Abstract
Helminth infection in pigs serves as an excellent model for the study of the interaction between human malnutrition and parasitic infection and could have important implications in human health. We had observed that pigs infected with Trichuris suis for 21 days showed significant changes in the proximal colon microbiota. In this study, interactions between worm burden and severity of disruptions to the microbial composition and metabolic potentials in the porcine proximal colon microbiota were investigated using metagenomic tools. Pigs were infected by a single dose of T. suis eggs for 53 days. Among infected pigs, two cohorts were differentiated that either had adult worms or were worm-free. Infection resulted in a significant change in the abundance of approximately 13% of genera detected in the proximal colon microbiota regardless of worm status, suggesting a relatively persistent change over time in the microbiota due to the initial infection. A significant reduction in the abundance of Fibrobacter and Ruminococcus indicated a change in the fibrolytic capacity of the colon microbiota in T. suis infected pigs. In addition, ∼10% of identified KEGG pathways were affected by infection, including ABC transporters, peptidoglycan biosynthesis, and lipopolysaccharide biosynthesis as well as α-linolenic acid metabolism. Trichuris suis infection modulated host immunity to Campylobacter because there was a 3-fold increase in the relative abundance in the colon microbiota of infected pigs with worms compared to naïve controls, but a 3-fold reduction in worm-free infected pigs compared to controls. The level of pathology observed in infected pigs with worms compared to worm-free infected pigs may relate to the local host response because expression of several Th2-related genes were enhanced in infected pigs with worms versus those worm-free. Our findings provided insight into the dynamics of the proximal colon microbiota in pigs in response to T. suis infection.
Introduction
Swine have been widely used as a model for human diseases due to anatomic, physiological, and immunological similarities between the two species [1]. Moreover, the biodiversity of the gut microbiota between pigs and humans is comparable [2], [3]. Diverse genetic resources in pigs are readily available, which frequently leads to a whole spectrum of phenotypic changes in response to infection with bacteria, viruses, and parasites common to humans as well as similar dietary patterns. For example, Ossabaw miniature pigs respond rapidly to high-fat, high-cholesterol atherogenic diets and display numerous classical characteristics of human metabolic syndrome [4] that are modulated by daily feeding of probiotics (Solano-Aguilar et al. personal communications). Likewise, helminth infections are common in all pig production systems around the world [5] and prevalent in humans from resource poor areas worldwide. The whipworm Trichuris suis in pigs is an example of a common helminth infection that results in generally mild symptoms, such as diarrhea, anorexia, and retarded growth commonly controlled by management and anthelmintic drugs, but is a re-emerging problem especially in organic and free-range pig production systems. Studies on T. suis infection in pigs have important implications to human health because they can be zoonotic [5] and therapeutic [6]. Morphological and biometric parameters between T. suis and T. trichiura overlap and cannot be differentiated. The latter infects approximately 1049 million people globally [7]. Evolutionary relatedness and similar predilection sites in the mucosa of the upper large intestine of both species suggest that the pig-T. suis system can serve as an excellent model of human malnutrition and parasitic infection [8]. Recently, the immune modulating properties of helminths have been exploited to treat autoimmune diseases including inflammatory bowel diseases (IBD) such as Crohn's disease (CD) [9] and ulcerative colitis (UC) [6]. The appeal of one therapeutic agent to manage diseases as diverse as allergy, multiple sclerosis, rheumatoid arthritis, psoriatic arthritis, and autism is a powerful stimulator of further study to describe mechanisms of action (Human Helminth Co-infections Clinical Trials Database (www.niaid.nih.gov). While many trials have documented positive clinical outcomes, T. suis therapy has nevertheless drawn criticism over concerns of the invasiveness of worms on human physiology [10] as well as potential gastrointestinal side effects [11]. It is known that the enteric microbiota plays a critical role in the pathogenesis of IBD [12]. For example, enterobacteria are observed more frequently in CD than in healthy control human subjects [13]. Several studies have suggested that probiotics within the genera Lactobacillus and Bifidobacterium may have favorable impact on the treatment of patients with CD by altering the gut microbiota and modulating the host immune system [14]. Recently, we demonstrated that a 21-day T. suis infection in pigs induced a profound change in both microbial composition and metabolic potential in the lumen of the proximal colon [2]. Changes in abundance of Succinivibrio and Mucispirillum were associated with parasite-induced alterations in carbohydrate and amino acid metabolism and niche disruptions in mucosal pathology [2]. However, major determinants of phylogenetic and functional composition of the porcine colon microbiota remain unknown. In this study, we investigated the relationship between adult T. suis worm burden and changes in the pig proximal colon luminal microbiota. The results indicated that T. suis-induced changes in the proximal colon microbiota were similar regardless of the persistence or host clearance of adult worms. In addition, the local host mucosal response was associated with worm burden and the intensity of Th2-related and allergy/asthma associated gene expression.
Results
Worm burden and changes in localized inflammation
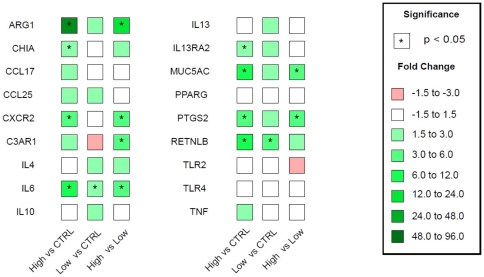
The adult T. suis worm burden and associated pathology in the proximal colon become more disparate in a group of out-bred pigs between seven and nine weeks after inoculation with some pigs showing fewer than 10 worms and an apparently normal mucosa and others showing hundreds of worms with localized inflammation, mucus production, and smooth muscle hypertrophy; worm clearance is virtually complete between weeks 9 and 11 [15]. We selected three infected pigs at 53 days after inoculation that had >300 adult worms (667±391 sd), five infected pigs with 0 worms, and three uninfected pigs to evaluate changes in local gene expression in the epithelial layer of the proximal colon and associated changes in the luminal microbiota. The presence of high numbers of adult worms (>300) significantly increased expression of arg1, chia, cxcr2, il6, il13ra2, muc5ac, ptgs2, and retnlb compared to uninfected control pigs and arg1, cxcr2, c3ar1, il6, muc5ac, and ptgs2 were higher in infected pigs with worms versus worm-free infected pigs (Fig. 1). Expression of ccl17, ccl25, c3ar1, and tnf were not significantly higher than uninfected controls, and levels of pprg, tlr2, and tlr4 were not increased among the groups.
Figure 1. Localized changes in gene expression in the proximal colon epithelium.
The relative changes in gene expression were determined by real-time RT-PCR (TaqMan) and compared between T. suis-infected pigs with a high worm burden (High) and low worm burden (worm-free or Low), and uninfected naïve control pigs (CTRL).
Changes in the proximal colon microbial composition in response to T. suis infection
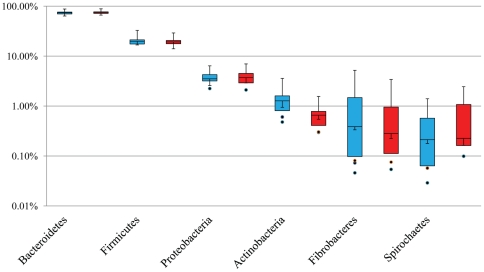
Taxonomic profiles of the porcine proximal colon microbiota were evaluated using both MetaPhyler [16] and MG RAST programs [17]. The core microbiota of the porcine proximal colon included 19 phyla, 39 classes, 93 families and 121 genera, identified by the MetaPhyler method (Table 1). Of note, the 5 most abundant phyla accounted for approximately 99% of all assigned sequence reads with Bacteroidetes (72.64%), Firmicutes (20.33%) and Proteobacteria (3.69%) as among the most abundant. The percentage composition at a phylum level derived from both MetaPhyler and MG-RAST were similar (Fig. 2). In addition, our findings on the phylum-level composition using whole-genome shotgun (WGS) reads were comparable to those obtained using bar-coded pyrosequencing of the V3–V5 regions of the 16S rRNA gene [2].
Table 1. Taxonomic profiles of the porcine colon microbiota.
| Phylum | Class | Family | Genus | |
| Total | 27 (29) | 64 (34) | 213 (223) | 372 (778) |
| Mean ±sd | 23.18±1.66 | 52.55±5.45 | 159.45±16.26 | 238.36±28.77 |
| (28.09±0.30) | (34.00±0.00) | (220.18±1.25) | (661.55±18.85) | |
| Core | 19 (28) | 39 (34) | 93 (217) | 121 (592) |
Numbers of taxa identified by MetaPhyler (MG-RAST) are listed (N = 11).
Figure 2. Phylum-level relative composition of the microbiome in the porcine proximal colon.
Boxes denote the inter-quartile range between the 1st and 3rd quartiles (25 and 75%, respectively, N = 11). Blue: detected using MetaPhyler; Red: detected using MG-RAST. Y-axis: log scale.
Pigs infected with T. suis for 53 days had a profound change in the proximal colon microbial composition. Of 27 phyla collectively identified by MetaPhyler, Fibrobacteres, Spirochaetes, Tenericutes, and Gemmatimonadetes were significantly decreased (P<0.05) by infection regardless of the worm burden in the colon. The abundance of Fibrobacteres was 2.65% in the parasite naive pigs compared to 0.38% in the infected pigs while the relative abundance of Spirochaetes displayed a similar reduction from 0.79% in controls to 0.17% in infected pigs. The abundance of both phyla in the proximal colon microbiota between infected pigs with worms and worm-free was indistinguishable.
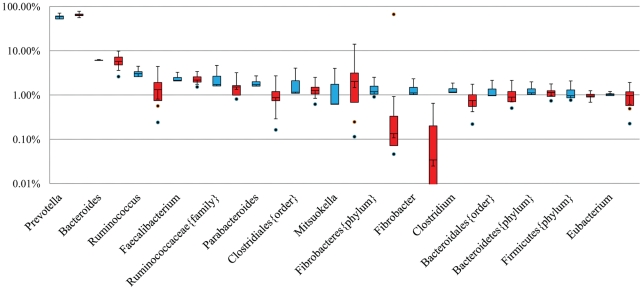
The abundance of 48 of the 372 genera collectively identified in the proximal colon microbiota by MetaPhyler was significantly affected by infection. The percentage of genera altered was similar to that observed in pigs infected with T. suis for 21 days [2]. Fibrobacter and a potentially novel genus within the phylum Fibrobacteres were among the most abundance genera significantly affected by infection (Fig. 3). The abundance of Treponema, Dorea, and a novel genus within the phylum Spirochaetes were also significantly decreased by infection (Table 2). The abundance of Ruminococcus was also reduced by 2-fold at 53 days (Fig. 3) after inoculation similar to that observed at 21 days after inoculation [2]. Interestingly, the relative abundance of Campylobacter was low but reliably detected in the porcine proximal colon microbiota. Its abundance in infected pigs with worms was 3-fold higher than in the parasite naïve pigs (Table 2); supporting the observation that T. suis infection increases the risk of Campylobacter infection in pigs [18]. There was, however, a notable 10-fold difference in Campylobacter abundance in the proximal colon microbiota in infected pigs with worm compared to infected worm-free pigs (P<0.05). As Table 2 shows, the incidence of Campylobacter in infected worm-free pigs was lower than in the parasite naive pigs (P<0.05).
Figure 3. Relative abundance of 15 genera in the porcine proximal colon microbiota detected using MetaPhyler.
Boxes denote the inter-quartile range between the 1st and 3rd quartiles (25 and 75%, respectively). Blue: Parasite naïve pigs (N = 3); Red: Infected pigs (N = 8). Symbol {} denotes a possible novel genus within the taxon indicated. For example, Ruminococcaceae{family} indicates a possible novel genus within the family Ruminococcaceae. A significant repression of relative abundance of the genus Fibrobacter and a possible novel genus in the phylum Fibrobacteres was detected in the proximal colon microbiota of Trichuris suis infected pigs. Y-axis: log scale.
Table 2. Genera significantly impacted by helminth infection in the porcine colon microbiota.
| Genus | Uninfected | Worm | Worm-free |
| Fibrobacter | 2.728±1.150a | 0.435±0.454b | 0.420±0.488b |
| Treponema | 1.468±0.195a | 0.382±0.432b | 0.289±0.346b |
| Spirochaeta | 0.280±0.082a | 0.080±0.072b | 0.077±0.077b |
| Dorea | 0.181±0.020a | 0.089±0.043b | 0.108±0.048b |
| Campylobacter | 0.108±0.045a | 0.340±0.573a | 0.032±0.012b |
| Brachyspira | 0.057±0.014a | 0.025±0.016b | 0.023±0.014b |
| Mycoplasma | 0.028±0.004a | 0.013±0.007b | 0.013±0.008b |
| Thermotoga | 0.024±0.008a | 0.012±0.007b | 0.013±0.007a |
| Actinobacillus | 0.022±0.001a | 0.014±0.004b | 0.013±0.005b |
| Francisella | 0.010±0.001a | 0.005±0.003b | 0.005±0.003b |
| Erysipelothrix | 0.010±0.003a | 0.005±0.003b | 0.005±0.002b |
Numbers denote mean ±SD of percentage composition (N = 4). Only genera significantly impacted detected by both MetaPhyler and MG RAST are listed. Different superscripted letters indicated significantly different at P<0.05 based on a modified t-test.
The protein repertoire and pathways impacted by T. suis infection
Trimmed sequence reads were de novo assembled using SOAPdenovo software [19]. The process resulted in 257,415 contigs assembled at a mean length 445.02 bp (±29.77 sd) per sample (N50 = 482 bp). Genes or open reading frames (ORFs) were predicted using FragGeneScan from these contigs. These ORFs were annotated against the Pfam database (v24.0). Collectively, a total of 5,157 Pfam Protein families were identified. The ten most abundant Pfam families in the parasite naive pigs were ABC transporter (PF00005, 0.9256%), TonB dependent receptor (PF00593, 0.6344%), TonB-dependent Receptor Plug Domain (PF07715, 0.5532%), ATPase family (PF00004, 0.5281%), AcrB/AcrD/AcrF family (PF00873, 0.4989%), Glycosyl transferase family 2 (PF00535, 0.4810%), Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase (PF02518, 0.4753%), Aminotransferase class I and II (PF00155, 0.4617%), Elongation factor Tu GTP binding domain (PF00009, 0.4508%), and Response regulator receiver domain (PF00072, 0.4423%). To gain insight into possible shifts in functionality and metabolic potentials in the proximal colon microbiota in response to a 53 day infection with T. suis, Gene Ontology (GO) terms associated with these Pfam protein families were identified. 103 of the 1390 GO terms were significantly affected by infection (P<0.05). As Table 3 shows, the infection seemingly had a broad impact on biological processes and molecular functions.
Table 3. Gene Ontology (GO) terms significantly affected by helminth infection.
| GO Term | Description | Uninfected | Worm | Worm-free |
| GO:0006412 | translation | 2.515±0.039a | 2.285±0.108b | 2.420±0.087a |
| GO:0005622 | intracellular | 1.774±0.061a | 1.650±0.079a | 1.647±0.053b |
| GO:0003735 | structural constituent of ribosome | 1.097±0.053a | 0.928±0.059b | 0.986±0.061a |
| GO:0005840 | ribosome | 1.083±0.050a | 0.918±0.057b | 0.973±0.061a |
| GO:0006096 | glycolysis | 0.339±0.004a | 0.350±0.007b | 0.354±0.009b |
| GO:0016769 | transferase activity | 0.265±0.007a | 0.278±0.010a | 0.286±0.007b |
| GO:0044237 | cellular metabolic process | 0.232±0.013a | 0.252±0.008b | 0.248±0.014a |
| GO:0016829 | lyase activity | 0.229±0.009a | 0.213±0.008b | 0.221±0.003a |
| GO:0009253 | peptidoglycan catabolic process | 0.165±0.003a | 0.186±0.020a | 0.187±0.008b |
| GO:0016740 | transferase activity | 0.125±0.006a | 0.156±0.017b | 0.144±0.014a |
| GO:0008484 | sulfuric ester hydrolase activity | 0.124±0.005a | 0.163±0.021b | 0.145±0.014a |
| GO:0004672 | protein kinase activity | 0.114±0.006a | 0.080±0.011b | 0.089±0.012b |
| GO:0006468 | protein amino acid phosphorylation | 0.102±0.007a | 0.073±0.016b | 0.080±0.011b |
| GO:0008134 | transcription factor binding | 0.082±0.008a | 0.070±0.006a | 0.069±0.004b |
| GO:0016620 | oxidoreductase activity | 0.079±0.001a | 0.071±0.006a | 0.072±0.004b |
| GO:0016114 | terpenoid biosynthetic process | 0.079±0.009a | 0.091±0.004a | 0.088±0.005b |
| GO:0016810 | hydrolase activity [C-N bonds] | 0.077±0.002a | 0.091±0.008b | 0.087±0.010a |
| GO:0008237 | metallopeptidase activity | 0.077±0.004a | 0.088±0.001b | 0.090±0.006b |
| GO:0009236 | cobalamin biosynthetic process | 0.074±0.015a | 0.125±0.012b | 0.103±0.013b |
| GO:0005529 | sugar binding | 0.074±0.005a | 0.091±0.011b | 0.089±0.008b |
| GO:0006526 | arginine biosynthetic process | 0.066±0.005a | 0.056±0.004b | 0.056±0.003b |
| GO:0045261 | ATP synthase complex [F(1)] | 0.063±0.004a | 0.073±0.010a | 0.076±0.005b |
| GO:0000902 | cell morphogenesis | 0.061±0.003a | 0.069±0.005a | 0.069±0.004b |
| GO:0004332 | fructose-bisphosphate aldolase activity | 0.060±0.002a | 0.053±0.003b | 0.055±0.002b |
| GO:0004356 | glutamate-ammonia ligase activity | 0.054±0.003a | 0.044±0.009a | 0.045±0.003b |
| GO:0003883 | CTP synthase activity | 0.050±0.001a | 0.040±0.005b | 0.042±0.003b |
| GO:0006221 | pyrimidine nucleotide biosynthesis | 0.050±0.001a | 0.040±0.005b | 0.042±0.003b |
103 out of the 1390 GO terms were significantly impacted based on a modified t-test. GO terms with relative abundance >0.05% were listed. Numbers denote the percentage of Pfam protein families assigned to each category (mean ±sd). Different superscript letters indicate P <0.05.
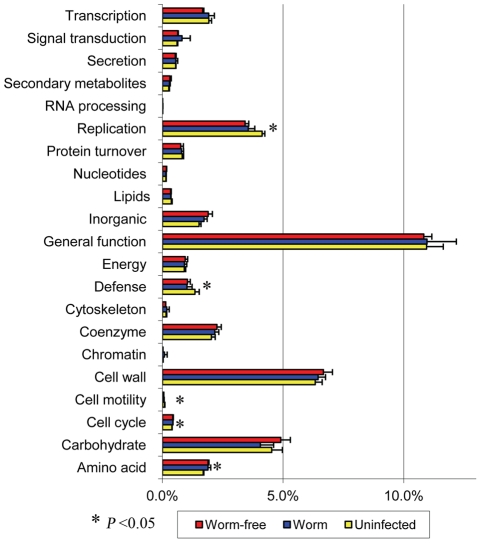
The protein repertoire of the porcine colon microbiota was also assessed by the eggNOG database annotation using the MG-RAST pipeline. Of 39,777 eggNOGs identified, several proteins involved in carbohydrate metabolism were among the most abundant in the parasite naïve pigs, such as α-L-fucosidase (NOG04067, 1.05%), glycoside hydrolase family 43 (GH43, 1.01%), α-L-Rhamnosidase (NOG10735, 0.66%), and pectate lyase (NOG44882, 0.56%). Indeed, xylosidase, glucosidase, α-galactosidase, polysaccharide biosynthesis protein, and carbohydrate binding protein, as well as the above-mentioned rhamnosidase and pectate lyase, were among the most abundant in the parasite naïve pigs. Trichuris suis infection had a significant influence on the functional composition in the proximal colon microbiota. For example, infection induced a significant reduction in the relative abundance of α-amylase (NOG71025), from 0.14% in the control uninfected pigs to 0.04% in the infected pigs. The abundance of GH43 followed a similar trend and was significantly reduced by infection. Overall, infection resulted in a significant change in the abundance of some key eggNOGs in the proximal colon microbiota regardless of worm status, suggesting a relatively persistent change over time in the microbiota due to the initial infection (Fig. 4). Of 22 NOG functional categories identified by MG-RAST, several classes such as amino acid transport and metabolism and replication, were significantly affected (Fig. 5). In addition, the number of sequences annotated to “defense mechanisms” was significantly reduced by infection, from 1.35% in controls to 1.03% in the infected pigs regardless of worm burden.
Figure 4. Differences in functional profiles and metabolic potentials of the porcine proximal colon microbiota between uninfected (CT) and infected groups (Worm and Worm-free).
Principal component analysis (PCA) was performed using the ade4 package in R based on relative abundance of 50 selected function classes assigned using the eggNOG database.
Figure 5. Functional categories affected by Trichuris suis infection in the porcine proximal colon microbiota annotated using the eggNOG database.
The class labeled “unknown”, which accounted for 61.34% of hits, were not included. * denotes significantly impacted by infection, regardless of the worm burden.
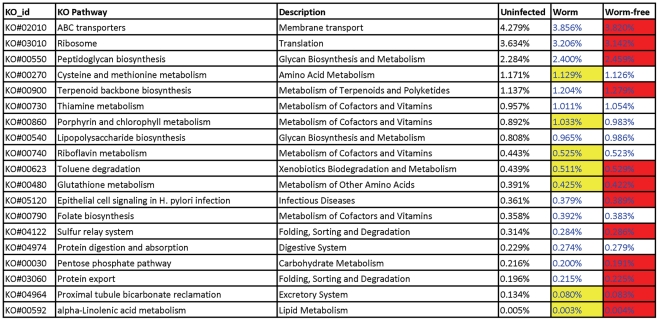
Metagenomic sequences were also annotated against Kyoto Encyclopedia of Genes and Genomes (KEGG) databases using the MG-RAST pipeline. A total of 7150 KEGG entries were identified. The five most abundant KEGG in the parasite naive pigs were DNA-directed RNA polymerase subunit β (K03046, 0.90%), β-glucosidase (K01188, 0.90%), carbamoyl-phosphate synthase large subunit (K01955, 0.89%), β-galactosidase (K01190, 0.77%), and excinuclease ABC subunit A (K03701, 0.73%). The relative abundance of approximately 7% of all KEGGs identified was significantly altered by infection. These KEGGs included starch phosphorylase (K00688), its abundance from 0.42% in the control uninfected pigs to 0.35% in the infected pigs. Similarly, the abundance of β-mannosidase (K01192) was also decreased as a result of infection. Approximately 10% of 297 KEGG Orthology (KO) pathways identified were affected by infection. As Fig. 6 shows, ABC transportors (KO#02010), peptidoglycan biosynthesis (KO#00550), lipopolysaccharide biosynthesis (KO#00540), alpha-linolenic acid metabolism (KO#00592) were among the 29 KO pathways affected by infection.
Figure 6. Select pathways were significantly affected infection with Trichuris suis in the porcine proximal colon microbiota.
Numbers denote the percentage of hits positively assigned to each pathway. Different colors (Red or Yellow) indicate a significant difference between the cohorts infected with Trichuris suis that had adults worms (Worm) and the infected cohort where adult worms had been expelled (Worm-free). The numbers with a Blue font denote significant difference in relative abundance between uninfected control and infected (Worm + Worm-free) groups.
Discussion
The gut microbiota plays a critical role in host nutrient metabolism as well as in the development of host immune systems [20], [21]. However, the dynamics of the gut microbiota in response to parasitic infections have only been examined recently [2], [22]. We showed previously that infection of pigs with T. suis for 21 days induced a profound change in proximal colon luminal microbiota with approximately ∼13% of all genera identified significantly affected by infection [2]. For example, there was a significant reduction in the relative abundance of important genera such as Oscillibacter and Succinivibrio. The changes in taxonomical profiles lead to alterations in the metabolic potential of the porcine colon microbiota, including repressing carbohydrate metabolism and lysine biosynthesis [2]. It is not clear if the worm directly altered the metabolic potential by local depletion of volatile organic compounds (VOC) that are co-factors in carbohydrate and lysine metabolism or affected metabolism upstream in the small intestine to alter the composition of metabolites in the proximal colon [2]. All pigs infected with T. suis for 21 days had oleic acid in the proximal colon that was not detected in parasite naïve pigs [2]. This observation suggested that the worm altered fatty acid absorption in the small intestine and that local increases in oleic acid could exert antibacterial properties to alter the local microbiome or lipolytic properties that are pro-inflammatory to the mucosa [2]. In the current study, we characterized the porcine colon microbiota at 53 days after inoculation with infective T. suis eggs. Specifically, we examined the effect of worm burden on the persistence of the altered proximal colon microbiota that was detected at 21 days post infection. Between seven and nine weeks after inoculation with infective T. suis eggs there is development of a self-cure reaction that is represented by some pigs having a persistent adult worm burden and localized inflammation, and others that have few or no adult worms and a normal mucosa [15]. This is typical of mammalian host resistance expressed as a skewed distribution of adult worm burden in genetically robust out-bred populations. While the percentage of the genera significantly affected was similar between the 21-day (13%) and 53-day infections (48 genera out of the 372 genera identified using MetaPhyler ≈13%), the difference in temporal composition profiles in the proximal colon microbiota was distinct. The most abundant genera significantly affected by a 21-day infection include Oscillibacter and Succinivibrio (both significantly decreased) as well as Paraprevotella (a six-fold increase in its relative abundance from 0.47% in the uninfected controls to 3.03% in the infected pigs). However, the relative abundance of these genera was not changed at 53 day post infection. The relative abundance of Spirochaeta and Dorea was significantly decreased in pigs infected with T. suis for 53 days, confirming the previous findings in the 21-day study [2]. Among the ten most abundant genera in the proximal colon microbiota of the parasite naive pigs (Fig. 3), Fibrobacter was one of the genera with its relative abundance significantly repressed by infection regardless of the worm burden (Table 2). In addition, the MetaPhyler results indicated a possible novel genus in the phylum Fibrobacteres in the porcine colon microbiota, which was also significantly reduced by infection. Sequence reads were analyzed independently using BLAT against the first sequenced genome from the phylum Fibrobacteres, Fibrobacter succinogenes S85 [23], a species that plays a critical role in fiber digestion in the rumen. Sequence alignment confirmed a significant reduction in abundance of Fibrobacter by infection. The lower level of Fibrobacter abundance derived from the DNA sequence alignment, compared to what was calculated from predicted protein sequences, suggests the presence of novel bacteria within the genus Fibrobacter or the phylum Fibrobacteres in the porcine proximal colon microbiota with sufficient sequence divergence from the S85 strain. Fibrobacter is known to possess a unique array of hemicellulose-degrading enzymes and is an efficient and prolific degrader of cellulose as its sole energy source [23]. The relative abundance of another group of important bacteria determining the fibrolytic capability in the rumen and hindgut, Ruminococcus, ranked the 3rd most abundant genus in the porcine proximal colon of parasite naive pigs, showed a 2-fold reduction in response to T. suis infection (Fig. 3). These data indicated that the fibrolytic capacity of the proximal colon microbiota may be impaired by T. suis infection.
Infection of pigs with T. suis is associated with exacerbation of campylobacteriosis [24]–[26], which is caused by bacteria such as Campylobacter jejuni and C. coli and results in a broad range of complications, including acute diarrhea. Infections by Campylobacter disrupt the absorptive capacity of host epithelial cells [27]. However, Campylobacter does not normally cause colonic infection in pigs without a concomitant T. suis infection [18]. IL-4, which is strongly up-regulated by helminth infection, enhanced internalization of intestinal pig epithelial cells by C. jejuni and subsequent bacterial invasion in a dose-dependent manner [26], suggesting this Th2 cytokine plays a critical role in the exacerbated pathology resulting from dual infections of pigs with T. suis and Campylobacter spp. Localized gene expression for IL-4 was not significantly increased in the proximal colon at 53 days after inoculation (Fig. 1), but was increased earlier in the course of infection [28] which could have facilitated Campylobacter invasion in situ. In this study, we observed a 3-fold increase in the relative abundance of Campylobacter in the T. suis infected pigs. However, in the infected worm-free pigs Campylobacter abundance was significantly decreased (Table 2). The T. suis-facilitated uptake and antigen processing of Campylobacter spp by lymphoglandular complexes in the pig colon and subsequent induction of local anti- Campylobacter antibody responses in the ileum and colon could explain the significant reduction in Campylobacter from the proximal colon of worm-free infected pigs [29]. Thus, clearance of adult T. suis from infected pigs may have a therapeutic effect against selected bacterial pathogens that is inhibited by adult worm persistence.
Parasitic nematodes activate potent Th2-associated immune responses that support resistance to infection and an asthma/allergy related response that, if left uncontrolled, can contribute to mucosal inflammation [30]. Enhanced gene expression of arg1 and chia represent markers of Th2-induced alternatively activated macrophages (AAM) that were diminished in the pig as the worms were cleared from the proximal colon (Fig. 1). The chemokine ligands ccl17 and ccl25 are related to AAM development and were increased in pigs with high numbers of worms, although not to significant levels of stimulation. The AAM plays a protective role against helminth parasites that invade the mucosa of the small intestine [31] and can regulate intestinal smooth muscle hyper-contractility in response to infection [32]. This worm-dependent modulation of AAM markers in T. suis-infected pigs was recently supported by the loss of expression of related markers in the proximal colon of T. muris-infected Balb/c mice soon after expulsion (Madden et al., personal communication). Local gene expression of muc5ac and retnlb in T. suis-infected pigs are related to products that contribute directly to resistance to T. muris in the colon of mice [33], [34]. Unregulated expression of retnlb, however, can also lead to mucosal inflammation during infection of mice with T. muris [35] as well as the expression of ptgs2 and c3ar1 [36] that are induced by asthma/allergy associated inflammation and were differentially expressed in pigs with high T. suis worm burden with increased mucosal pathology.
Epithelial cells responses to infection are protective against T. muris [37], [38] and the expression of cxcr2 in T. suis infected pigs may relate to epithelial cell signaling as well as the expression of il13ra2 for its role in both epithelial and smooth muscle signaling (Madden et al., personal communication) and localized control of inflammation [30], [39]. The increased gene expression of il10 and il13 in infected pigs that had cleared adult T. suis, although not statistically significant, indicated a trend toward and an anti-inflammatory response in the proximal colon that supported the appearance of a normal mucosa in these pigs.
The host ability to control infection with T. suis and modulate the level of localized inflammation is dependent on adult worm burden and changes in the intestinal microbiome and related metabolic changes during the course of infection [2]. The regulatory mechanisms involved are important to the rapid removal of the worm that reduces the spread of infection and reduces inflammation as well as the control of bacterial pathogens like Campylobacter spp. that contribute to secondary disease and represent a zoonotic threat to humans. There is also the importance of understanding these events to maximize the therapeutic potential of this nematode as a modulator of inflammatory diseases in humans. What remains is to distinguish the worm, microbiome, and host factors that skew these responses in favor of healthy outcomes.
Materials and Methods
Animals and parasitology
Infection protocols and sampling were essentially similar to those reported previously [2]. Briefly, 14 female piglets (Cross bred of Landrace X Yorkshire X Poland China) at three months of age were maintained indoors on sealed concretes with free access to a balanced ration and water. No antibiotics were used during the study. A single dose of infective T. suis eggs (2×104 egg/pig) was inoculated per os (N = 9). The infection was allowed to progress for 53 days after inoculation. Five other pigs of the same age were orally dosed with PBS and served as parasite naive controls. All pigs were sacrificed at the same date when the infection reached 53 days. Animal management and handling were conducted based on a protocol specifically approved by the USDA-ARS Beltsville Area Animal Care and Use Committee (Protocol #10-011), following Institutional Animal Care and Use Committees (IACUC) guidelines. Luminal fecal contents were collected from the proximal colon at ∼30 cm from the ileal/caecal junction. Colon tissue samples were also collected at ∼30 cm from this junction. The pH of the contents was measured using a hand-held pH meter for semi-solid materials. Both fecal and tissue samples were snap frozen in liquid nitrogen prior to storage at −80°C until metagenomic DNA and total RNA were extracted. Colon pathology was examined by virtual and microscopic observation [18]. Trichuris suis worms at this stage of the infection can be visually counted on the surface of the mucosa. The pigs were free of inadvertent Ascaris suum infection based on the absence of worms from the small intestines and white spot lesions on the liver.
Quantitative reverse transcriptase (RT)-PCR
Total RNA samples extracted from the epithelial cell layer of the proximal colon that was separated manually by peeling it away from the muscularis of T. suis-infected (three pigs with worms and five with no detected worms) and five uninfected pigs [28], including all pigs used for the microbiome study. Briefly, frozen tissue sections removed from pigs at necropsy and placed immediately in liquid nitrogen followed by storage at −80C until use. Tissues were subsequently homogenized in Trizol (Invitrogen, Grand Island, NY) and RNA was extracted from homogenized samples according to the manufacturer's instruction. The extracted RNA was treated with DNase in the presence of RNase inhibitor. RNA integrity, quantity, and genomic DNA contamination were assessed using the Experion RNA Analysis Chips (Bio-Rad). cDNA was synthesized using iScript cDNA Synthesis kit from Bio-Rad. The sequence of probes and primers and running conditions of RT-PCR were obtained from the DGIL Porcine Immunology and Nutrition Database http://www.ars.usda.gov/Services/docs.htm?docid=6065. Primers and high-performance liquid chromatography-purified, 5′,6-carboxy-4,7,2′,7′-tetrachlorofluorescein-, 3′ Black Hole Quencher-1-labeled fluorescent probes were synthesized (Biosource, Camarillo, CA). Real-time RT-PCR was performed using 15 ng/well of cDNA in 15 μl on an ABI 7900 sequence detector system (Applied Biosystems, Foster City, CA). Data for gene expression were normalized to the housekeeping gene RPL32 and converted to ΔCT [40]–[41].
Metagenomic DNA extraction and sequencing
Metagenomic DNA was extracted from fecal samples using a QIAamp DNA stool kit (Qiagen, Valenica, CA) with modifications to the protocol described [22], [42]. DNA integrity was verified using a Bioanalyzer 2100 (Agilent, Palo Alto, CA). Metagenomic DNA concentration was quantified by fluorometry. Approximately 1.0 µg of high-quality DNA was processed using an Illumina TruSeq DNA sample prep kit following manufacturer's instruction (Illumina, San Diego, CA, USA). Final individual libraries were validated, pooled based on their respective 6-bp adaptors and sequenced at 100 bp/sequence read using an Illumina HiSeq 2000 sequencer. Approximately 47,958,917±10,634,382 (mean ±sd) raw sequence reads per sample were generated for this study. Sequence reads were deposited to the MG-RAST and are publically accessible at the metagenomic analysis server (http://metagenomics.anl.gov/) (accession# 4474250.3 to 4474257.3, 4474259.3, 4474261.3, and 4474262.3).
Data analysis and statistics
Metagenomic DNA samples extracted from the proximal colon microbiota of three parasite naïve and eight infected pigs (4 with adult worms and 4 worm-free) were sequenced. Raw sequence reads from the WGS approach were first trimmed using SolexaQA, a Perl-based software package calculating quality statistics from FASTQ files generated by Illumina sequencers [43]. Reads of host origin were then removed using Bowtie [44]. The resultant quality reads were then analyzed using MetaPhyler [16]. The relative abundance data from MetaPhyler were analyzed based on a modified t-test [45]
Raw sequence reads were uploaded into a MG-RAST server [46] for quantitative views of the microbial populations in the lumen of the pig proximal colon based on WGS sequence data. The data were then analyzed following the MG-RAST pipeline (v3.0) including quality filtering, dereplication to remove possible sequencing artifacts, and removal of host contaminants. Open reading frames (ORF) were then predicted using FragGeneScan [47], a recently developed program combining sequencing error models and codon usages in a hidden Markov model to improve the prediction of protein-coding region in short reads. The microbial classification was then obtained using the lowest common ancestor method in the pipeline. Sequence counts positively assigned to a given taxon at the phylum-, class-, family-, and genus- levels were normalized. Compositional differences between MetaPhyler and MG-RAST annotation platforms were analyzed using an unpaired t- test.
Quality WGS sequences were de novo assembled using SOAPdenovo software [19]. ORF were predicted from all contigs greater than 200 bp using FragGeneScan (v1.14). Functional annotation was further performed according to the KEGG and Pfam (v24.0) databases. Pfam 24.0 seed alignments were downloaded, and a database of core profile HMMs was compiled using the HMMSCAN software package (v3.0), which was used to annotate predicted proteins.
Acknowledgments
We'd like to thank Ashley Sperling and Alicia Beavers for their excellent technical assistance. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U. S. Department of Agriculture. The USDA is an equal opportunity provider and employer.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: SW and WL were supported by Award R01HG005978 from the National Human Genome Research Institute (NHGRI), Award R01RR025030 from National Center for Research Resources (NCRR) and CAMERA project funded by Gordon and Betty Moore Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NHGRI, NCRR or the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dawson HD. McAnulty PA, Dayan, A.D, Ganderup, N.C, Hastings, K.L, editors. A comparative assessment of the pig, mouse and human genomes. The minipig in biomedical research: CRC Press. 2011. pp. 323–342.
- 2.Li RW, Wu S, Li W, Navarro K, Couch RD, et al. Infection and Immunity; 2012. Alterations in the colon microbiota induced by the gastrointestinal nematode Trichuris suis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu J, Mahowald MA, Ley RE, Lozupone CA, Hamady M, et al. Evolution of symbiotic bacteria in the distal human intestine. PLoS biology. 2007;5:e156. doi: 10.1371/journal.pbio.0050156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neeb ZP, Edwards JM, Alloosh M, Long X, Mokelke EA, et al. Metabolic syndrome and coronary artery disease in Ossabaw compared with Yucatan swine. Comparative medicine. 2010;60:300–315. [PMC free article] [PubMed] [Google Scholar]
- 5.Roepstorff A, Mejer H, Nejsum P, Thamsborg SM. Helminth parasites in pigs: new challenges in pig production and current research highlights. Veterinary parasitology. 2011;180:72–81. doi: 10.1016/j.vetpar.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 6.Summers RW, Elliott DE, Urban JF, Thompson RA, Weinstock JV. Trichuris suis therapy for active ulcerative colitis: a randomized controlled trial. Gastroenterology. 2005;128:825–832. doi: 10.1053/j.gastro.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Stephenson LS, Holland CV, Cooper ES. 121 Suppl. Parasitology; 2000. The public health significance of Trichuris trichiura. pp. S73–95. [DOI] [PubMed] [Google Scholar]
- 8.Boes J, Helwigh AB. 121 Suppl. Parasitology; 2000. Animal models of intestinal nematode infections of humans. pp. S97–111. [DOI] [PubMed] [Google Scholar]
- 9.Summers RW, Elliott DE, Qadir K, Urban JF, Thompson R, et al. Trichuris suis seems to be safe and possibly effective in the treatment of inflammatory bowel disease. The American journal of gastroenterology. 2003;98:2034–2041. doi: 10.1111/j.1572-0241.2003.07660.x. [DOI] [PubMed] [Google Scholar]
- 10.Van Kruiningen HJ, West AB. Potential danger in the medical use of Trichuris suis for the treatment of inflammatory bowel disease. Inflammatory bowel diseases. 2005;11:515. doi: 10.1097/01.mib.0000160369.47671.a2. [DOI] [PubMed] [Google Scholar]
- 11.Bager P, Kapel C, Roepstorff A, Thamsborg S, Arnved J, et al. Symptoms after ingestion of pig whipworm Trichuris suis eggs in a randomized placebo-controlled double-blind clinical trial. PloS one. 2011;6:e22346. doi: 10.1371/journal.pone.0022346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shanahan F. Inflammatory bowel disease: immunodiagnostics, immunotherapeutics, and ecotherapeutics. Gastroenterology. 2001;120:622–635. doi: 10.1053/gast.2001.22122. [DOI] [PubMed] [Google Scholar]
- 13.Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, et al. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Famularo G, Mosca L, Minisola G, Trinchieri V, De Simone C. Probiotic lactobacilli: a new perspective for the treatment of inflammatory bowel disease. Current pharmaceutical design. 2003;9:1973–1980. doi: 10.2174/1381612033454207. [DOI] [PubMed] [Google Scholar]
- 15.Kringel H, Roepstorff A. Trichuris suis population dynamics following a primary experimental infection. Veterinary parasitology. 2006;139:132–139. doi: 10.1016/j.vetpar.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Liu B, Gibbons T, Ghodsi M, Treangen T, Pop M. Accurate and fast estimation of taxonomic profiles from metagenomic shotgun sequences. BMC Genomics. 2011;12(Suppl 2):S4. doi: 10.1186/1471-2164-12-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, et al. The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mansfield LS, Urban JF The pathogenesis of necrotic proliferative colitis in swine is linked to whipworm induced suppression of mucosal immunity to resident bacteria. Veterinary immunology and immunopathology. 1996;50:1–17. doi: 10.1016/0165-2427(95)05482-0. [DOI] [PubMed] [Google Scholar]
- 19.Li R, Yu C, Li Y, Lam TW, Yiu SM, et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 20.Festi D, Schiumerini R, Birtolo C, Marzi L, Montrone L, et al. Gut microbiota and its pathophysiology in disease paradigms. Digestive diseases. 2011;29:518–524. doi: 10.1159/000332975. [DOI] [PubMed] [Google Scholar]
- 21.Young VB. Current opinion in gastroenterology; 2011. The intestinal microbiota in health and disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li RW, Wu S, Li W, Huang Y, Gasbarre LC. Metagenome plasticity of the bovine abomasal microbiota in immune animals in response to Ostertagia ostertagi infection. PloS one. 2011;6:e24417. doi: 10.1371/journal.pone.0024417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suen G, Weimer PJ, Stevenson DM, Aylward FO, Boyum J, et al. The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PloS one. 2011;6:e18814. doi: 10.1371/journal.pone.0018814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mansfield LS, Gauthier DT, Abner SR, Jones KM, Wilder SR, et al. Enhancement of disease and pathology by synergy of Trichuris suis and Campylobacter jejuni in the colon of immunologically naive swine. The American journal of tropical medicine and hygiene. 2003;68:70–80. [PubMed] [Google Scholar]
- 25.Shin JL, Gardiner GW, Deitel W, Kandel G. Does whipworm increase the pathogenicity of Campylobacter jejuni? A clinical correlate of an experimental observation. Canadian journal of gastroenterology = Journal canadien de gastroenterologie. 2004;18:175–177. doi: 10.1155/2004/298064. [DOI] [PubMed] [Google Scholar]
- 26.Parthasarathy G, Mansfield LS. Recombinant interleukin-4 enhances Campylobacter jejuni invasion of intestinal pig epithelial cells (IPEC-1). Microbial pathogenesis. 2009;47:38–46. doi: 10.1016/j.micpath.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 27.Zheng J, Meng J, Zhao S, Singh R, Song W. Campylobacter-induced interleukin-8 secretion in polarized human intestinal epithelial cells requires Campylobacter-secreted cytolethal distending toxin- and Toll-like receptor-mediated activation of NF-kappaB. Infection and immunity. 2008;76:4498–4508. doi: 10.1128/IAI.01317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kringel H, Iburg T, Dawson H, Aasted B, Roepstorff A. A time course study of immunological responses in Trichuris suis infected pigs demonstrates induction of a local type 2 response associated with worm burden. International journal for parasitology. 2006;36:915–924. doi: 10.1016/j.ijpara.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 29.Mansfield LS, Gauthier DT. Lymphoglandular complexes are important colonic sites for immunoglobulin A induction against Campylobacter jejuni in a swine disease model. Comparative medicine. 2004;54:514–523. [PubMed] [Google Scholar]
- 30.Wilson MS, Ramalingam TR, Rivollier A, Shenderov K, Mentink-Kane MM, et al. Colitis and intestinal inflammation in IL10-/- mice results from IL-13Ralpha2-mediated attenuation of IL-13 activity. Gastroenterology. 2011;140:254–264. doi: 10.1053/j.gastro.2010.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anthony RM, Urban JF AlemF, Jr, Hamed HA, Rozo CT, et al. Memory T(H)2 cells induce alternatively activated macrophages to mediate protection against nematode parasites. Nature medicine. 2006;12:955–960. doi: 10.1038/nm1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao A, Urban JF, Anthony RM, Sun R, Stiltz J, et al. Th2 cytokine-induced alterations in intestinal smooth muscle function depend on alternatively activated macrophages. Gastroenterology 135: 217–225. 2008;e211 doi: 10.1053/j.gastro.2008.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hasnain SZ, Evans CM, Roy M, Gallagher AL, Kindrachuk KN, et al. Muc5ac: a critical component mediating the rejection of enteric nematodes. The Journal of experimental medicine. 2011;208:893–900. doi: 10.1084/jem.20102057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Artis D, Wang ML, Keilbaugh SA, He W, Brenes M, et al. RELMbeta/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13596–13600. doi: 10.1073/pnas.0404034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nair MG, Guild KJ, Du Y, Zaph C, Yancopoulos GD, et al. Goblet cell-derived resistin-like molecule beta augments CD4+ T cell production of IFN-gamma and infection-induced intestinal inflammation. Journal of immunology. 2008;181:4709–4715. doi: 10.4049/jimmunol.181.7.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khodoun M, Strait R, Orekov T, Hogan S, Karasuyama H, et al. Peanuts can contribute to anaphylactic shock by activating complement. The Journal of allergy and clinical immunology. 2009;123:342–351. doi: 10.1016/j.jaci.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cliffe LJ, Humphreys NE, Lane TE, Potten CS, Booth C, et al. Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science. 2005;308:1463–1465. doi: 10.1126/science.1108661. [DOI] [PubMed] [Google Scholar]
- 38.Zaiss DM, Yang L, Shah PR, Kobie JJ, Urban JF, et al. Amphiregulin, a TH2 cytokine enhancing resistance to nematodes. Science. 2006;314:1746. doi: 10.1126/science.1133715. [DOI] [PubMed] [Google Scholar]
- 39.Morimoto M, Zhao A, Madden KB, Dawson H, Finkelman FD, et al. Functional importance of regional differences in localized gene expression of receptors for IL-13 in murine gut. Journal of immunology. 2006;176:491–495. doi: 10.4049/jimmunol.176.1.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dawson HD, Beshah E, Nishi S, Solano-Aguilar G, Morimoto M, et al. Localized multigene expression patterns support an evolving Th1/Th2-like paradigm in response to infections with Toxoplasma gondii and Ascaris suum. Infection and immunity. 2005;73:1116–1128. doi: 10.1128/IAI.73.2.1116-1128.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (N Y) 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 42.Li RW, Connor EE, Li C, Baldwin Vi RL, Sparks ME. Characterization of the rumen microbiota of pre-ruminant calves using metagenomic tools. Environmental microbiology. 2012;14:129–139. doi: 10.1111/j.1462-2920.2011.02543.x. [DOI] [PubMed] [Google Scholar]
- 43.Cox MP, Peterson DA, Biggs PJ. SolexaQA: At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11 doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS computational biology. 2009;5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Glass EM, Wilkening J, Wilke A, Antonopoulos D, Meyer F. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harbor protocols 2010: pdb. 2010;prot5368 doi: 10.1101/pdb.prot5368. [DOI] [PubMed] [Google Scholar]
- 47.Rho M, Tang H, Ye Y. FragGeneScan: predicting genes in short and error-prone reads. acids research. 2010;38:e191. doi: 10.1093/nar/gkq747. [DOI] [PMC free article] [PubMed] [Google Scholar]