Abstract
High-performance metabolic profiling (HPMP) by Fourier-transform mass spectrometry coupled to liquid chromatography gives relative quantification of thousands of chemicals in biologic samples but has had little development for use in toxicology research. In principle, the approach could be useful to detect complex metabolic response patterns to toxicologic exposures and to detect unusual abundances or patterns of potentially toxic chemicals. As an initial study to develop these possible uses, we applied HPMP and bioinformatics analysis to plasma of humans, rhesus macaques, marmosets, pigs, sheep, rats and mice to determine: 1) whether more chemicals are detected in humans living in a less controlled environment than captive species, and 2) whether a subset of plasma chemicals with similar inter-species and intra-species variation could be identified for use in comparative toxicology. Results show that the number of chemicals detected was similar in humans (3221) and other species (range 2537 to 3373). Metabolite patterns were most similar within species and separated samples according to family and order. A total of 1485 chemicals were common to all species; 37% of these matched chemicals in human metabolomic databases and included chemicals in 137 out of 146 human metabolic pathways. Probability-based modularity clustering separated 644 chemicals, including many endogenous metabolites, with inter-species variation similar to intra-species variation. The remaining chemicals had greater inter-species variation and included environmental chemicals as well as GSH and methionine. Together, the data suggest that HPMP provides a platform that can be useful within human populations and controlled animal studies to simultaneously evaluate environmental exposures and biological responses to such exposures.
Keywords: metabolomics, plasma, mass spectrometry, probability-based modularity clustering, exposome
1. Introduction
A recent biomonitoring survey of Americans by the Centers for Disease Control and Prevention (CDC) found traces of 212 environmental chemicals, including chemicals from plastics, pesticides and flame retardants (CDC 2011). Such exposures appear to mostly reflect common commercial uses, do not exceed relevant safety thresholds and have little if any evidence for associated adverse health effects. Furthermore, the chemicals in the CDC study were selected because of postulated high health concerns (e.g., bioaccumulative/persistent, plasticizers, and chemicals with ongoing exposure) so that the findings suggest that chemicals with short environmental or biological half-lives are likely to have even lower internal-dose exposure profiles and even lower potential for adverse health outcomes.
Despite the extensive scientific foundation of the methods and the re-assuring nature of such findings, contemporary risk assessment and surveillance strategies were devised as an affordable approach to minimize population risks and have several limitations. For instance, detection of an exposure of concern is possible only if it occurs at a frequency sufficient to be detected within the number of samples measured; if 20,000 people are studied in the US and they are representatively distributed among the 50 states and across the age and sex distributions of the population, then significant exposures to a single chemical may have to affect >1% of a relevant subpopulation, such as middle-age men in a single geographical region, e.g. the Mississippi Delta, to be detected. Second, because of cost limitations, chemicals are prioritized for targeted analysis based upon evidence of hazard and potential risk. Unknown or unidentified risks are excluded by this targeted approach. For instance, effects of low-level (0.001%) contaminants of commercial chemicals may not be readily detected in toxicology studies but be environmentally relevant if they are stable and substantial amounts of the agent are produced. Similarly, chemicals with a short environmental half-life are converted to other chemicals; it is very difficult to know whether these include minor but persistent products that bioaccumulate. Third, the selection of chemicals for analysis depends upon in vitro toxicity models, animal testing and extrapolation. While sound in principle, this approach cannot include the full range of possible adverse health effects due to complex genetic, epigenetic and co-exposure interactions in humans. Fourth, the approach assumes that individuals have similar sensitivities to environmental exposures, i.e., that inclusion of a safety factor is adequate to account for deviation of individuals from the norm. Knowledge from therapeutic drug use shows that even an extensively consumed drug like acetaminophen, used safely by hundreds of millions of people, causes toxicity at therapeutic doses in some individuals (Bonkovsky et al. 1994; Kwan et al. 1995). Thus, even though environmental health risk assessment and surveillance are effective in providing cost-effective means to minimize risk, there are continuing needs to improve this process.
New chemical analysis and profiling technologies can potentially provide cost-effective approaches to: a) more thoroughly evaluate biological responses of cell and animal studies for extrapolation to humans; b) more broadly monitor exposures on a personal basis; and c) more effectively detect adverse exposure effects within the context of other pathophysiologic processes under real-life conditions. Such technologies and innovative applications include advanced biosensors (Rea et al. 2011), microfluidics (Kraly et al. 2009), ultra-high resolution chemical separations (Nordstrom et al. 2006), and improved sensitivity mass spectrometry (Soltow et al. 2011), nuclear magnetic resonance spectrometry (Inouye et al. 2010) and high-resolution imaging (Miura et al. 2010). Development of such approaches has the promise to provide more comprehensive, rapid and less expensive methods to identify existing and newly emerging exposures of concern (Want et al. 2010).
High-performance metabolic profiling (HPMP) is a high-throughput chemical analysis developed as a practical approach for use in personalized medicine (Johnson et al. 2010). The method uses the high resolution and mass accuracy of Fourier-transform mass spectrometry (Marshall and Hendrickson 2008) to support measurement of up to 7000 chemicals in 20 µl samples in 20 min (Soltow et al. 2011), thereby making this method potentially affordable for routine measurement of endogenous metabolites and metabolic patterns for disease diagnosis and health management. In application of this technology to biological samples, there is an ambiguity in terminology because “metabolomics” is used as a general term for all chemicals, yet all chemicals in biological samples are not biologically relevant “metabolites”. In the present report, we use “chemical” in a non-specific way to refer to any chemical detected. HPMP was developed as a high-throughput approach to study metabolism and disease, and we use the term “endogenous metabolite” in the present study to refer to these biochemicals, as represented by KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa 2002; Kanehisa and Goto 2000) human metabolic pathways. Other chemicals include those derived from diet, enteric flora, pharmaceuticals and environmental sources, many of which are currently unidentified. The present study is based upon the observation that HPMP analyses include environmental chemicals, e.g., insecticides, fungicides and plasticizers, raising the possibility that HPMP could be adopted as part of a universal exposure surveillance strategy for health and environmental exposures (Soltow et al. 2011). In principle, combinations of chemical separation and ionization strategies could provide an approach to survey the environmental chemical space (Howard and Muir 2011) measured as the abundance or patterns of chemicals in routine blood or urine samples from individuals during normal healthcare visits (Soltow et al. 2011; Weis et al. 2005).
The current study was designed to gain information about potential use of HPMP in exposure surveillance through study of plasma from seven mammalian species (human, rhesus macaque, common marmoset, pig, sheep, rat, mouse). We performed HPMP on plasma and applied bioinformatic approaches to determine the fraction of plasma chemicals that are common among the mammalian species, characterize these in terms of matches to metabolic databases and identify ones with low inter-species variation that could be suitable to support biological response monitoring along with environmental chemical surveillance.
2. Materials and Methods
2.1. Materials
Acetonitrile (HPLC grade), formic acid (puriss. p.a. 98%), water (HPLC grade) and caffeine were obtained from Sigma–Aldrich (St. Louis) and the reverse phase test mix (Cat#: 47641-U) was from Supelco Analytical (Bellefonte, PA). Trimethyl-[13C3]-caffeine (Cat#: CLM-514-0) and [15N]-L-tyrosine (Cat#: NLM-590-0) were obtained from Cambridge Isotope Laboratories, Inc (Andover, PA).
2.2. Plasma Samples
For the purpose of characterization of species and/or sex on phylogenetic differences in metabolic pathways, 2 sample sets (Sample Set 1 and 2) were analyzed at different times. In Sample Set 1, archival samples from seven mammalian species were studied (Supplemental Material, Table 1). Samples from animal species were obtained under standard experimental conditions, respectively differing in diet and environment according to husbandry practices for the different species (Supplemental Material, Table 1). Samples were stored at −80° C until analysis, in all cases for <2 y; at present there is no systematic knowledge concerning the long-term stability of the thousands of chemicals in the samples. Sample Set 2 (Supplemental Material, Table 1) was designed to determine whether metabolic differences according to sex could be discriminated from phylogenetic differences by HPMP.
2.3. High-Performance Metabolic Profiling (HPMP)
Details have been previously published (Johnson et al. 2010; Soltow et al. 2011). Briefly, plasma (50 µl) was treated with acetonitrile (2:1) and an internal standard mix (Soltow et al. 2011) and then centrifuged at 13,000 × g for 5 min to remove protein. Extracts were placed in a refrigerated autosampler and 10 µl volumes were analyzed in duplicate with DC-FTMS platforms, one using an anion exchange (AE) column (Hamilton PRPX-110S, 2.1 mm ×10 cm, Reno) and the other using a C18 column (17 mm × 2.1 mm, Pompton Plains), both with aC18 precolumn (Higgins Analytical Targa column, Mtn View). Samples were fractionated with a formate gradient, ionized with electrospray ionization in the positive mode, and detected with an LTQ-FT spectrometer (Thermo, San Jose) with m/z from 85–850. Data were extracted using apLCMS (Yu et al. 2009) as m/z features, where an m/z feature is defined by m/z (mass/charge), RT (retention time) and ion intensity (integrated ion intensity for the peak). Endogenous metabolites were annotated using Madison Metabolomics Consortium Database (MMCD) (Markley et al. 2007), Metlin Mass Spectrometry Database (Smith et al. 2005) and MS/MS.
2.4. Bioinformatics
Hierarchical Clustering Analysis (HCA) (Pirouette software; Infometrix, Bothell, WA) was used as a data reduction method to visualize phylogenic similarities of metabolic spectra among the 7 species. Principal Component Analysis (PCA) (Pirouette software; Infometrix, Bothell, WA) with autoscaling was used as a data reduction method to visualize discriminatory factors, shown in two-dimensional (2-D) and three-dimensional (3-D) PCA score plots. Metabolite pathway mapping was performed using KEGG (Kanehisa 2002; Kanehisa and Goto 2000) to match high-resolution m/z data to known endogenous metabolites and metabolic pathways. The large number of m/z features (>3000), most of which did not match endogenous metabolites in the metabolic databases, precluded confirmation of chemical identities of most features. However, over 90% of metabolites in the KEGG human metabolic database have unique m/z at the resolution used, and MS/MS and coelution studies confirmed identity of many features (e.g., amino acids, intermediary metabolites, environmental chemicals). One-way ANOVA and Tukey test were used to compare the relative intensities of chemicals in human plasma to those of 6 different species. Significance was expressed at p <0.05.
2.5. Probability-based modularity clustering
Probability-based modularity clustering was used to determine associations of chemicals within species and between species. This is a graph-based technique for automated clustering that provides numerical optimization across all possible clusterings. The method is based upon modularity clustering (Newman 2006), an approach which maximizes the "modularity function", a direct measure of the quality of a particular clustering of nodes in a graph. A detailed discussion of maximization and formulation of modularity function is available (Stone and Ayroles 2009). Briefly, in an undirected graph with nodes and edges, modularity clustering determines the community structure of by constructing adjacency between two nodes and, to define the closeness of nodes. A modularity function is defined
| (1) |
where is a partition of the nodes into groups and. Thus, measures the within-cluster sum of adjacency, measures the sum of adjacency over all edges attached to nodes in cluster, and measures the sum of all edge adjacency in the graph. The term / relates to the empirical probability that both ends of a randomly selected edge lie in cluster, and the term relates to the empirical probability that one specific end of a randomly selected edge lies in cluster. Hence, the modularity function of partition is the difference between the partition's empirical within edge probability and the partition's hypothetical randomly-placed edge probability. Stone and Ayroles (Stone and Ayroles 2009) used this correlation-based adjacency measure to cluster genes and, based upon this, we used a probability-based adjacency measure to cluster chemicals for phylogenetic groups. We used the adjacency between chemicals to measure contribution of individual chemicals to similarity among groups. Specifically, for (human, non-human primate, rodentia, artiodactyla), is the -th group effect. The adjacency between chemicals and, denoted by, is defined as the p-value of the test:
With this adjacency measure, high p-values lead to failure to reject the null hypothesis, which indicates that high p-values relate to the conclusion of the four species being similar. Adjacency close to 0 leads to rejection of the null hypothesis, meaning that metabolite and result in quite dissimilar, or divergent, effects of the relative intensities among species. By maximizing the modularity function as in equation (1), the adjacency constructed as above produces a partition of clusters that separates chemicals that are non-differentiating (have the highest average adjacency among groups) from those that are differentiating (have the lowest average adjacency among groups). A detailed discussion of iterative and automatic maximization of involving no cut-off value is available (Newman 2006). Within each cluster, chemicals are ordered according to adjacency within the cluster, with the upper left chemicals showing best associations among groups and the lower right chemicals showing greatest divergence among groups. For this analysis, chemicals with >30% zero values were excluded, leaving 1418 of the common chemicals for reported results. Results were similar using for all species individually, but the current analysis was used because of the small n for some species.
3. Results
3.1. HPMP of different species
A complete table of results for m/z features for individuals is available from the authors upon request. Results for each order, family and species are summarized in Figure 1A and B. Each order had a similar total number of chemicals ranging from 3382 to 3723. Species from two families, pig and sheep, had relatively fewer features, but the number of features/individual was similar so this may reflect the smaller number of individuals studied. Of interest, the number of chemicals detected in human plasma was similar to that in the experimental animals even though the human subjects were not in a controlled research environment and expected to have more variable diet and environmental exposure.
Figure 1.
A. Phylogenetic relationship of 7 mammalian species with associated number of chemicals detected by HPMP for each order, family and species. B. Venn diagram of the chemicals common among families. C. Hierarchical clustering analysis (HCA) of plasma metabolic profiles after variance scaling shows similarity of individuals of the same species.
The distribution of features among the orders is shown as a Venn diagram in Figure 1B. Of the total 3820 m/z features, less than half (1485) was present in all species (Supplemental Material, Table 2). A greater number of common features was found between primates and rodents than between artiodactyls and either primates or rodents.
3.2. Similarities of metabolic profiles using HCA
HCA was performed on the data for the 3820 chemicals in individual plasma samples to characterize similarity of metabolic spectra. The results (Figure 1C) showed that individuals within a species were more similar to other individuals within that species than to individuals from other species. However, species generally were not more similar within family than between family, e.g., humans and rhesus were more similar to pig and sheep than to marmoset. Thus, the similarities in metabolic character of plasma reflected in HCA only partially recapitulate phylogeny.
3.3. PCA of HPMP data for plasma from 7 mammalian species
Data reduction by PCA showed better classification according to taxonomic rank. A 3-D PCA score plot (Figure 2A), rotated to visualize maximal separation, showed that the 7 mammalian species separated by phyogenetic order (primata, actiodactyla, rodentia) using PC1, 2 and 3, which represents 63% of total variation. In Figure 2B, a 2-D PCA score plot representing 56% of total variation showed nearly complete separation according to species. This sample set did not have equal distribution according to sex, so a second set of samples was examined to determine whether individuals were separated in a PCA score plot according to sex. This set of samples contained 5 females and 5 males from human, marmoset and mouse. The 2-D PCA score plot (Figure 2C) showed that separation was largely due to species and not to sex. According to the results, the first three principal components of a PCA analysis of 3820 chemicals separates according to species with additional separation according taxonomic family and order but not according to sex.
Figure 2.
A. Three-dimensional PCA score plot, in which the first three principal components (PCs) explained 62% of total variation, is arbitrarily rotated to visualize discrimination according to species, family and order. B. Two-dimensional PCA score plot with PC1 and PC2 corresponding to Panel D. C. 2-D PCA score plot for Sample Set 2 to evaluate whether samples from different species (human, marmoset and mouse) were discriminated according to sex. In A and B, each symbol shows a different class: diamond, primate (human, H, filled; rhesus, Rh, open; marmoset, Ma, with line); circle, artiodactyla (pig, P, filled; sheep, S, open); triangle, rotentia (rat, Ra, filled; mouse, Mo, open). In C, males, filled symbols; females open symbols; human, diamond; marmoset, square; mouse, triangle.
3.4. Pathway analysis of 1485 chemicals common to 7 mammalian species
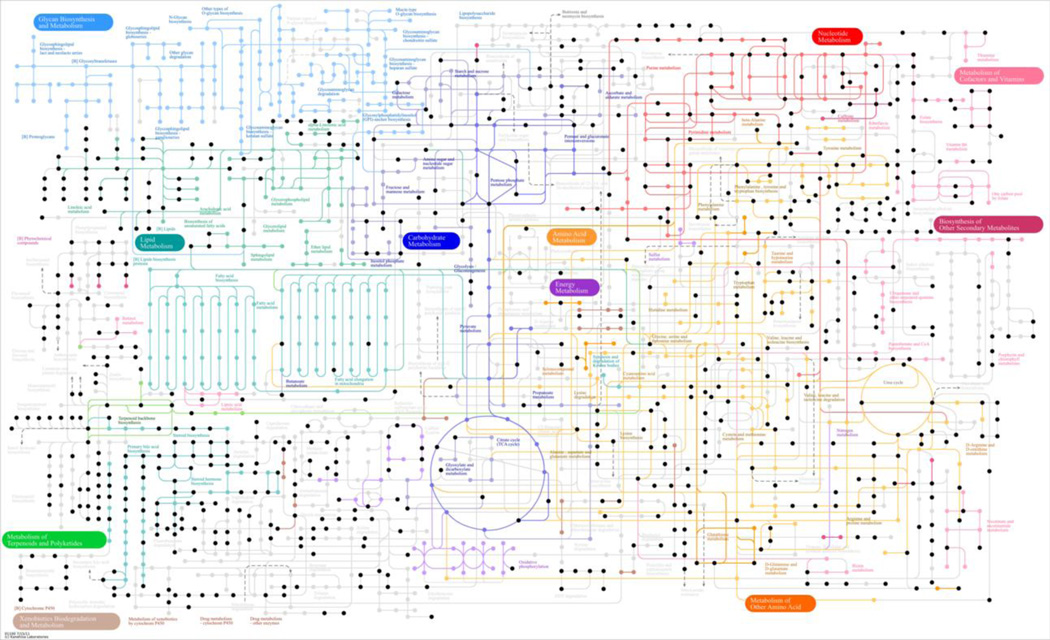
Metabolic databases provide the capability to search for chemicals that match the high-accuracy m/z data. Searches against Metlin (Smith et al. 2005) and MMCD (Madison Metabolomics Consortium Database; (Markley et al. 2007) showed that less than half of the m/z match known human endogenous metabolites (Draper et al. 2009; Soltow et al. 2011). KEGG pathway analysis similarly showed that only 673 of the m/z matched known metabolites (Figure 3). Although these endogenous metabolites represent only about 20% of the total chemicals detected, they included metabolites in 137 pathways of the total 146 pathways in the KEGG human reference metabolic pathways. Examination of some selected chemicals (alanine, threonine, tyrosine, methionine, GSH, pirimicarb) showed significant differences in signal intensities between species (Supplemental Material, Figure 1). Thus, the results suggest that HPMP could provide a useful approach for comparative studies evaluating endogenous metabolites and other common chemicals.
Figure 3.
KEGG metabolic pathway matches for m/z features that are common to mammalian species. High mass accuracy m/z for 1485 common chemicals revealed 666 matches as shown in black, including matches for metabolites in 137 of the total 146 KEGG human reference metabolic pathways. Corresponding metabolite names are available at http://www.genome.jp/kegg-bin/show_pathway?13154256908831/hsa01100.args.
3.5. Probability-Based Modularity Clustering of 1485 chemicals common to 7 mammalian species
We used probability-based modularity clustering as a means to analyze chemicals according to the significance of their association with other chemicals (Newman 2006). By setting criteria to classify chemicals according to significance of within-species correlations and significance of between-species correlations, a distribution was established in which modules of chemicals that have similar variation among species (644 chemicals, Module 1, Figure 4) are discriminated from chemicals that have greater variation among species than within species (774 chemicals, Module 2, Figure 4). In each module, the heatmap provides a visualization of the p-value of correlations between chemicals so that chemicals are classified for how well they fit the classification; additional graphical discrimination between chemicals in Modules 1 and 2 is provided in Figure 5. Module 1 was enriched in endogenous metabolites, such as leucine/isoleucine, citrulline, cystine and other amino acids and metabolites for which we have previously confirmed identities by co-elution and MS/MS criteria (Johnson et al. 2010; Soltow et al. 2011). Module 2 was enriched in m/z matching environmental chemicals, such as pirimicarb and di-N-butyl phthalate, and also included some endogenous metabolites (GSH, methionine and glutamine). We have previously confirmed identities of the latter endogenous metabolites (Johnson et al. 2010; Soltow et al. 2011). To confirm identity of some of the presumed environmental chemicals, co-elution and MS/MS studies were performed on selected m/z that matched pirimicarb, triethyl phosphate, di-N-butyl phthalate and rotenone. Results confirmed these identifications except for rotenone (Supplemental Material, Figure 2). The MS/MS for the m/z corresponding to rotenone had an ion dissociation pattern that matched a minor contaminant present in the commercial rotenone preparation.
Figure 4.
Probability-based modularity clustering of 1485 common chemicals according to intra- and inter-species variation. Module 1 (M1) includes chemicals with similar characteristics in all 7 mammalian species while Module 2 (M2) contains chemicals that have different characteristics among the species. M1 showed a preponderance of endogenous metabolites while M2 showed a preponderance of other chemicals, including known environmental chemicals. These data suggest that HPMP can be used both for surveillance of environmental exposures by focusing on chemicals in M2 and for study of biologic responses to environmental exposures by focusing on chemicals in M1.
Figure 5.
The relationship between inter-species variation and within-error variation for selected chemicals in Module 1 (M1) and 2 (M2) of probability-based modularity clustering analysis shown in Fig 3. Trajectories above the diagonal line, represented by the arrows in A–C, indicate that within-species variation is large relative to interspecies variation. In contrast, trajectories below the diagonal line, represented by arrows in D–F, show that interspecies variation is large relative to within-species variation. This difference between chemicals in Module 1 and Module 2 provides a basis to use HPMP for two purposes, direct evaluation of environmental exposures (Module 2), and biological responses to environmental exposures (Module 1). A: leucine/isoleucine (M1), B: citrulline (M1), C: cystine (M1), D: glutamine (M2), E: pirimicarb (M2), F: triethylphosphate (M2).
4. Discussion
Toxicology testing is the primary approach to hazard identification (Toxicity testing in the 21st century, Natl Acad Sci Press) (Judson et al. 2008; Judson et al. 2009; Judson et al. 2010), and high-throughput in vitro screening capabilities now allow rapid evaluation of a range of adverse effects in cell and molecular systems. Coupling knowledge of such hazards to comparative research in animal models has contributed significantly to understanding of human risks by helping understand mechanisms and identifying potential exposures of concern (Bergen and Mersmann 2005; Soucek and Gut 1992). For example, sequencing of the genomes of many species (Soucek et al. 2003) along with detoxification and toxicokinetic studies (Butt et al. 2010; Soucek and Gut 1992) has facilitated extrapolations of human risks (Peng et al. 2010; Soucek and Gut 1992). There are, however, a number of limitations to this approach, and contemporary comparative biology increasingly relies upon new information-rich technologies and systems biology approaches (Bergen and Mersmann 2005; Roos et al. 2011; Travlos et al. 2011).
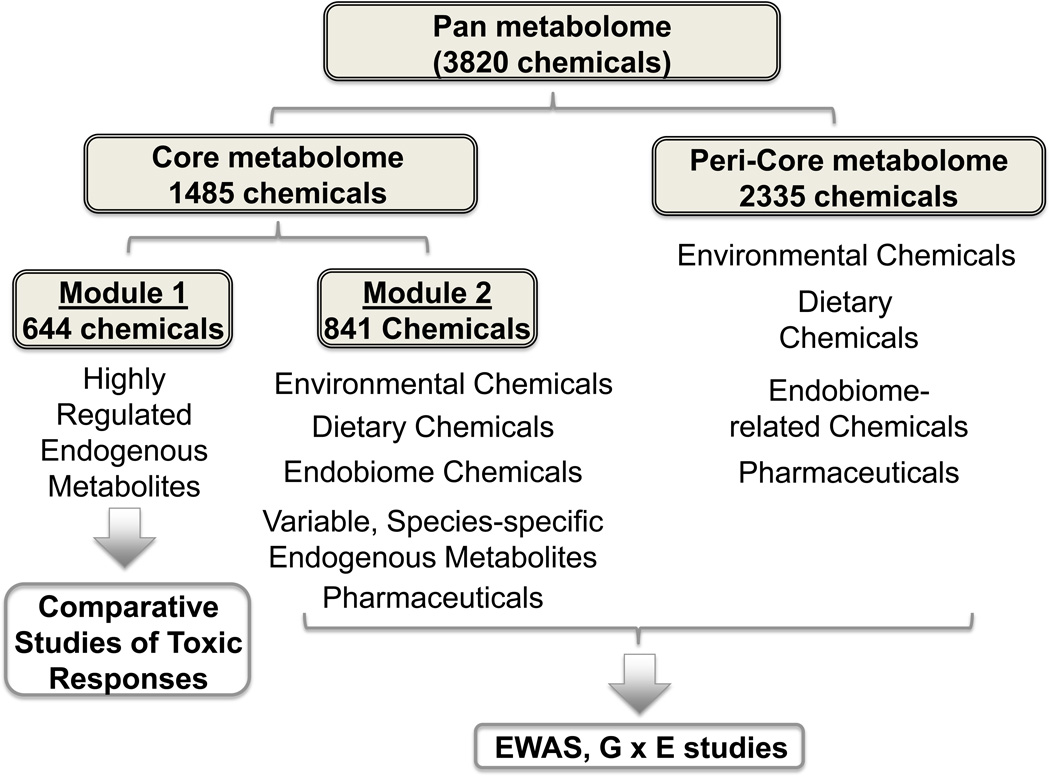
The present research shows that HPMP could potentially be used to improve comparisons between species in comparative toxicology research. The analyses of plasma from 7 species resulted in detection of 3820 total m/z, which can be considered a representation of the “pan” metabolome of mammals (Figure 6). These data confirm previous findings (Draper et al. 2009; Soltow et al. 2011) that humans and other mammalian species have thousands of chemicals in plasma, with over half not represented by chemicals in human metabolic databases such as Metlin (Smith et al. 2005) and MMCD (Markley et al. 2007). It should be noted that the full complement of the “pan” metabolome cannot be estimated from this analysis because plasma does not represent other biologic compartments and there is no way to evaluate the fraction of the total metabolome that is detected by the methods used. In the context of the current study, this pan metabolome can be considered to consist of a “core” metabolome of 1485 chemicals that are common to all species and a “peri-core” metabolome (i.e., chemicals surrounding the core metabolome) that includes the other 2335 chemicals (Figure 6). The core metabolome includes 644 chemicals with inter-species variation similar to intra-species variation, indicating that chemicals from this subset are likely to be useful to study toxic responses (Figure 6). Specifically, toxicity can occur as a consequence of exposure to chemicals that are relatively rapidly eliminated from an organism and, as a result, cannot be detected at later times. Comparative toxicity studies of Module 1 chemicals could support identification of biomarkers and/or provide response patterns useful for related epidemiological research of such toxicities in humans.
Figure 6.
Characterization of HPMP data for comparative toxicology and biomonitoring of exposures. HPMP analyses detected 3820 m/z, which reflects the total metabolome, designated here as the “pan” metabolome. The pan metabolome consists of a “core” metabolome that this is common to the 7 mammalian species and a “peri-core” metabolome that includes all of the chemicals surrounding the core metabolome. The core metabolome includes Module 1, consisting of chemicals that have inter-species variation similar to intra-species variation, and Module 2, consisting of chemicals that have greater inter-species variation than intra-species variation. Module 1 includes endogenous metabolites and has characteristics suitable for comparative studies of biologic responses to toxic exposure. Module 2 includes environmental chemicals, variable endogenous metabolites, chemicals derived from the diet, chemicals derived from the microbiome, and pharmaceuticals. Module 2 and peri-core metabolites can be used to support environment-wide association studies (EWAS) and gene-environment (G × E) studies. Identification and classification of chemicals in Module 2 and the peri-core metabolome will greatly facilitate interpretation of EWAS and G × E study results.
An important priority to enhance use of HPMP data involves identification and annotation of the 1485 chemicals to allow subclassification according to endogenous metabolites, dietary chemicals, microbiome-related chemicals, therapeutic chemicals and environmental chemicals (Figure 6). Such identification and classification is labor-intensive but is critical to support environment-wide association studies (EWAS) (Patel et al. 2010) and gene-environment (G × E) associations (Petronis 2010) (Figure 6). Importantly, the m/z data also can be used to support metabolome-wide association studies (MWAS) of health and disease phenotypes and genome-metabolome (G × M) association studies even without identification and classification. In such cases, however, post-hoc identification will still be needed to fully understand mechanisms.
The presence of environmental chemicals in research animals highlights a limitation of low-dose exposure research. Environmental exposures from water and air may not substantially differ in research animals from that to which humans are exposed. Research animals also consume food formulated by agricultural industries that also produce human food, and the animal food is processed using commercial operations sharing many characteristics of human food production. Furthermore, research animals are exposed to plastics and other chemicals to which humans are exposed. Thus, despite the concept that research animals are maintained under highly controlled environmental conditions, the present data show that plasma of research animals contains a number and quantities of environmental chemicals similar to humans and therefore cannot be considered “unexposed”.
Accumulating evidence indicates that genetic factors represent only a fraction of total risk for chronic and age-related diseases (Rappaport 2011; Willett 2002; Willett et al. 2011). The majority of risk is linked to environmental or gene-environment interactions, emphasizing the need for systematic study of the exposome (Faisandier et al. 2011; Rappaport and Smith 2010). A pilot EWAS of type 2 diabetes (Patel et al. 2010) using CDC data on environmental chemicals suggested that environmental factors can be found with effect sizes comparable to loci found by GWAS (Hindorff et al. 2009). Other studies also link low-level environmental exposures to health risks, including cancer (Soutar et al. 2000; Whitrow et al. 2003), respiratory diseases (Whitehead et al. 2011; Wright et al. 2001), endocrine disruption (Reif et al. 2010; Vis et al. 2010), reproductive failure (Bunderson-Schelvan et al. 2011; Pant et al. 2007) and neurodegenerative diseases (Perrone-Capano and Di Porzio 2000; Sherer et al. 2001). While current approaches using analytically rigorous methods to measure subsets of known hazardous chemicals in populations at greatest risk is cost-effective in minimizing population risk to known hazards, such approaches can be inadequate to detect unanticipated chemicals, such as exemplified by the widespread distribution of perfluorooctanoic acid (PFOA), a chemical not recognized to be released into the environment (Frisbee et al. 2010; Steenland et al. 2010). Cumulative effects from exposure to multiple similar chemicals (Boas et al. 2010; Pant et al. 2008), and long-term and multigenerational effects due to genomic and epigenomic mechanisms (Perrone-Capano and Di Porzio 2000; Ziech et al. 2010), also highlight a need to characterize lifelong exposure histories (Faisandier et al. 2011; Rappaport 2011; Ziech et al. 2010). However, surveillance of all possible exposures is effectively impossible because greater than 80,000 agents are registered with EPA for commercial use. Furthermore, more than 2,800 chemicals have annual production volumes exceeding 454 tons/year (Muir and Howard 2006). If each such agent has 10 minor contaminants and each agent and contaminant is converted to 10 products in the biosphere, one would need to devise means to identify and perform toxicity screening for nearly a half-million additional chemicals to appropriately support associated risk assessment and surveillance.
The present data suggest an alternative approach could be developed using HPMP for MWAS studies that complements this risk assessment and surveillance approach and addresses some of its key limitations. The approach would start with analysis of representative samples from a large number of individuals (e.g. 10,000–20,000) for whom associated demographic and health phenotyping information is available. Such analyses could be accomplished within a single reference laboratory with routine, automated analysis using one to four high-resolution mass spectrometers (5000–10,000 samples per year per spectrometer). These samples would be collected, stored and analyzed in duplicate or triplicate, along with pooled reference plasma samples as available from the National Institute of Standards (McGaw et al. 2010), with standard analytic procedures and internal standards (Soltow et al. 2011). The high-resolution mass spectral data would be entered into a cumulative database structure with accurate mass m/z, chromatographic retention time (RT) and intensity information. This data would provide a resource for association studies, such as discovery of associations of chemicals with geographical regions, dietary patterns and phenotypic information present in the dataset. Known chemicals would be annotated in this database. For unknowns, the accurate mass m/z is sufficient to predict elemental composition in many cases, and along with RT, this m/z would provide information for post-hoc identification and validation studies. Associated MS/MS information could facilitate post-hoc identification and validation. Although initially limited by the sample size, extent of associated disease information and knowledge of chemical identities, this reference data would allow any newly designed study, whether from a specific geographical region or selected population, to be conducted within a framework allowing comparison to a more standardized reference population. Importantly, the data would include information on unknown and unidentified chemicals detected in human plasma, providing a basis to investigate exposure distributions by geography and other demographic factors. Addition of data from representative samples on an annual basis would allow detection of exposure trends, even for unidentified chemicals. Inclusion of disease outcome data would allow detailed EWAS and G × E studies, and repeat analyses on the same individuals would allow systematic study of lifelong exposures as conceptualized in the exposome.
5. Conclusions
This study shows that high-performance metabolic profiling (HPMP) detects a spectrum of low-level environmental chemicals in plasma of research animals that is comparable to that which is found in plasma of humans. Of 3820 chemicals detected, 1485 were common to all 7 mammalian species, suggesting a general utility of HPMP for comparative environmental health research. Analysis of the 1485 common chemicals by probability-based clustering identified a subset of 644 that included endogenous metabolites with characteristics useful for comparative studies of biologic responses to toxicologic exposure. The other >2000 chemicals can be used to provide a non-targeted surveillance of chemical exposures for MWAS of health and disease phenotypes and genome-metabolome (G × M) association studies. Together, the data suggest that HPMP provides a platform that can be useful within human populations and controlled animal studies to simultaneously evaluate environmental exposures and biological responses to such exposures.
Highlights.
Six mammal species had low-abundance environmental chemicals similar to humans
Of 3820 chemicals detected, 1485 common metabolites were present in the seven species
Metabolites with low interspecies variability identified for bioeffect monitoring
Analysis detected >2000 chemicals suitable for EWAS and G × E studies
Supplementary Material
Acknowledgements
This research was supported in part by NIH Grants ES016731, ES011195, AG038746, RR025008, HD46501; RR00165, HL083019, HL070892, RR023356
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that they have no competing financial interests.
References
- Bergen WG, Mersmann HJ. Comparative aspects of lipid metabolism: impact on contemporary research and use of animal models. The Journal of nutrition. 2005;135:2499–2502. doi: 10.1093/jn/135.11.2499. [DOI] [PubMed] [Google Scholar]
- Boas M, Frederiksen H, Feldt-Rasmussen U, Skakkebaek NE, Hegedus L, Hilsted L, Juul A, Main KM. Childhood exposure to phthalates: associations with thyroid function, insulin-like growth factor I, and growth. Environ Health Perspect. 2010;118:1458–1464. doi: 10.1289/ehp.0901331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkovsky HL, Kane RE, Jones DP, Galinsky RE, Banner B. Acute hepatic and renal toxicity from low doses of acetaminophen in the absence of alcohol abuse or malnutrition: evidence for increased susceptibility to drug toxicity due to cardiopulmonary and renal insufficiency. Hepatology. 1994;19:1141–1148. [PubMed] [Google Scholar]
- Bunderson-Schelvan M, Pfau JC, Crouch R, Holian A. Nonpulmonary outcomes of asbestos exposure. J Toxicol Environ Health B Crit Rev. 2011;14:122–152. doi: 10.1080/10937404.2011.556048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt CM, Muir DC, Mabury SA. Elucidating the pathways of poly- and perfluorinated acid formation in rainbow trout. Environ Sci Technol. 2010;44:4973–4980. doi: 10.1021/es100702a. [DOI] [PubMed] [Google Scholar]
- CDC. Fourth National Report on Human Exposure to Environmental Chemicals 2011, Updated Tables. National Center for Environmental Health, Centers for Disease Control and Prevention; 2011. [Google Scholar]
- Draper J, Enot DP, Parker D, Beckmann M, Snowdon S, Lin W, Zubair H. Metabolite signal identification in accurate mass metabolomics data with MZedDB, an interactive m/z annotation tool utilising predicted ionisation behaviour 'rules'. BMC Bioinformatics. 2009;10:227. doi: 10.1186/1471-2105-10-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faisandier L, Bonneterre V, De Gaudemaris R, Bicout DJ. Occupational exposome: A network-based approach for characterizing Occupational Health Problems. J Biomed Inform. 2011;44:545–552. doi: 10.1016/j.jbi.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Frisbee SJ, Shankar A, Knox SS, Steenland K, Savitz DA, Fletcher T, Ducatman AM. Perfluorooctanoic acid, perfluorooctanesulfonate, and serum lipids in children and adolescents: results from the C8 Health Project. Arch Pediatr Adolesc Med. 2010;164:860–869. doi: 10.1001/archpediatrics.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard PH, Muir DC. Identifying New Persistent and Bioaccumulative Organics Among Chemicals in Commerce II: Pharmaceuticals. Environ Sci Technol. 2011;45:6938–6946. doi: 10.1021/es201196x. [DOI] [PubMed] [Google Scholar]
- Inouye M, Kettunen J, Soininen P, Silander K, Ripatti S, Kumpula LS, Hamalainen E, Jousilahti P, Kangas AJ, Mannisto S, Savolainen MJ, Jula A, Leiviska J, Palotie A, Salomaa V, Perola M, Ala-Korpela M, Peltonen L. Metabonomic, transcriptomic, and genomic variation of a population cohort. Mol Syst Biol. 2010;6:441. doi: 10.1038/msb.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Yu T, Strobel FH, Jones DP. A practical approach to detect unique metabolic patterns for personalized medicine. Analyst. 2010;135:2864–2870. doi: 10.1039/c0an00333f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, Reif DM, Rotroff DM, Shah I, Richard AM, Dix DJ. In vitro screening of environmental chemicals for targeted testing prioritization: the ToxCast project. Environ Health Perspect. 2010;118:485–492. doi: 10.1289/ehp.0901392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix D, Houck K, Elloumi F, Martin M, Cathey T, Transue TR, Spencer R, Wolf M. ACToR--Aggregated Computational Toxicology Resource. Toxicology and applied pharmacology. 2008;233:7–13. doi: 10.1016/j.taap.2007.12.037. [DOI] [PubMed] [Google Scholar]
- Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, Dellarco V, Henry T, Holderman T, Sayre P, Tan S, Carpenter T, Smith E. The toxicity data landscape for environmental chemicals. Environ Health Perspect. 2009;117:685–695. doi: 10.1289/ehp.0800168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. The KEGG database. Novartis Found Symp. 2002;247:91–101. discussion 101–103, 119–128, 244–152. [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraly JR, Holcomb RE, Guan Q, Henry CS. Review: Microfluidic applications in metabolomics and metabolic profiling. Analytica chimica acta. 2009;653:23–35. doi: 10.1016/j.aca.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan D, Bartle WR, Walker SE. Abnormal serum transaminases following therapeutic doses of acetaminophen in the absence of known risk factors. Digestive diseases and sciences. 1995;40:1951–1955. doi: 10.1007/BF02208663. [DOI] [PubMed] [Google Scholar]
- Markley JL, Anderson ME, Cui Q, Eghbalnia HR, Lewis IA, Hegeman AD, Li J, Schulte CF, Sussman MR, Westler WM, Ulrich EL, Zolnai Z. New bioinformatics resources for metabolomics. Pac Symp Biocomput. 2007:157–168. [PubMed] [Google Scholar]
- Marshall AG, Hendrickson CL. High-resolution mass spectrometers. Annu Rev Anal Chem (Palo Alto Calif) 2008;1:579–599. doi: 10.1146/annurev.anchem.1.031207.112945. [DOI] [PubMed] [Google Scholar]
- Miura D, Fujimura Y, Yamato M, Hyodo F, Utsumi H, Tachibana H, Wariishi H. Ultrahighly sensitive in situ metabolomic imaging for visualizing spatiotemporal metabolic behaviors. Analytical chemistry. 2010;82:9789–9796. doi: 10.1021/ac101998z. [DOI] [PubMed] [Google Scholar]
- Muir DC, Howard PH. Are there other persistent organic pollutants? A challenge for environmental chemists. Environ Sci Technol. 2006;40:7157–7166. doi: 10.1021/es061677a. [DOI] [PubMed] [Google Scholar]
- McGaw EA, Phinney KW, Lowenthal MS. Comparison of orthogonal liquid and gas chromatography-mass spectrometry platforms for the determination of amino acid concentrations in human plasma. J Chromatogr A. 2010;1217:5822–5831. doi: 10.1016/j.chroma.2010.07.025. [DOI] [PubMed] [Google Scholar]
- Newman ME. Modularity and community structure in networks. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom A, O'Maille G, Qin C, Siuzdak G. Nonlinear data alignment for UPLC-MS and HPLC-MS based metabolomics: quantitative analysis of endogenous and exogenous metabolites in human serum. Analytical Chemistry. 2006;78:3289–3295. doi: 10.1021/ac060245f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant N, Kumar R, Mathur N, Srivastava SP, Saxena DK, Gujrati VR. Chlorinated pesticide concentration in semen of fertile and infertile men and correlation with sperm quality. Environ Toxicol Pharmacol. 2007;23:135–139. doi: 10.1016/j.etap.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Pant N, Shukla M, Kumar Patel D, Shukla Y, Mathur N, Kumar Gupta Y, Saxena DK. Correlation of phthalate exposures with semen quality. Toxicology and applied pharmacology. 2008;231:112–116. doi: 10.1016/j.taap.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Patel CJ, Bhattacharya J, Butte AJ. An Environment-Wide Association Study (EWAS) on type 2 diabetes mellitus. PloS one. 2010;5:e10746. doi: 10.1371/journal.pone.0010746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Oo ML, Andersen JK. Synergistic effects of environmental risk factors and gene mutations in Parkinson's disease accelerate age-related neurodegeneration. Journal of neurochemistry. 2010;115:1363–1373. doi: 10.1111/j.1471-4159.2010.07036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone-Capano C, Di Porzio U. Genetic and epigenetic control of midbrain dopaminergic neuron development. Int J Dev Biol. 2000;44:679–687. [PubMed] [Google Scholar]
- Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- Rappaport SM. Implications of the exposome for exposure science. J Expo Sci Environ Epidemiol. 2011;21:5–9. doi: 10.1038/jes.2010.50. [DOI] [PubMed] [Google Scholar]
- Rappaport SM, Smith MT. Epidemiology. Environment and disease risks. Science. 2010;330:460–461. doi: 10.1126/science.1192603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea G, Polticelli F, Antonacci A, Lambreva M, Pastorelli S, Scognamiglio V, Zobnina V, Giardi MT. Computational Biology, Protein Engineering, and Biosensor Technology: a Close Cooperation for Herbicides Monitoring. In: Larramendy ML, Solloneski S, editors. Herbicides, Theory and Applications, InTech. 2011. pp. 93–120. [Google Scholar]
- Reif DM, Martin MT, Tan SW, Houck KA, Judson RS, Richard AM, Knudsen TB, Dix DJ, Kavlock RJ. Endocrine profiling and prioritization of environmental chemicals using ToxCast data. Environ Health Perspect. 2010;118:1714–1720. doi: 10.1289/ehp.1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos R, Andersson PL, Halldin K, Hakansson H, Westerholm E, Hamers T, Hamscher G, Heikkinen P, Korkalainen M, Leslie HA, Niittynen M, Sankari S, Schmitz HJ, van der Ven LT, Viluksela M, Schrenk D. Hepatic effects of a highly purified 2,2',3,4,4',5,5'-heptachlorbiphenyl (PCB 180) in male and female rats. Toxicology. 2011;284:42–53. doi: 10.1016/j.tox.2011.03.013. [DOI] [PubMed] [Google Scholar]
- Sherer TB, Betarbet R, Greenamyre JT. Pesticides and Parkinson's disease. Scientific World Journal. 2001;1:207–208. doi: 10.1100/tsw.2001.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- Soltow QA, Strobel FH, Mansfield KG, Wachtman L, Park Y, Jones DP. High-performance metabolic profiling with dual chromatography-Fourier-transform mass spectrometry (DC-FTMS) for study of the exposome. Metabolomics. 2011 doi: 10.1007/s11306-011-0332-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek P, Gut I. Cytochromes P-450 in rats: structures, functions, properties and relevant human forms. Xenobiotica. 1992;22:83–103. doi: 10.3109/00498259209053106. [DOI] [PubMed] [Google Scholar]
- Soucek P, Gut I, Trneny M, Skovlund E, Grenaker Alnaes G, Kristensen T, Borresen-Dale AL, Kristensen VN. Multiplex single-tube screening for mutations in the Nijmegen Breakage Syndrome (NBS1) gene in Hodgkin's and non-Hodgkin's lymphoma patients of Slavic origin. Eur J Hum Genet. 2003;11:416–419. doi: 10.1038/sj.ejhg.5200972. [DOI] [PubMed] [Google Scholar]
- Soutar CA, Robertson A, Miller BG, Searl A, Bignon J. Epidemiological evidence on the carcinogenicity of silica: factors in scientific judgement. Ann Occup Hyg. 2000;44:3–14. doi: 10.1016/s0003-4878(99)00047-2. [DOI] [PubMed] [Google Scholar]
- Steenland K, Fletcher T, Savitz DA. Epidemiologic evidence on the health effects of perfluorooctanoic acid (PFOA) Environ Health Perspect. 2010;118:1100–1108. doi: 10.1289/ehp.0901827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EA, Ayroles JF. Modulated modularity clustering as an exploratory tool for functional genomic inference. PLoS Genet. 2009;5:e1000479. doi: 10.1371/journal.pgen.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travlos GS, Hard GC, Betz LJ, Kissling GE. Chronic progressive nephropathy in male F344 rats in 90-day toxicity studies: its occurrence and association with renal tubule tumors in subsequent 2-year bioassays. Toxicol Pathol. 2011;39:381–389. doi: 10.1177/0192623310388432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vis DJ, Westerhuis JA, Hoefsloot HC, Pijl H, Roelfsema F, van der Greef J, Smilde AK. Endocrine pulse identification using penalized methods and a minimum set of assumptions. Am J Physiol Endocrinol Metab. 2010;298:E146–E155. doi: 10.1152/ajpendo.00048.2009. [DOI] [PubMed] [Google Scholar]
- Want EJ, Wilson ID, Gika H, Theodoridis G, Plumb RS, Shockcor J, Holmes E, Nicholson JK. Global metabolic profiling procedures for urine using UPLC-MS. Nat Protoc. 2010;5:1005–1018. doi: 10.1038/nprot.2010.50. [DOI] [PubMed] [Google Scholar]
- Weis BK, Balshaw D, Barr JR, Brown D, Ellisman M, Lioy P, Omenn G, Potter JD, Smith MT, Sohn L, Suk WA, Sumner S, Swenberg J, Walt DR, Watkins S, Thompson C, Wilson SH. Personalized exposure assessment: promising approaches for human environmental health research. Environ Health Perspect. 2005;113:840–848. doi: 10.1289/ehp.7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead T, Metayer C, Buffler P, Rappaport SM. Estimating exposures to indoor contaminants using residential dust. J Expo Sci Environ Epidemiol. 2011 doi: 10.1038/jes.2011.11. [DOI] [PubMed] [Google Scholar]
- Whitrow MJ, Smith BJ, Pilotto LS, Pisaniello D, Nitschke M. Environmental exposure to carcinogens causing lung cancer: epidemiological evidence from the medical literature. Respirology. 2003;8:513–521. doi: 10.1046/j.1440-1843.2003.00497.x. [DOI] [PubMed] [Google Scholar]
- Willett WC. Dietary fat plays a major role in obesity: no. Obes Rev. 2002;3:59–68. doi: 10.1046/j.1467-789x.2002.00060.x. [DOI] [PubMed] [Google Scholar]
- Willett WC, Colditz GA, Hiatt RA. Combating environmental causes of cancer. N Engl J Med. 2011;364:2266. doi: 10.1056/NEJMc1103912. author reply 2267–2268. [DOI] [PubMed] [Google Scholar]
- Wright AL, Holberg CJ, Taussig LM, Martinez FD. Factors influencing the relation of infant feeding to asthma and recurrent wheeze in childhood. Thorax. 2001;56:192–197. doi: 10.1136/thorax.56.3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Park Y, Johnson JM, Jones DP. apLCMS--adaptive processing of high-resolution LC/MS data. Bioinformatics. 2009;25:1930–1936. doi: 10.1093/bioinformatics/btp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziech D, Franco R, Pappa A, Malamou-Mitsi V, Georgakila S, Georgakilas AG, Panayiotidis MI. The role of epigenetics in environmental and occupational carcinogenesis. Chem Biol Interact. 2010;188:340–349. doi: 10.1016/j.cbi.2010.06.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.