, Segall JE, Hofmann K, Zwahlen C, Peter M. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J. 2002;21:1565–1576. doi: 10.1093/emboj/21.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JE. Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- Chantalat S, Courbeyrette R, Senic-Matuglia F, Jackson CL, Goud B, Peyroche A. A novel Golgi membrane protein is a partner of the ARF exchange factors Gea1p and Gea2p. Mol Biol Cell. 2003;14:2357–2371. doi: 10.1091/mbc.E02-10-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson CL, Chabre M. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature. 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell. 2007;18:2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- De Matteis MA, Luini A. Exiting the Golgi complex. Nat Rev Mol Cell Biol. 2008;9:273–284. doi: 10.1038/nrm2378. [DOI] [PubMed] [Google Scholar]
- de Wit MC, de Coo IF, Halley DJ, Lequin MH, Mancini GM. Movement disorder and neuronal migration disorder due to ARFGEF2 mutation. Neurogenetics. 2009;10:333–336. doi: 10.1007/s10048-009-0192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehring DA, Adler AS, Hosseini A, Hicke L. A C-terminal sequence in the guanine nucleotide exchange factor Sec7 mediates Golgi association and interaction with the Rsp5 ubiquitin ligase. J Biol Chem. 2008;283:34188–34196. doi: 10.1074/jbc.M806023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNitto JP, Delprato A, Gabe Lee MT, Cronin TC, Huang S, Guilherme A, Czech MP, Lambright DG. Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol Cell. 2007;28:569–583. doi: 10.1016/j.molcel.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011;12:362–375. doi: 10.1038/nrm3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online. 2009;11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzusoff A, Redding K, Crosby J, Fuller RS, Schekman R. Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futai E, Hamamoto S, Orci L, Schekman R. GTP/GDP exchange by Sec12p enables COPII vesicle bud formation on synthetic liposomes. EMBO J. 2004;23:4146–4155. doi: 10.1038/sj.emboj.7600428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauss R, Trautwein M, Sommer T, Spang A. New modules for the repeated internal and N-terminal epitope tagging of genes in Saccharomyces cerevisiae. Yeast. 2005;22:1–12. doi: 10.1002/yea.1187. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- Glick BS, Nakano A. Membrane traffic within the Golgi apparatus. Annu Rev Cell Dev Biol. 2009;25:113–132. doi: 10.1146/annurev.cellbio.24.110707.175421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld JU, Salchert K, Koncz C, Jurgens G. A conserved domain of the arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell. 2000;12:343–356. doi: 10.1105/tpc.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima T, Ferguson KM, Sternweis PC, Ross EM, Smigel MD, Gilman AG. The effect of activating ligands on the intrinsic fluorescence of guanine nucleotide-binding regulatory proteins. J Biol Chem. 1987;262:752–756. [PubMed] [Google Scholar]
- Hofmann I, Thompson A, Sanderson CM, Munro S. The Arl4 family of small G proteins can recruit the cytohesin Arf6 exchange factors to the plasma membrane. Curr Biol. 2007;17:711–716. doi: 10.1016/j.cub.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Honda A, Al-Awar OS, Hay JC, Donaldson JG. Targeting of Arf-1 to the early Golgi by membrin, an ER-Golgi SNARE. J Cell Biol. 2005;168:1039–1051. doi: 10.1083/jcb.200409138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA. Toward a model for Arf GTPases as regulators of traffic at the Golgi. FEBS Lett. 2009;583:3872–3879. doi: 10.1016/j.febslet.2009.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn RA, Randazzo P, Serafini T, Weiss O, Rulka C, Clark J, Amherdt M, Roller P, Orci L, Rothman JE. The amino terminus of ADP-ribosylation factor (ARF) is a critical determinant of ARF activities and is a potent and specific inhibitor of protein transport. J Biol Chem. 1992;267:13039–13046. [PubMed] [Google Scholar]
- Klarlund JK, Guilherme A, Holik JJ, Virbasius JV, Chawla A, Czech MP. Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science. 1997;275:1927–1930. doi: 10.1126/science.275.5308.1927. [DOI] [PubMed] [Google Scholar]
- Klarlund JK, Tsiaras W, Holik JJ, Chawla A, Czech MP. Distinct polyphosphoinositide binding selectivities for pleckstrin homology domains of GRP1-like proteins based on diglycine versus triglycine motifs. J Biol Chem. 2000;275:32816–32821. doi: 10.1074/jbc.M002435200. [DOI] [PubMed] [Google Scholar]
- Klebe C, Prinz H, Wittinghofer A, Goody RS. The kinetic mechanism of Ran--nucleotide exchange catalyzed by RCC1. Biochemistry. 1995;34:12543–12552. doi: 10.1021/bi00039a008. [DOI] [PubMed] [Google Scholar]
- Klemm RW, Ejsing CS, Surma MA, Kaiser HJ, Gerl MJ, Sampaio JL, de Robillard Q, Ferguson C, Proszynski TJ, Shevchenko A, et al. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J Cell Biol. 2009;185:601–612. doi: 10.1083/jcb.200901145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolanus W, Nagel W, Schiller B, Zeitlmann L, Godar S, Stockinger H, Seed B. Alpha L beta 2 integrin/LFA-1 binding to ICAM-1 induced by cytohesin-1, a cytoplasmic regulatory molecule. Cell. 1996;86:233–242. doi: 10.1016/s0092-8674(00)80095-1. [DOI] [PubMed] [Google Scholar]
- Krauss M, Jia JY, Roux A, Beck R, Wieland FT, De Camilli P, Haucke V. Arf1-GTP-induced tubule formation suggests a function of Arf family proteins in curvature acquisition at sites of vesicle budding. J Biol Chem. 2008;283:27717–27723. doi: 10.1074/jbc.M804528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Li CC, Chiang TC, Wu TS, Pacheco-Rodriguez G, Moss J, Lee FJ. ARL4D recruits cytohesin-2/ARNO to modulate actin remodeling. Mol Biol Cell. 2007;18:4420–4437. doi: 10.1091/mbc.E07-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippe R, Miaczynska M, Rybin V, Runge A, Zerial M. Functional synergy between Rab5 effector Rabaptin-5 and exchange factor Rabex-5 when physically associated in a complex. Mol Biol Cell. 2001;12:2219–2228. doi: 10.1091/mbc.12.7.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lowery J, Szul T, Seetharaman J, Jian X, Su M, Forouhar F, Xiao R, Acton TB, Montelione GT, Lin H, et al. A novel C-terminal motif within the Sec7 domain of guanine nucleotide exchange factors regulates ARF binding and activation. J Biol Chem. 2011 doi: 10.1074/jbc.M111.230631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolea F, Chun J, Chen DW, Clarke I, Summerfeldt N, Dacks JB, Melancon P. Arf3 is activated uniquely at the trans-Golgi network by brefeldin A-inhibited guanine nucleotide exchange factors. Mol Biol Cell. 2010;21:1836–1849. doi: 10.1091/mbc.E10-01-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margarit SM, Sondermann H, Hall BE, Nagar B, Hoelz A, Pirruccello M, Bar-Sagi D, Kuriyan J. Structural evidence for feedback activation by Ras. GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- Marino-Ramirez L, Hu JC. Isolation and mapping of self-assembling protein domains encoded by the Saccharomyces cerevisiae genome using lambda repressor fusions. Yeast. 2002;19:641–650. doi: 10.1002/yea.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- Morinaga N, Tsai SC, Moss J, Vaughan M. Isolation of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP ribosylation factor (ARF) 1 and ARF3 that contains a Sec7-like domain. Proc Natl Acad Sci USA. 1996;93:12856–12860. doi: 10.1073/pnas.93.23.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouratou B, Biou V, Joubert A, Cohen J, Shields DJ, Geldner N, Jurgens G, Melancon P, Cherfils J. The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genomics. 2005;6:20. doi: 10.1186/1471-2164-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Peyroche A, Paris S, Jackson CL. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature. 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- Ramaen O, Joubert A, Simister P, Belgareh-Touze N, Olivares-Sanchez MC, Zeeh JC, Chantalat S, Golinelli-Cohen MP, Jackson CL, Biou V, et al. Interactions between conserved domains within homodimers in the BIG1, BIG2, and GBF1 Arf guanine nucleotide exchange factors. J Biol Chem. 2007;282:28834–28842. doi: 10.1074/jbc.M705525200. [DOI] [PubMed] [Google Scholar]
- Rambourg A, Clermont Y, Kepes F. Modulation of the Golgi apparatus in Saccharomyces cerevisiae sec7 mutants as seen by three-dimensional electron microscopy. Anat Rec. 1993;237:441–452. doi: 10.1002/ar.1092370402. [DOI] [PubMed] [Google Scholar]
- Sheen VL, Ganesh VS, Topcu M, Sebire G, Bodell A, Hill RS, Grant PE, Shugart YY, Imitola J, Khoury SJ, et al. Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nature Genet. 2004;36:69–76. doi: 10.1038/ng1276. [DOI] [PubMed] [Google Scholar]
- Spang A, Herrmann JM, Hamamoto S, Schekman R. The ADP ribosylation factor-nucleotide exchange factors Gea1p and Gea2p have overlapping, but not redundant functions in retrograde transport from the Golgi to the endoplasmic reticulum. Mol Biol Cell. 2001;12:1035–1045. doi: 10.1091/mbc.12.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalder D, Barelli H, Gautier R, Macia E, Jackson CL, Antonny B. Kinetic studies of the Arf activator Arno on model membranes in the presence of Arf effectors suggest control by a positive feedback loop. J Biol Chem. 2011;286:3873–3883. doi: 10.1074/jbc.M110.145532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Kahn RA, Botstein D, Hoyt MA. ADP ribosylation factor is an essential protein in Saccharomyces cerevisiae and is encoded by two genes. Mol Cell Biol. 1990;10:6690–6699. doi: 10.1128/mcb.10.12.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujita K, Itoh T, Kondo A, Oyama M, Kozuka-Hata H, Irino Y, Hasegawa J, Takenawa T. Proteome of acidic phospholipid-binding proteins: spatial and temporal regulation of Coronin 1A by phosphoinositides. J Biol Chem. 2010;285:6781–6789. doi: 10.1074/jbc.M109.057018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswarlu K, Oatey PB, Tavare JM, Cullen PJ. Insulin-dependent translocation of ARNO to the plasma membrane of adipocytes requires phosphatidylinositol 3-kinase. Curr Biol. 1998;8:463–466. doi: 10.1016/s0960-9822(98)70181-2. [DOI] [PubMed] [Google Scholar]
- Volpicelli-Daley LA, Li Y, Zhang CJ, Kahn RA. Isoform-selective effects of the depletion of ADP-ribosylation factors 1–5 on membrane traffic. Mol Biol Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.