How cells organize into structured tissues has been a long-standing question for the field of developmental biology (1). Whereas many of the key cellular and extracellular regulatory components have become identified over the past decades, including soluble growth factors and insoluble matrix factors in the environment and receptors on the cells themselves, fundamental principles by which molecular regulatory processes govern resulting cell–cell interactions remain largely unclear. More recently, a nascent medical technology field of tissue engineering has arisen that can be viewed essentially as applied developmental biology—with bioengineers seeking to control for their own purposes cell population organization into functional tissue structures, often employing biomaterials approaches (2). Success in this technology field, correspondingly, depends similarly on elucidating how cell organizational processes are governed by interactions of cell receptors with cognate ligands in their surroundings. In this issue of PNAS, a collaborative effort combining biomaterials and molecular cell biology methodologies provides an important advance in both the fields of developmental biology and tissue engineering (3).
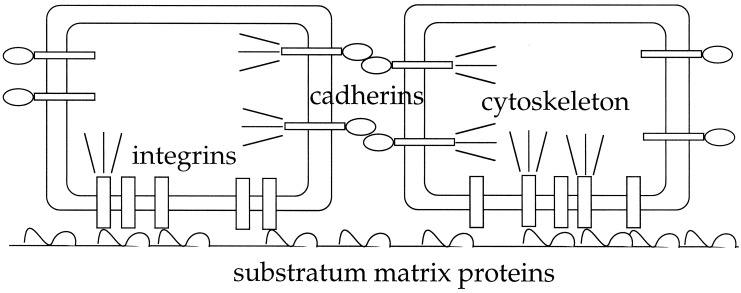
A phenomenon central to cell organization into tissue structures is cell motility, including active cell locomotion through or over matrices and stress-induced matrix remodeling by cells. Both motility-related processes depend on transmission of intracellular cytoskeleton-generated forces to the surrounding environment, whether adjacent cells, extracellular matrix, or a synthetic material. Force transmission as well as generation are regulated by signaling pathways governed by a variety of environmental stimuli, but in any situation are directly mediated by receptors in the cell plasma membrane that link to the cytoskeleton (see Fig. 1). One important class of these receptors are the integrins, αβ heterodimeric transmembrane proteins that recognize and bind mainly to components of the extracellular matrix (although they can additionally recognize and bind to heterotypic cell counterreceptors in certain cases; ref. 4). A second, similarly important class is the cadherins, calcium-dependent transmembrane proteins that interact homotypically with their counterparts on neighboring cells (5). As cells are stimulated by, for instance, growth factors in their environment to generate cytoskeletal forces, one can imagine these forces being transmitted to the surrounding matrix and proximal cells, causing redistribution of cell spatial locations leading ultimately to a new multicellular structural arrangement. A key question, both for basic scientific understanding of tissue morphogenesis and for technological creation of materials enabling desired tissue morphogenesis, is whether there might be simplifying principles by which at least some aspects of resultant multicellular organization can be predicted from quantifiable characteristics of the integrin and cadherin-mediated cell/matrix and cell/cell interactions, respectively.
Figure 1.
Schematic illustration of receptors in the cell plasma membrane that link to the cytoskeleton for force transmission. Integrins, αβ heterodimeric transmembrane proteins, bind the extracellular matrix. Cadherins, calcium-dependent transmembrane proteins, bind homotypically to their counterparts on adjacent cells.
One way to approach this problem is from an engineering design perspective, characterizing the cell/matrix and cell/cell interactions in terms of representative quantitative parameters, and then attempting to manipulate the outcome behavior by varying these parameters systematically. This approach is used by Ryan et al. (3), cleverly bringing together a variety of relevant techniques from materials science, molecular cell biology, and biophysics. To vary the strength of cell/substratum adhesion, a series of copolymers incorporating different ratios of protein-adsorbing poly(desamino-tyrosyl-tyrosine ethyl esther) (DTE) and protein-resistant poly(ethylene glycol) (PEG) polymers were synthesized. The greater the PEG proportion, the lower the cell/substratum adhesivity was found, as measured by a simple washing assay. Because the cells were cultured in protein-containing serum, the cell/substratum adhesion was mediated indirectly by the copolymer via proteins adsorbed onto the substratum. To vary the strength of cell/cell cohesion, three cell lines were used, each expressing different types or levels of a cadherin: cell line LR1 expressed R-cadherin, and cell lines LN5 and LN2 expressed N-cadherin at high and low levels, respectively. Cell/cell cohesion was quantified by means of an assay in which a compressive force balancing the tendency of a cell aggregate to round up was determined. It was found that the three cell lines exhibited diverse degrees of cohesion, with LR1 > LN5 > LN2.
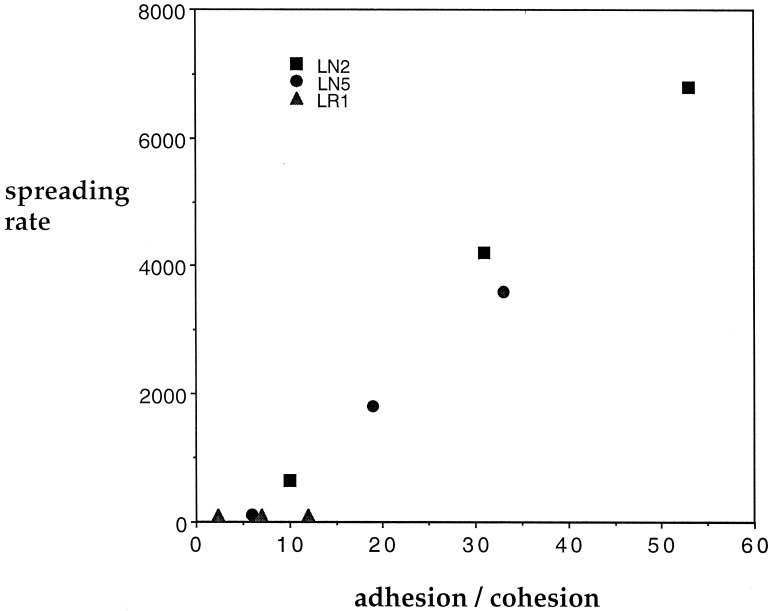
With panels of three cell/cell cohesion values and five cell/substratum adhesion values resulting from the sets of DTE/PEG ratios and cadherin expression properties, the extents of spreading of cell aggregates across the substrata were subsequently measured. Fig. 2 shows a compilation of the particular cases for which spreading rates were noted explicitly. From this figure, it can be seen that the spreading rate appears to be clearly dependent on the ratio of cell/substratum adhesion to cell/cell cohesion, offering what can be considered as a “bioengineering design principle” that quantitatively captures the chief conclusion by Ryan et al., that “either decreasing substratum adhesivity or increasing cell-cell cohesivity dramatically slowed the spreading rate of cell aggregates.” This finding is an exciting extension of the original seminal concept described by Steinberg of cell differential adhesion as a tissue organizing property (6). It will be very interesting to now have this new principle examined across a broader range of substratum conditions (e.g., more rigorously altering the level and types of matrix proteins) and cell types. Most likely, of course, the particular correlation curves obtained in any such broader test will exhibit disparate slopes—reflecting differences in cytoskeleton-generated forces on different protein surfaces and for different cell types—but the crucial test would be whether the adhesion-to-cohesion ratio could account for spreading rate data within any given set of cell and substratum protein types.
Figure 2.
Spreading rate as a function of the cell/substratum adhesion to cell/cell cohesion ratio (values taken from ref. 3 with units not specified here). LR1, cell line expressing R-cadherin; LN5, cell line expressing high levels of N-cadherin; LN2, cell line expressing low levels of N-cadherin. Cohesion was quantified as the compressive force balancing the rounding of a cell aggregate.
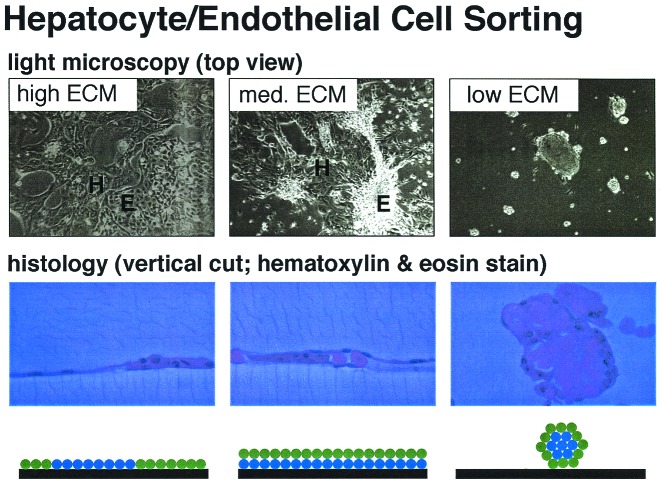
An intriguing next step is applicability to explaining structural organization of multiple cell types, as they interact homotypically with themselves and heterotypically with each other via their cadherins, and each with the substratum via their integrins (7). An ability to understand and control spatial distributions of multiple cell populations will enable rational approaches to tissue engineering applications, where proper multicellular organization in three-dimensional structures is desired. An especially important tissue engineering objective is the need to build vascularized tissue constructs from blood vessel endothelial cells and connective tissue cells by providing them with effective cues using ligand-coated biomaterials scaffolds and soluble growth factors. The cell/substratum adhesion aspect of this problem has been addressed with reasonable success from a quantitative engineering design perspective (8), but the cell/cell cohesion aspect has been tougher to quantify and manipulate. The results from the work of Ryan et al. are encouraging of attempts to control organization of multiple cell types by rationally altering the scaffold substratum, because Fig. 2 suggests that one ought to be able to create substratum conditions for which the adhesion-to-cohesion ratio for one cell type favors its spreading whereas the adhesion-to-cohesion ratio for a second cell type disfavors its spreading. Indeed, such a finding has been reported (9) for rational control of the spatial organization of endothelial cells and hepatocytes in liver tissue engineering applications. As can be seen in Fig. 3, for three different levels of collagen coating on a culture dish surface, three different categories of organization of these two cell types can be obtained. At high collagen levels, both the endothelial cells and hepatocytes spread across the substratum, forming side-by-side two-dimensional structures. At intermediate collagen levels, the endothelial cells form a layer on the substratum, and the hepatocytes form a layer on top of the endothelial cells. At low collagen levels, the hepatocytes aggregate into an inner sphere surrounded by a circumferential shell of endothelial cells that attaches to the substratum. Presumably, hepatocyte cell cohesion is greater than endothelial cell cohesion, and the net structure outcome is governed by the adhesion-to-cohesion ratios for the hepatocytes and endothelial cells as their respective cell/substratum adhesion strengths are changed. Formulation of this behavior in the framework of an engineering design principle, as emphasized by Ryan et al., should facilitate construction of scaffolds that generate three-dimensional vascularized liver tissue structures in vitro (10).
Figure 3.
Different levels of type 1 collagen coating on a culture dish result in different organization of endothelial cells and hepatocytes. High collagen levels cause both cell types to spread across the substratum (Left). On intermediate collagen levels, endothelial cells form a layer on the substratum whereas hepatocytes form a layer on top of the endothelial cells (Center). Low levels of collagen result in an inner layer of hepatocyte aggregate surrounded by endothelial cells (Right). Photographs courtesy of Mark Powers, Massachusetts Institute of Technology.
It must be emphasized that underlying the biophysical processes of cell/substratum adhesion and cell/cell cohesion is a network of biochemical signaling pathways that regulate cytoskeletal force generation as well as integrin- and cadherin-mediated force transmission. Certainly, the operation of these processes as cell organizational governors can be much more complex than the simple relationships discussed here. For instance, increased levels of the α5 integrin subchain in N-cadherin-expressing myoblasts have been observed to increase aggregation rather than decrease it (11), the opposite of what might be initially expected. At least partial explanation appears to lie in an additional effect of integrin signaling, beyond an increase in cell/substratum adhesion, by which membrane extension activity is reduced, leading to a lower propensity for cell locomotion. However, this sort of pleiotropic effect can, in fact, be included in the overall engineering design analysis, provided that analysis of cell motility includes the full range of biophysical processes—including membrane extension—rather than solely cell/substratum adhesion (12, 13). That is, maximally useful engineering design principles for cell organization into tissue structures will require the most comprehensive models for cell motility behavior, including all of the biophysical processes along with critical biochemical signaling regulation.
Footnotes
See companion article on page 4323.
References
- 1.Trinkaus J P. Cells Into Organs: The Forces That Shape the Embryo. Englewood Cliffs, NJ: Prentice–Hall; 1984. [Google Scholar]
- 2.Langer R, Vacanti J P. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 3.Ryan P L, Foty R A, Kohn J, Steinberg M S. Proc Natl Acad Sci USA. 2001;98:4323–4327. doi: 10.1073/pnas.071615398. . (First Published March 27, 2001; 10.1073/pnas.071615398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hynes R O. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 5.Gumbiner B M. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- 6.Steinberg M S. Proc Natl Acad Sci USA. 1962;48:1577–1582. doi: 10.1073/pnas.48.9.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martz E, Phillips H M, Steinberg M S. J Cell Sci. 1974;16:401–419. doi: 10.1242/jcs.16.2.401. [DOI] [PubMed] [Google Scholar]
- 8.Palecek S P, Loftus J C, Ginsberg M H, Lauffenburger D A, Horwitz A F. Nature (London) 1997;385:537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 9.Powers M J, Griffith L G. Microsc Res Tech. 1998;43:379–384. doi: 10.1002/(SICI)1097-0029(19981201)43:5<379::AID-JEMT4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 10.Griffith L G, Wu B, Cima M J, Powers M J, Chaignaud B, Vacanti J P. Ann N Y Acad Sci. 1997;831:382–397. doi: 10.1111/j.1749-6632.1997.tb52212.x. [DOI] [PubMed] [Google Scholar]
- 11.Huttenlocher A, Lakonishok M, Kinder M, Wu S, Truong T, Knudsen K A, Horwitz A F. J Cell Biol. 1998;141:515–526. doi: 10.1083/jcb.141.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maheshwari G, Wells A, Griffith L G, Lauffenburger D A. Biophys J. 1999;76:2814–2823. doi: 10.1016/S0006-3495(99)77435-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox A, Sastry S K, Huttenlocher A. Mol Biol Cell. 2001;12:265–277. doi: 10.1091/mbc.12.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]