Abstract
The earthworm Eisenia fetida is one of the most used species in standardized soil ecotoxicity tests. Endpoints such as survival, growth and reproduction are eco-toxicologically relevant but provide little mechanistic insight into toxicity pathways, especially at the molecular level. Here we applied a toxicogenomic approach to investigate the mode of action underlying the reversible neurotoxicity of hexanitrohexaazaisowurtzitane (CL-20), a cyclic nitroamine explosives compound. We developed an E. fetida-specific shotgun microarray targeting 15119 unique E. fetida transcripts. Using this array we profiled gene expression in E. fetida in response to exposure to CL-20. Eighteen earthworms were exposed for 6 days to 0.2 μg/cm2 of CL-20 on filter paper, half of which were allowed to recover in a clean environment for 7 days. Nine vehicle control earthworms were sacrificed at day 6 and 13, separately. Electrophysiological measurements indicated that the conduction velocity of earthworm medial giant nerve fiber decreased significantly after 6-day exposure to CL-20, but was restored after 7 days of recovery. Total RNA was isolated from the four treatment groups including 6-day control, 6-day exposed, 13-day control and 13-day exposed (i.e. 6-day exposure followed by 7-day recovery), and was hybridized to the 15K shot-gun oligo array. Statistical and bioinformatic analyses suggest that CL-20 initiated neurotoxicity by non-competitively blocking the ligand-gated GABAA receptor ion channel, leading to altered expression of genes involved in GABAergic, cholinergic, and Agrin-MuSK pathways. In the recovery phase, expression of affected genes returned to normality, possibly as a result of autophagy and CL-20 dissociation/metabolism. This study provides significant insights into potential mechanisms of CL-20-induced neurotoxicity and the recovery of earthworms from transient neurotoxicity stress.
Introduction
Changes in environmental quality and health can be perceived by organisms living in close contact with the environment. These organisms are often sensitive to xenobiotics introduced to the environment and can serve as sentinel species for the environment. As sentinel species, earthworms have been considered one of the best bioindicators or biomonitors for soil ecosystems owing to their essentially important roles in soil pedogenesis, soil structure, fertility and terrestrial food chain (1). Earthworms have also been used extensively for assessing environmental risk and chemical toxicity in laboratory and field settings (2). Two closely related earthworm families, Eisenia spp. and Lumbricus spp., are commonly used as test organisms in standardized soil-based toxicity tests as well as filter paper contact tests (3). Previously, we developed a cDNA microarray and used it to assess gene expression response in E. fetida exposed to explosive compounds 2,4,6-trinitrotoluene (TNT) and hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) (4–6). In a similar effort, Owen et al. (2008) made a L. rubellus-specific microarray (7). These represent a recent spike in the use of genomic tools to discern toxicological mechanisms and discover novel biomarkers in environmentally relevant species (8–10).
In the present study, we investigated the reversible neurotoxicity of the cyclic nitroamine compound 2,4,6,8,10,12-hexanitro-2,4,6,8,10,12-hexaazaisowurtzitane, commonly known as HNIW or CL-20(CAS# 1358 5-90-4; molecular formula C6H6N12O12). As a relatively new nitroamine energetic compound, CL-20 is superior to conventional high-energy propellants and explosives (11). For instance, CL-20 is estimated 20% more powerful than HMX (octahydro-1,3,5,7-tetranitro-1,3,5,7-tetrazocine) (12). On the other hand, the potential adverse environmental impact of CL-20 is of concern as it has been demonstrated to elicit acute and chronic toxicities in both invertebrates (e.g., earthworms (13;14) and potworms (15;16)) and vertebrates (e.g., Japanese quail (17)). Although CL-20 does not affect the growth of higher plants such as alfalfa (Medicago sativa) and perennial ryegrass (Lolium perenne), it can accumulate in plants and may pose a potential risk of biomagnification across the food chain (18;19).
The motivation of the present study stems from our previous observations that CL-20, taken up through dermal contact (20), induces neurotoxicity in E. fetida (13), as reflected in reduced whole body muscarinic acetylcholine receptor (mAChR) levels and giant nerve fiber conduction velocity (14). Interestingly, earthworms are capable of recovering rapidly from the neurotoxicity once removed from the stressor CL-20 (13;14). To further explore molecular mechanisms underlying the reversible neurotoxicity, we employed a toxicogenomics approach to interrogating transcriptional response to CL-20 exposure and recovery. We hypothesized that the neurochemical and electrophysiological changes would be accompanied by alterations in the expression of genes related to neurotransmission pathways. To test the hypothesis, a species-specific, high density shotgun oligo array was developed from nerve tissues and used to profile the transcriptome in E. fetida. Differentially expressed genes were inferred by statistical analysis and were further validated using quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) assays. Bioinformatics analysis of differentially expressed genes indicated that GABAergic and other neuro transmission pathways were affected by CL-20.
Experimental Procedures
Development of nerve tissue-specific Eisenia fetida microarray
A neurological 15K-oligo earthworm array was developed in a shot-gun fashion prior to this study (see Supplementary Text for detailed descriptions of the development procedure). Briefly, it involved (1) nerve tissue dissection and total RNA extraction; (2) preparation of E. fetida cDNA sample for high throughput next generation sequencing; (3) sequence assembly and oligo array design; and (4) annotation of unique sequences. This array design has been submitted to the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) as platform GPL9416. A total of 15119 non-redundant 60-mer oligo probes, each targeting a unique transcript, were spotted on this array, of which 8688 probed targets have meaningful annotation (see Supplementary Table 1).
Earthworm exposure to CL-20
A continuous earthworm (E. fetida) culture was maintained as previously described (6). Prior to exposure, mature earthworms bearing a visible clitellum picked out from the soil-bedded culture were purged overnight on moistened filter paper. A total of 36 healthy-looking earthworms weighing 350~450 mg were selected and split into two groups, control and treated. Exposures were conducted in 120-mL glass vials lined with filter paper. One mL of CL-20 dissolved in methanol (concentration = 25 mg/L, confirmed by HPLC analysis) or methanol alone (vehicle control) was spread onto the filter paper and allowed to evaporate in a ventilated chemical hood for 24 hr. Two mL of distilled water was added to each vial to moisten the dried filter paper before moving an earthworm into the vial. Earthworms of the treated group received 6-d exposure to CL-20 (nominal concentration 0.2 μg/cm2) on moistened filter paper, whereas those of the control group to methanol-spiked filter paper. After the 6-d exposure, half of the earthworms in each group were sacrificed, while the other half were transferred to new clean vials for a 7-d recovery, resulting in 4 treatment groups (9 worms/group) composed of 6-d control, 6-d treated, 13-d control and 13-d treated (6-d exposure followed by 7-d recovery). Earthworms were not fed throughout the 13-d duration. At the termination (day 6 or 13), earthworms were measured for conduction velocity of the medial giant fiber (MGF) using a non-invasive electrophysiological technique (13;14). Earthworms were then transferred into 6-mL capped cryogenic vials, snap-frozen in liquid nitrogen, and fixed in RNA later®-ICE (4 ml/worm/vial, Ambion, Austin, TX).
Array hybridization
Total RNA was extracted from the RNAlater®-ICE-fixed whole worm body using RNeasy Mini Kits (Qiagen). Six out of nine earthworms per treatment were used to accommodate three 8×15K custom arrays. Sample labeling and microarray processing were performed according to Agilent’s protocol “One-Color Microarray-Based Gene Expression Analysis”(version 1.0). Agilent one-color spike-in RNA mix was diluted 5000-fold and a 5-μL aliquot of the diluted spike-in mix was added to 500 ng of total RNA sample prior to labeling. The labeling reactions were performed using the Agilent Low RNA Input Linear Amplification Kit in the presence of cyanine 3-CTP. The labeled cRNA was hybridized to the custom-designed shot-gun E. fetida 15K-oligo array (one sample/array) at 65°C for 17 hours using Agilent’s Gene Expression Hybridization Kit. After washing, each array was scanned at three different PMT (Photo-Multiplier Tube) gain levels (350, 450 and 510) using a GenePix 4200AL scanner (Molecular Devices, Sunnyvale, CA).
Microarray data analysis
Raw gene expression data were extracted from scanned array images as spot and background signal intensity using GenePix Pro 6.1 (Molecular Devices). A spot was flagged out if its raw median signal intensity was below its background level (median + standard deviation), or if it was saturated (=65000). A gene was flagged out if over 50% of the 24 samples had missing values for this gene. The filtered data was background subtracted and log-transformed. The spike-in RNA mix with known composition and RNA copy numbers was used to construct standard linear regression curves, from which the unknown earthworm RNA concentrations were derived. Then, the log-transformed RNA concentrations were normalized to the median value on the same array. The Class Comparison Between Groups of Arrays Tool as implemented in the BRB-ArrayTools v3.6 package (21) was employed to infer differentially expressed genes using a multivariate permutation test. We performed all 924 possible permutations for each comparison in the multivariate permutation test to provide 80% confidence that the number of false-positive genes was no more than 10. The test statistics used are random variance t-statistics for each gene (22). Although t-statistics were used, the multivariate permutation test is non-parametric and does not require the assumption of Gaussian distributions.
Quantitative reverse transcription PCR (qRT-PCR)
PCR primer sets were designed using ABI PRISM Primer Express v 2.0 (Applied Biosystems, Carlsbad, CA) for a select group of genes identified from analysis of microarray data and functional annotation. β-actin and 18S ribosomal RNA, both unaffected by CL-20, were chosen as the reference genes (4). Oligonucleotide primers were synthesized by Eurofins MWG Operon (Huntsville, AL). The same total RNA samples used for microarray hybridization were first reverse transcribed into cDNA in a 20-μl reaction containing 1 μg total RNA, random primers and SuperScript™ III reverse transcriptase (Invitrogen) following the manufacture’s instruction. Synthesized cDNA was diluted to 10 ng/μl. qRT-PCR was performed on an ABI Sequence Detector 7900 (Carlsbad, CA). Each 20-μl reaction was run in quadruplicate and contained 2 μl of synthesized cDNA templates along with 500 nM primers and 10 μl of 2×S YBR® Green PCR Master Mix (ABI). Cycling parameters were 95°C for 15 min to activate the DNA polymerase, then 45 cycles of 95°C for 15 s and 60°C for 1 min. Melting dissociation curves were performed to verify that single products without primer-dimers were amplified. Raw fluorescence data were exported as clipped files. Starting concentrations of mRNAs and PCR efficiencies for each sample were calculated by an assumption-free linear regression on the Log(fluorescence) per cycle number data using LinRegPCR (23). The mRNA concentration of each selected gene was normalized to the geometric mean of 18S and actin (4;24). The Student’s t test was performed to determine statistical difference between treatments in the initial copy number of amplified RNA genes.
Results
Earthworm toxicity test
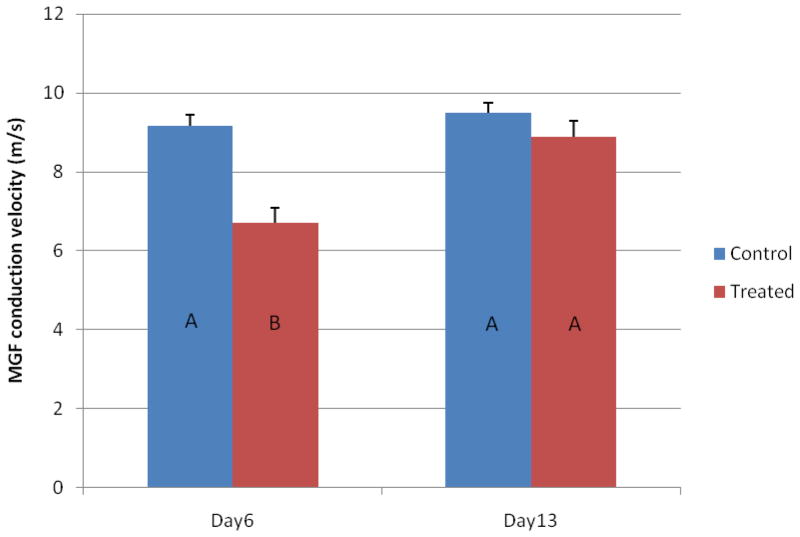
A total of 36 earthworms were used in the filter paper contact toxicity test. Half of the earthworms were exposed to CL-20 (0.2 μg/cm2) for 6 days whereas the other half exposed to the solvent control. Half of the CL-20 treated earthworms were allowed to recover for 7 days. Two mortalities occurred in CL-20 treated worms, one at day 6 and the other at day 13, leaving seven survivors at the end of the test. To define neurotoxicity states, an on-invasive electrophysiological measurement was made of each worm immediately prior to takedown. Statistical analysis of the results (Figure 1) indicates that the MGF conduction velocity in ventral nerve cord was significantly impaired after 6-d of exposure to CL-20 (2-tailed t-test, p = 7.4 × 10−6), but it was restored by the end of the 7-d recovery phase (2-tailed t-test, p = 0.21). These results are in agreement with our previous observations (13;14).
Figure 1.
Electrophysiological measurements of the medial giant fiber (MGF) conduction velocity in the control and CL-20-treated worms at Day 6 and Day 13. Results are shown as mean (column) + standard error (error bar). Sample numbers are as follows: 18 (Day 6 control), 17 (Day 6 treated), 9 (Day 13 control), and 7 (Day 13 treated). Nine worms from the control and the treated groups were sacrificed immediately after the Day 6 measurement and two treated worms died leaving 7 treated worms alive at Day 13. Different letters (“A” and “B”) in the columns indicate statistically significant difference among the four groups (ANOVA, p < 0.001).
Microarray experiment
Total RNA extracted from 6 worms per treatment group was hybridized to the 15K-feature earthworm array with one sample per array. Twenty-four samples were distributed on 3 slides (8 arrays/slide) using a block design to reduce errors associated with variations within and between slides (see Supplementary Figure 1). Each slide was scanned three times, i.e., at PMT 350, 450 and 510. Performance evaluation using the spike-in control RNAs showed that the highest PMT level produced both higher background noise and more saturated spots. The regression curve had the most narrow linearity range (2 log-units) at PMT 510, while 3 log-units at PMT 450 and 3.5 log-units at PMT 350 (see Supplementary Figure 2). Therefore, the dataset acquired at PMT 510 was not used for further statistical analysis. The raw and the normalized data of the 24 arrays scanned at PMT 350 and 450 have been deposited in the GEO database as GSE23948.
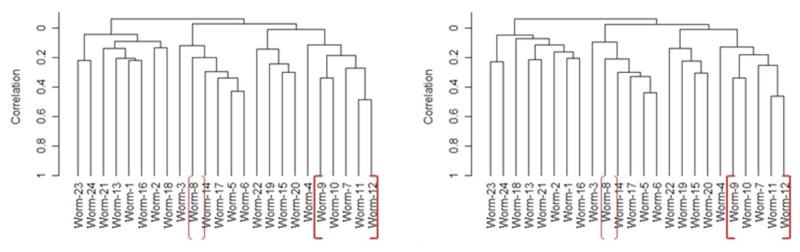
We conducted three two-class comparisons, i.e., 6-d control vs. 6-d treated, 13-d control vs. 13-d treated, and 6-d control vs. 13-d control, using BRB-ArrayTools (see Experimental Procedures for analysis details). We compared the two control treatments in order to look into the effect of 7 more days of starvation on gene expression because the worms were not fed throughout the whole 13-d experiment. Significant genes inferred for the three comparison categories were pooled together, resulting in 223 unique genes for the PMT 350 dataset and 221 unique genes for the PMT 450 dataset. Then, these two sets of significant genes were further combined into a list of 301 non-redundant genes, among which 117 had functional annotation, and 140 were identified from both datasets (Supplementary Table 2). Hierarchical clustering using expression data of these significant genes from the two datasets shows clear separation of the 6-d treated group (except one worm) from the other three groups (Figure 2), reinforcing the electrophysiological results (Figure 1).
Figure 2.
Dendrogram for clustering earthworm samples using array expression data of 301 significant genes. Centered correlation and average linkage were chosen in the clustering algorithm. Treatment: Worm #1 to 6 = 6-d control, #7 to 12 = 6-d treated, #13 to 18 = 13-d control, #19 to 24 = 13-d treated. Left panel: PMT350 dataset; Right panel: PMT450 dataset.
qRT-PCR validation of selected transcripts
We picked 16 transcripts for qRT-PCR validation, 12 of which are putatively related to neurological pathways (Table 1). Two reference genes (18S ribosomal RNA and β-actin) were chosen as normalizers. Specifically, six transcripts putatively code for genes or gene domains involved in the agrin-MuSK (muscle-specific kinase)-rapsyn signaling pathway, three in the cholinergic pathway, two in the GABAergic pathway, and one in the adrenergic pathway. Two transcripts (acetoacetyl-CoA synthetase and N-terminal acetyl transferase) involved in metabolic pathways were also selected for validation. EWContig14 (dopamine β-monooxygenase, also called dopamine β-hydroxylase or DBH) and two fibrinolysis-related transcripts, New Contig1108 (kringle) and EWContig15 (ferritin), were chosen to compare the effects between TNT and CL-20 because our previous study showed that they were significantly affected by TNT (4).
Table 1.
Target transcripts selected for expression validation using qRT-PCR. Shown below are sequences of designed primer sets and annotation of target genes.
| Target transcript | Putative pathway | Putative functional gene/domain | E-value | Forward primer | Reverse primer |
|---|---|---|---|---|---|
| Contig26414 | Agrin-MuSK | IPR012680 Laminin G, subdomain 2 (Agrin) | 4.7E-11 | GGACTGGAACCGACGGATAA | TCACAATGTCGATGGACGTTCT |
| EW1_F1P01_F11 | Agrin-MuSK | IPR002350 Protease inhibitor l1, Kazal (Follistatin/Agrin) | 3.5E-06 | CCGACGCAGATGGAACCTT | GAGGCGTGCAAAACCAACA |
| EW1_F1P05_D12 | Agrin-MuSK | IPR002110 Ankyrin repeat; IPR000082 SEA (Agrin or I-kB protein) | 1.7E-16 | CCAGCTAGACCTCGATACCAAAA | GTTTCTTTCTCCCCAAGCTGAA |
| Contig25790 | Agrin-MuSK | IPR015745 Protein kinase C (Serine/Threonine protein kinase) | 3.1E-14 | AGGACAGCCACCGTTTGATG | ATCGACTCCCTGGACAACGAT |
| Contig09562 | Agrin-MuSK | IPR000024 Frizzled CRD [cysteine rich domain] region (MuSK) | 2.6E-17 | GCAGGTGTAGGGCGGTCTT | AGGGAACTGGGAACATTCGA |
| Contig18180 | Agrin-MuSK | PTHR11210 RING finger; IPR001841 Zinc finger, RING-type (Rapsyn) | 2.2E-50 | AACCCCCCATGCCACAGT | CTGCCGAAACCACATAATGGAT |
| Contig11538 | Cholinergic | PTHR11559:SF33 Acetylcholinesterase (AChE) | 2.5E-04 | CTTCGTCGTGGCCTTTAGAAGT | TGCGGAACTCCCATCCAT |
| Contig24498 | Cholinergic | nAChR, Pfam02932 Neur_chan_memb e | 3.0E-06 | TTCGTAGGATCACGCTCGTCTA | CGCCTTCCTTCGCATCTG |
| NewContig1522 | Cholinergic | nAChR, Pfam02931 Neur_chan_LBD f | 1.9E-08 | TCATGTTCCACTGCAGGATCTC | TCCGCGATTGGTCCAGAA |
| Contig10303 | GABAergic | PTHR10969:SF4 GABAA receptor-associated protein 2 (GABAA receptor) | 5.6E-08 | TCGTTTGCGTATTATCCAGATGAG | ACATTGATGACCTGGACAAAAGAA |
| EW1_F1P09_E05 | GABAergic | PTHR10969:SF4 GABAA receptor-associated protein 2 (GABAA receptor) | 3.7E-16 | CTATAATTACAGGAGCTCGATCAGGAT | AAGTGAGTTTTCTAGATGACGGAGAGA |
| Contig12744 | Mevalonate | PTHR11968:SF40 Acetoacetyl-CoA synthetase-like | 7.4E-23 | GCTGCCAACTGCCAATTCA | ATGACGCTAGGAACGTGTCTTG |
| Contig27065 | Acetylation | PTHR23091 N-terminal acetyltransferase | 2.9E-39 | GCCGATGCCAAGTCTTCTGTA | GCTGTCGTGAGGATCGTAGTGA |
| EWContig14 a | Adrenergic | Dopamine β-hydroxylase (DBH) | 3.0E-17 | GACGAAGGACGCAGATCAGG | TCCTTGTAATAGGCGAGCGG |
| NewContig1108 | Fibrinolysis | IPR000001 Kringle | 7.4E-18 | CGCTCTCGCATTACCAGTGTT | GCGGAGATTCGAGTCTAAGTTCA |
| EWContig15b | Fibrinolysis | Ferritin | 3.0E-13 | ACGTCATGTTCCTCAGACTCTCG | GGAAGACAAGAAACCCCGAAAG |
| Normalizer-1c | Protein synthesis | 18S ribosomal RNA | 0.0E-00 | TTGATTACGTCCCTGCCCTTTG | GGTCCAATCCGAGGATCTCACTA |
| Normalizer-2d | Cell motility, etc. | β-Actin | 1.0E-99 | TTGAAGGTCGTCTCGTGGATAC | CGGTCAGGTCATCACCATCG |
Consensus of 3 Sanger EST sequences, i.e., EW2_R1P11_G03, EW2_F1P03_H02, and EW1_F1P04_A02.
Consensus of 5 ESTs, i.e., EW2_R1P05_C11, EW2_R1P05_G05, EW2_R1P08_B06, EW2_R1P10_C02, and EW2_R1P02_G02.
See GenBank accession no. X79872 for the full sequence.
Consensus of 3 ESTs, i.e., EW1_F1P05_E03, EW2_R1P04_C12, and EW2_R1P06_F12.
Neurotransmitter-gated ion-channel transmembrane region.
Neurotransmitter-gated ion-channel ligand binding domain.
qRT-PCR results are in good agreement with microarray results on all nine genes for which both assays had data available (Table 2). Particularly, qRT-PCR validated the differential expression of seven transcripts identified from microarray data. These seven transcripts were EW1_F1P01_F11 (putatively encoding the Kazaldomain of Agrin), EW1_F1P05_D12 (Agrin, SEA), Contig25790 (Ser/Thr protein kinase), Contig24498 (nAChR, Neur_chan_memb), Contig10303 (GABAA receptor or GABAA-R), Contig12744 (Acetoacetyl-CoA synthetase), and NewContig1108 (kringle). However, there exist some statistical discrepancies between microarray and qRT-PCR results with regard to Contig18180 (Rapsyn, Zinc finger) and NewContig1522 (nAChR, Neur_chan_LBD). According to the microarray data, Contig18180 was significantly up-regulated in 6-d CL-20-treated worms or 13-d control worms if compared to 6-d control worms, and NewContig1522 was significantly down-regulated in 6-d treated worms if compared to 6-d control worms, but qRT-PCR results were unable to support the statistical significance for these comparisons.
Table 2.
Quantitative reverse-transcriptase PCR validation results in comparison with microarray results. Shown below are Student’s t statistics and fold change.
| Target transcript | Functional annotation | qRT-PCR | Microarray | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P (2-tailed t test) | fold change | P (2-tailed t test) | fold change | ||||||||||
| C6 v T6 | C13 v T13 | C6 v C13 | T6/C6 | T13/C13 | C13/C6 | C6 v T6 | C13 v T13 | C6 v C13 | T6/C6 | T13/C13 | C13/C6 | ||
| Contig26414 a | Agrin, Laminin G_2 | 0.719 | 0.715 | 0.372 | 0.95 | 1.06 | 1.12 | ||||||
| EW1_F1P01_F11 b | Agrin, Kazal/Follistatin | 0.004 | 0.122 | 0.166 | 21.8 | 3.84 | 4.64 | 0.008 | 0.270 | 0.241 | 8.88 | 1.86 | 4.95 |
| EW1_F1P05_D12 | Agrin, SEA | 0.030 | 0.975 | 0.028 | 1.43 | 1.01 | 1.50 | 0.005 | 0.986 | 0.029 | 1.75 | 1.00 | 1.88 |
| Contig25790 b | Ser/Thr protein kinase | 0.004 | 0.403 | 0.150 | 1.76 | 0.78 | 1.31 | 0.001 | 0.839 | 0.004 | 1.71 | 1.09 | 1.52 |
| Contig09562 a | MuSK, Frizzled CRD | 0.004 | 0.642 | 0.003 | 1.68 | 1.09 | 2.00 | ||||||
| Contig18180 b | Rapsyn, RING-type Zinc finger | 0.841 | 0.853 | 0.107 | 0.97 | 0.96 | 1.36 | 0.000 | 0.432 | 0.014 | 1.33 | 0.90 | 1.34 |
| Contig11538 a | AChE, Acetylcholinesterase | 0.940 | 0.464 | 0.410 | 0.97 | 0.77 | 1.41 | ||||||
| Contig24498 | nAChR, Neur_chan_memb | 0.002 | 0.659 | 0.001 | 1.51 | 1.05 | 1.63 | 0.186 | 0.012 | 0.009 | 1.53 | 0.96 | 1.69 |
| NewContig1522 | nAChR, Neur_chan_LBD | 0.803 | 0.315 | 0.275 | 0.91 | 0.76 | 1.46 | 0.013 | 0.347 | 0.308 | 0.72 | 1.15 | 1.15 |
| Contig10303 b | GABAA receptor | 0.015 | 0.595 | 0.001 | 1.63 | 1.16 | 1.91 | 0.001 | 0.899 | 0.002 | 3.04 | 1.04 | 2.98 |
| EW1_F1P09_E05 a | GABAA receptor | 0.008 | 0.590 | 0.013 | 1.77 | 1.18 | 1.73 | ||||||
| Contig12744 b | Acetoacetyl-CoA synthetase | 0.003 | 0.224 | 0.695 | 1.85 | 1.42 | 1.07 | 0.002 | 0.064 | 0.514 | 2.12 | 1.70 | 1.10 |
| Contig27065 a | N-terminal acetyltransferase | 0.620 | 0.845 | 0.089 | 1.09 | 1.04 | 1.51 | ||||||
| EWContig14 c | Dopamine β-hydroxylase | 0.168 | 0.632 | 0.819 | 42.5 | 1.44 | 1.12 | ||||||
| NewContig1108 | Kringle | 0.041 | 0.722 | 0.290 | 0.45 | 0.82 | 1.64 | 0.044 | 0.421 | 0.995 | 0.36 | 0.61 | 1.62 |
| EWContig15 c | Ferritin | 0.142 | 0.627 | 0.245 | 170 | 1.19 | 1.41 | ||||||
No array data is presented because no array probe was designed due to the target sequence < the cut-off length of 118 bp, or the designed array probe targeted the antisense strand.
Identified as differentially expressed gene from microarray data using BRB-ArrayTools.
Color coding: yellow = significant at α = 0.01; orange = significant at α = 0.05; red = significantly up-regulated; green = significantly down-regulated. Worm treatment: C6 = Control 6-d; T6 = Treated 6-d; C13 = Control 13-d; T13 = Treated 13-d.
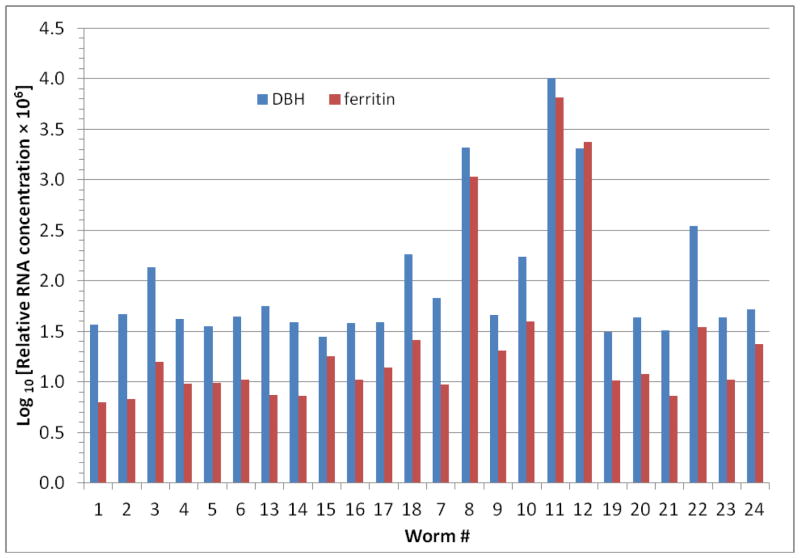
It is worth noting that half of the worms exposed to CL-20 without recovery (worm #8, 11 and 12) had 36~178- and 108~664-fold up-regulation in the expression of DBH and ferritin, separately, if compared to the average expression level in the 6-d control worms (Figure 3), though no statistically significant difference existed between these two treatment groups (Table 2). Like all other 14 selected transcripts, they showed no significant difference in expression measured by both microarray and qRT-PCR between the control and the treated after the 7-d recovery (Table 2). This, along with the far smaller number (~70) of differentially expressed transcripts at day 13 than that (~200) at day 6 (Supplementary Table 2), serves as additional evidence supporting the reversibility of CL-20-induced neurotoxicity. These results are consistent with the electrophysiological results (Figure 1).
Figure 3.
Relative expression of dopamine β-hydroxylase (DBH) and ferritin in all 24 worms measured by qRT-PCR. Gene expression is shown as Log 10 [Relative RNA concentration × 106]. The relative RNA concentration was normalized to the geometric mean of 18S and actin. Worm treatment: #1~6 = 6-d control; #13~18 = 13-d control; #7~12 = 6-d treated; #19~24 = 13-d treated.
Discussion
Toxic damage to the nervous system occurs by three basic mechanisms: (1) direct damage and death of neurons and glial cells; (2) interference with electrical transmission; and (3) interference with chemical neurotransmission (25). The latter two mechanisms may cause temporary neurological impairment or neurocognitive deficits and are often reversible when the triggering neurotoxicant is metabolized or removed from the body. In this study, the electrophysiological endpoint was measured non-invasively by applying light tactile stimulation to evoke giant nerve fiber spikes that synchronously trigger motoneuron spikes in each segment, and then determining the velocity that the spikes were conducted along the segmental body. The measured conduction velocity is a complex function that can be potentially modified by diverse toxicant actions on membrane excitability, gap junction properties, or synaptic function (26).
Reversible neurotoxicity has been observed for a wide variety of chemicals in invertebrates (26–29). For instance, Kyriakides et al. (1990) reported that triethyltin at 10−5 M caused a reversible neuronal membrane depolarization in leech neurons by an increase of intracellular free Ca2+ probably via both release from intracellular stores and inhibition of Ca2+ reuptake (29). Cerstiaens et al. (2003) observed reversible effects of kynureniens, endogenous metabolites of tryptophan, on the motor function of adult flesh flies (27). These studies, however, did not pinpoint the exact molecular targets associated with reversible neurotoxicity.
Recently, Kharrat et al. (2008) demonstrated that the marine phycotoxin gymnodinine-A (GYM-A) blocked nicotinic currents in Xenopus skeletal myocytes evoked by constant iontophoretical acetylcholine pulses in a reversible manner and that GYM-A was confirmed by competition binding assays to broadly target muscle-and neuron-type nicotinic AChRs with high affinity (28). RDX, an explosive compound structurally similar to CL-20, was found to bind to the picrotoxin convulsant site in GABAA-R chloride channel, causing a reduction of GABAergic inhibitory transmission in the rat amygdala and eliciting seizures (30), a reversible symptom in humans and animals for which recovery often occurs within a few days or weeks (31). Meanwhile, several microarray gene expression studies on brain tissues of rats, birds and fish all suggest that RDX, at either seizure-inducing or non-seizure-inducing doses, leads to heightened neuronal excitability through glutamate excitoxicity and down-regulation of many other neurotransmission-related genes (32–35). Despite discovery of multiple potential toxicity targets and modes of action, the authors did not go further to propose an eventual scheme for RDX neuro toxicity by connecting gene expression evidence with results from biochemical and electrophysiological assays.
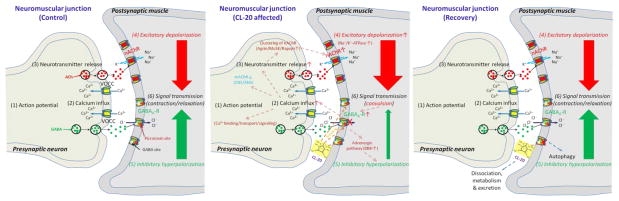
In the present study, we have obtained gene expression evidence that, when integrated with our previous findings and other literature reports, not only identifies CL-20-affected targets, but also sheds some light towards sequential molecular events underlying the reversible neurotoxicity of CL-20 (Figure 4). It is well known that two types of synapses are vulnerable to a wide variety of neurotoxicants: those between neurons and those between a neuron and a muscle cell or gland (e.g., neuromuscular junction or NMJ) (36). Synaptic dysfunction at NMJs, a well-studied neurotoxicity mechanism, is postulated as being responsible for CL-20 induced neurotoxicity, without excluding the role of impairment of synaptic functions between ganglia in the central (CNS) and peripheral (PNS) nervous systems. The latter remains unclear at this moment.
Figure 4.
Postulated mode of action for CL-20-induced reversible neurotoxicity at the dual innervated neuromuscular junction (NMJ) in the earthworm Eisenia fetida. The numbered events (1) to (6) are the basic sequential molecular events occurring at the NMJ upon the arrival of a neuronal impulse (action potential) at the pre-synaptic neuron terminal. Abbreviations: ACh = acetylcholine; GABA = γ-aminobutyric acid; VOCC = voltage-operated calcium (Ca2+) channel; nAChR = nicotinic acetylcholine receptor; GABAA-R = GABA-A receptor; MuSK = muscle-specific kinase; DBH = dopamine β-hydroxylase.
The somatic muscle cells of the earthworm possess dual innervations, an excitatory (depolarizing) one with acetylcholine as the major neurotransmitter, and an inhibitory (hyperpolarizing) one with GABA as the major neurotransmitter (37–39). Similar to RDX (30;40) but unlike many acetylcholinesterase (AChE)-inhibiting organophosphorus pesticides, CL-20-induced neurotoxicity does not appear to involve inhibition of AChE as the expression of an AChE-coding gene (Contig11538 in Table 2) was not affected by CL-20.
Our observation of two putative GABAA-R transcripts being affected by CL-20 suggests that the GABAA-R may be the primary target of CL-20. This hypothesis is supported by at least two lines of evidence obtained in vitro for RDX in the rat amygdala: (1) RDX binds to the picrotoxin convulsant site of the GABAA-R, a Cl− ion channel macromolecular complex, with a significant affinity; and (2) RDX reduces the frequency and amplitude of spontaneous GABAA-R-mediated inhibitory currents and the amplitude of GABA-evoked postsynaptic currents (30). Furthermore, a comparative meta-analysis of RDX impact on gene expression among five phylogenetically disparate species including rat, fathead minnow, earthworm, Northern bobwhite quail and coral shows that all species (even those with no organized nervous system) exhibit toxic effects consistent with a common molecular initiating event, i.e., binding to the GABAA-R or GABAA-like receptor (41). This can be explained by the evolutionary conservation of insect GABA receptors as evidenced by the fact that they share the same allosteric modulatory sites that characterize the mammalian GABAA-Rs (38;42). Unlike their vertebrate counterparts that are restricted to the brain and thus enjoy the protection of the blood-brain barrier, the invertebrate GABA receptors exist in both neuromuscular synapses and within ganglia and hence are much more vulnerable to exogenous agents (38).
Besides the GABAergic synapse, we also observed CL-20 effects on the cholinergic synapse. In the cholinergic system, two opposing pathways on the pre-and post-synaptic sides at the NMJ need to collaborate to induce a high-density accumulation of AChRs:a clustering pathway that works through agrin, MuSK (a transmembrane tyrosine kinase) and rapsyn, and a dispersing pathway that works through acetylcholine and the AChR (43). Pharmacological characterization of AChRs in the somatic earthworm muscle cells has revealed that they cannot be classified as either muscarinic or nicotinic (44) but rather nicotinic-like (39;45). Expression of the channel transmembrane domain of nicotinic AChR (nAChR) gene was significantly up-regulated by CL-20 (Table 2). In addition to nAChR, several genes involved in the Agrin-MuSK pathway (e.g., Agrin, Ser/Thr protein kinase, and MuSK) were also stimulated by CL-20 exposure (Table 2), consistent with our previous observation of agrin up-regulation in RDX-exposed earthworms (5). Expression of these affected genes returned to normality after a 7-d recovery. However, neurotransmitter receptor binding assays have shown that RDX has no affinity to either nicotinic or muscarinic AChRs in rat (30), suggesting that the effects of CL-20 on the cholinergic system may be cascading impact downstream of the initial antagonism to GABAA-R.
As previously demonstrated, the reversible neurotoxicity exerted by sub-lethal concentrations of CL-20 to E. fetida was reflected in the concurrent decrease (during exposure) and recuperation (during recovery) of MGF conduction velocity (13) and total mAChR level (14). Since AChRs at NMJs are exclusively nicotinic and mAChRs are located in the CNS and autonomic nervous system (ANS) (46), it is possible that mAChR down-regulation is caused by GABAA-R antagonism that leads to depolarization-induced changes in K+ current (activation of the K+-channel) in the CNS/ANS, as picrotoxin causes decreases in mAChRs in rat brain (47).
The adrenergic system might also be affected by CL-20 because DBH (converting dopamine to norepinephrine), one of the enzymes that catalyze catecholamine synthesis, was up-regulated several orders of magnitude in 50% of the 6-d treated earthworms (Table 2 and Figure 3). Like GABA, norepinephrine hyperpolarizes the membrane of earthworm somatic cells, possibly by stimulating the Na+/K+ pump via α-adrenergic receptors (48;49). In conjunction with the up-regulation of GABAA-R, the up-regulation of DBH was probably a concerted action to counter the blocking of the GABAA-R chloride ionophore by CL-20 that reduced GABAergic inhibition (30). The “all-or-none” effect pattern for DBH suggests that there might exist an unknown trigger that was activated in half of the treated worms.
Taken together, we propose that CL-20 initiates neurotoxicity by non-competitively blocking GABAA-Rs, leading to altered expression of genes involved in GABAergic, cholinergic, Agrin-MuSK, and adrenergic pathways at NMJs (Figure 4). Similar to RDX, the results presented here suggest that CL-20 may bind to thepicrotoxin site inside the GABAA ionophore, causing a decrease in mean channel open time and Cl− channel permeability, and reducing inhibitory postsynaptic currents (50). The decreased chloride permeability can intensify action potential and stimulate intracellular Ca2+ influx through various voltage-gated Ca2+ channels (51;52) leading to the following molecular events: (1) increased release of neurotransmitter acetylcholine; (2) up-regulation of the nAChR clustering (Agrin-MuSK) pathway; (3) stimulation of nAChR synthesis and membrane depolarization via increased Na+/K+-ATPase activity and Na+ influx (33;39); (4) down-regulation of mAChR in the CNS/ANS (14;47); (5) expression alteration of genes involved in Ca2+ binding, transport and signaling pathways (32;33;35); and (6) up-regulation of GABAA-R and DBH expression (53), perhaps in response to the enhanced excitatory depolarization as a feedback circuit. When exposed earthworms are moved to an uncontaminated environment, CL-20 in worm body could be gradually destructed through (1) autophagic degradation (traffic to autophagosomes) of defective CL-20-bound GABAA-Rs (54); (2) dissociation of CL-20 from the binding site; and (3) metabolism of the dissociated CL-20 by cytochrome P450 or P450-like enzymes (5;32). As a consequence of these and possibly many other processes, CL-20 is excreted from the NMJs, and the effective neurotransmission synapses are restored to their pre-exposure level, eventually resulting in a recovery or reversibility of CL-20-induced neurotoxicity. The aforementioned events are either observed in the present study or reported by others for a variety of species ranging from earthworms and fish to birds and rats. Nevertheless, we recognize that there are still wide knowledge gaps in understanding how these events are exactly connected to each other and what differences exist between different organisms and among structurally similar chemicals, both of which require more in-depth investigations.
A direct comparison can be made between TNT and CL-20 on the basis of ferritin- and DBH-coding gene expression measured in earthworms exposed to each compound. In sharp contrast to CL-20, TNT, a reactive oxygen/nitrogen species producer, significantly inhibited the expression of both genes (4). CL-20-triggered up-regulation of both genes in half of the exposed worms, whereas the non-triggered worms (worm #7, 9 and 10) exhibited expression levels similar to that in control worms (Table 2 and Figure 3). The neurological dysfunction observed in TNT-exposed earthworms is believed to be due to cascading/secondary effects, following the initial response to oxidative stress. On the contrary, CL-20 directly targets neurotransmission systems, causing neurotoxicity and other cascading effects (e.g., oxidative stress through ferritin). Gust et al. reported similar effects of RDX on genes involved in oxidative stress in fathead minnow brain tissue (35).
Starvation effects on cholinergic and GABAergic pathways observed in this study are in agreement with a report where starvation activated MAPK in C. elegans feeding muscle through a muscarinic AChR pathway (55). However, there is currently insufficient evidence, preventing us from postulating a detailed mechanism of action for starvation.
It is widely accepted that a complex neurotransmission and signaling system regulates rhythmic motor activities of the alimentary tract such as contraction and locomotion. In addition to the GABAergic, cholinergic and adrenergic neurotransmissions mentioned above, the peripheral stomatogastric nervous system and neurons innervating the body wall of earthworms host a wide variety of other neurotransmitters such as dopamine, serotonin (5-HT), proctolin, Eisenia-tetradekapeptide, FMRFamide, neuropeptide-Y, histamine and rhodopsin (37;56;57). For instance, dopaminergic sensory cells distributed in the epidermis of earthworms convey sensory information to the ventral nerve cord and activate the dorsal giant fibers and motoneurons controlling rapid reflexes (26;58). Partly owing to the incomplete transcriptome coverage of our shot-gun array, we are just beginning to understand how earthworms respond to and recover from damages caused by neurotoxicants. Further studies are warranted to investigate the role of interactions among different neurotransmission systems in the reversible neurotoxicity.
Supplementary Material
Acknowledgments
Funding Sources
This work was supported by the U.S. Army Environmental Quality/Installation Basic Research Program.
We thank Robert E. Boyd, Willie Brown and B. Lynn Escalon for their technical assistance. Permission was granted by the Chief of Engineers to publish this information.
Footnotes
ASSOCIATED CONTENT
Supporting Information. The following material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Text: Detailed description of the development procedure of an earthworm nerve tissue-specific 15K-oligo array.
Supplementary Table 1: An Excel file that presents annotation information of 30245 assembled contigs, including the BLASTX and InterProScan search results, as well as the number of unique targets with BLASTX and/or IPR hits.
Supplementary Table 2: The comparison categories, t-statistics, fold-change, probe/target identification and sequence, annotation and expression data of 301 statistically significant genes inferred by BRB-ArrayTools (including five duplicated probes/genes on the array).
Supplementary Figure 1. A block design for the hybridization of 24 Eisenia fetida RNA samples (4 treatments × 6 worm replicates) on 24 E. fetida oligo arrays (3 slides × 8 array).
Supplementary Figure 2. The linear regression range of ten spike-in RNAs. The X-axis represents the log-transformed known concentrations of the spike-ins, whereas the Y-axis the log-transformed signal intensity of hybridized control spots on the array. Shown in the figure as an example are the results of array 1 on slide 1.
References
- 1.Edwards CA. The importance of earthworms as key representatives of the soil fauna. In: Edwards CA, editor. Earthworm Ecology. CRC Press; Boca Raton: 2004. pp. 3–11. [Google Scholar]
- 2.Sanchez-Hernandez JC. Earthworm biomarkers in ecological risk assessment. Rev Environ Contam Toxicol. 2006;188:85–126. doi: 10.1007/978-0-387-32964-2_3. [DOI] [PubMed] [Google Scholar]
- 3.Reinecke AJ, Reinecke SA. Earthworms as test organisms in ecotoxicological assessment of toxicant impacts on ecosystems. In: Edwards CA, editor. Earthworm Ecology. CRC Press; Boca Raton: 2004. pp. 299–320. [Google Scholar]
- 4.Gong P, Guan X, Inouye LS, Pirooznia M, Indest KJ, Athow RS, Deng Y, Perkins EJ. Toxicogenomic analysis provides new insights into molecular mechanisms of the sublethal toxicity of 2,4,6-trinitrotoluene in Eisenia fetida. Environ Sci Technol. 2007;41(23):8195–8202. doi: 10.1021/es0716352. [DOI] [PubMed] [Google Scholar]
- 5.Gong P, Guan X, Inouye L, Deng Y, Pirooznia M, Perkins E. Transcriptomic analysis of RDX and TNT interactive sublethal effects in the earthworm Eisenia fetida. BMC Genomics. 2008;9(Suppl 1):S15. doi: 10.1186/1471-2164-9-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pirooznia M, Gong P, Guan X, Inouye LS, Yang K, Perkins EJ, Deng Y. Cloning, analysis and functional annotation of expressed sequence tags from the earthworm Eisenia fetida. BMC Bioinformatics. 2007;8 (Suppl 7):S7. doi: 10.1186/1471-2105-8-S7-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owen J, Hedley BA, Svendsen C, Wren J, Jonker MJ, Hankard PK, Lister LJ, Sturzenbaum SR, Morgan AJ, Spurgeon DJ, Blaxter ML, Kille P. Transcriptome profiling of developmental and xenobiotic responses in a keystone soil animal, the oligochaete annelid Lumbricus rubellus. BMC Genomics. 2008;9:266. doi: 10.1186/1471-2164-9-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iguchi T, Watanabe H, Katsu Y. Application of ecotoxicogenomics for studying endocrine disruption in vertebrates and invertebrates. Environ Health Perspect. 2006;114(Suppl 1):101–105. doi: 10.1289/ehp.8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miracle AL, Ankley GT. Ecotoxicogenomics: linkages between exposure and effects in assessing risks of aquatic contaminants to fish. Reprod Toxicol. 2005;19 (3):321–326. doi: 10.1016/j.reprotox.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Snape JR, Maund SJ, Pickford DB, Hutchinson TH. Ecotoxicogenomics: the challenge of integrating genomics into aquatic and terrestrial ecotoxicology. Aquat Toxicol. 2004;67(2):143–154. doi: 10.1016/j.aquatox.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Badgujar DM, Talawar MB, Asthana SN, Mahulikar PP. Advances in science and technology of modern energetic materials: an overview. J Hazard Mater. 2008;151(2–3):289–305. doi: 10.1016/j.jhazmat.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 12.Keshavarz MH, Motamedoshariati H, Moghayadnia R, Nazari HR, Azarniamehraban J. A new computer code to evaluate detonation performance of high explosives and their thermochemical properties, part I. J Hazard Mater. 2009;172(2–3):1218–1228. doi: 10.1016/j.jhazmat.2009.07.128. [DOI] [PubMed] [Google Scholar]
- 13.Gong P, Inouye LS, Perkins EJ. Comparative neurotoxicity of two energetic compounds, hexanitrohexaazaisowurtzitane and hexahydro-1,3,5-trinitro-1,3,5-triazine, in the earthworm Eisenia fetida. Environ Toxicol Chem. 2007;26(5):954–959. doi: 10.1897/06-361r.1. [DOI] [PubMed] [Google Scholar]
- 14.Gong P, Basu N, Scheuhammer AM, Perkins EJ. Neurochemical and electrophysiological diagnosis of reversible neurotoxicity in earthworms exposed to sublethal concentrations of CL-20. Environ Sci Pollut Res Int. 2010;17 (1):181–186. doi: 10.1007/s11356-009-0117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dodard SG, Sunahara GI, Kuperman RG, Sarrazin M, Gong P, Ampleman G, Thiboutot S, Hawari J. Survival and reproduction of enchytraeid worms, Oligochaeta, in different soil types amended with energetic cyclic nitramines. Environ Toxicol Chem. 2005;24 (10):2579–2587. doi: 10.1897/054-188r.1. [DOI] [PubMed] [Google Scholar]
- 16.Kuperman RG, Checkai RT, Simini M, Phillips CT, Anthony JS, Kolakowski JE, Davis EA. Toxicity of emerging energetic soil contaminant CL-20 to potworm Enchytraeus crypticus in freshly amended or weathered and aged treatments. Chemosphere. 2006;62 (8):1282–1293. doi: 10.1016/j.chemosphere.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Bardai G, Sunahara GI, Spear PA, Martel M, Gong P, Hawari J. Effects of dietary administration of CL-20 on Japanese quail Coturnix coturnix japonica. Arch Environ Contam Toxicol. 2005;49(2):215–222. doi: 10.1007/s00244-004-0231-9. [DOI] [PubMed] [Google Scholar]
- 18.Gong P, Sunahara GI, Rocheleau S, Dodard SG, Robidoux PY, Hawari J. Preliminary ecotoxicological characterization of a new energetic substance, CL-20. Chemosphere. 2004;56 (7):653–658. doi: 10.1016/j.chemosphere.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Rocheleau S, Lachance B, Kuperman RG, Hawari J, Thiboutot S, Ampleman G, Sunahara GI. Toxicity and uptake of cyclic nitramine explosives in ryegrass Lolium perenne. Environ Pollut. 2008;156(1):199–206. doi: 10.1016/j.envpol.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 20.Gong P, Escalon BL, Hayes CA, Perkins EJ. Uptake of hexanitrohexaazaisowurtzitane (CL-20) by the earthworm Eisenia fetida through dermal contact. Sci Total Environ. 2008;390(1):295–299. doi: 10.1016/j.scitotenv.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Simon R, Lam A, Li MC, Ngan M, Menenzes S, Zhao Y. Analysis of Gene Expression Data Using BRB-Array Tools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 22.Wright GW, Simon RM. A random variance model for detection of differential gene expression in small microarray experiments. Bioinformatics. 2003;19 (18):2448–2455. doi: 10.1093/bioinformatics/btg345. [DOI] [PubMed] [Google Scholar]
- 23.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339 (1):62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 24.Vandesompele J, de PK, Pattyn F, Poppe B, Van RN, De PA, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basu N. Cleveland CJ, editor. Neurotoxicity. Encyclopedia of Earth. 2008 www.eoearth.org/article/neurotoxicity.
- 26.Drewes CD. Sublethal effects of environmental toxicants on Oligochaete escape reflexes. American Zoologist. 1997;37:346–353. [Google Scholar]
- 27.Cerstiaens A, Huybrechts J, Kotanen S, Lebeau I, Meylaers K, De LA, Schoofs L. Neurotoxic and neurobehavioral effects of kynurenines in adult insects. Biochem Biophys Res Commun. 2003;312(4):1171–1177. doi: 10.1016/j.bbrc.2003.11.051. [DOI] [PubMed] [Google Scholar]
- 28.Kharrat R, Servent D, Girard E, Ouanounou G, Amar M, Marrouchi R, Benoit E, Molgo J. The marine phycotoxin gymnodimine targets muscular and neuronal nicotinic acetylcholine receptor subtypes with high affinity. J Neurochem. 2008;107(4):952–963. doi: 10.1111/j.1471-4159.2008.05677.x. [DOI] [PubMed] [Google Scholar]
- 29.Kyriakides MA, Sawyer RT, Allen SL, Simpson MG. Mechanism of action of triethyltin on identified leech neurons. Toxicol Lett. 1990;53 (3):285–295. doi: 10.1016/0378-4274(90)90233-c. [DOI] [PubMed] [Google Scholar]
- 30.Williams LR, roniadou-Anderjaska V, Qashu F, Finne H, Pidoplichko V, Bannon DI, Braga MF. RDX binds to the GABA(A) receptor-convulsant site and blocks GABAA receptor-mediated currents in the amygdala: a mechanism for RDX-induced seizures. Environ Health Perspect. 2011;119(3):357–363. doi: 10.1289/ehp.1002588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Agency for Toxic Substances and Disease Registry (ATSDR) Toxicological Profile for RDX. U.S. Department of Health and Human Services, Public Health Service; Atlanta, GA: 2010. [Google Scholar]
- 32.Bannon DI, Dillman JF, Hable MA, Phillips CS, Perkins EJ. Global gene expression in rat brain and liver after oral exposure to the explosive hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX) Chem Res Toxicol. 2009;22 (4):620–625. doi: 10.1021/tx800444k. [DOI] [PubMed] [Google Scholar]
- 33.Gust KA, Pirooznia M, Quinn MJ, Jr, Johnson MS, Escalon L, Indest KJ, Guan X, Clarke J, Deng Y, Gong P, Perkins EJ. Neurotoxicogenomic investigations to assess mechanisms of action of the munitions constituents RDX and 2,6-DNT in Northern bobwhite (Colinus virginianus) Toxicol Sci. 2009;110(1):168–180. doi: 10.1093/toxsci/kfp091. [DOI] [PubMed] [Google Scholar]
- 34.Gust KA, Brasfield SM, Stanley JK, Wilbanks MS, Chappell P, Perkins EJ, Lotufo GR, Lance RF. Genomic investigation of year-long and multigenerational exposures of fathead minnow to the munitions compound RDX. Environ Toxicol Chem. 2011;30(8):1852–1864. doi: 10.1002/etc.558. [DOI] [PubMed] [Google Scholar]
- 35.Gust KA, Wilbanks MS, Guan X, Pirooznia M, Habib T, Yoo L, Wintz H, Vulpe CD, Perkins EJ. Investigations of transcript expression in fathead minnow (Pimephales promelas) brain tissue reveal toxicological impacts of RDX exposure. Aquat Toxicol. 2011;101(1):135–145. doi: 10.1016/j.aquatox.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Atchison WD. Effects of neurotoxicants on synaptic transmission: lessons learned from electrophysiological studies. Neurotoxicol Teratol. 1988;10(5):393–416. doi: 10.1016/0892-0362(88)90001-3. [DOI] [PubMed] [Google Scholar]
- 37.Csoknya M, Takacs B, Koza A, Denes V, Wilhelm M, Hiripi L, Kaslin J, Elekes K. Neurochemical characterization of nervous elements innervating the body wall of earthworms (Lumbricus, Eisenia): immunohistochemical and pharmacological studies. Cell Tissue Res. 2005;321 (3):479–490. doi: 10.1007/s00441-005-1134-4. [DOI] [PubMed] [Google Scholar]
- 38.Lunt GG. GABA and GABA receptors in invertebrates. Seminars in Neuroscience. 1991;3(3):251–258. [Google Scholar]
- 39.Volkov EM, Nurullin LF, Volkov ME, Nikolsky EE, Vyskocil F. Mechanisms of carbacholine and GABA action on resting membrane potential and Na+/K+-ATPase of Lumbricus terrestris body wall muscles. Comp Biochem Physiol A Mol Integr Physiol. 2011;158 (4):520–524. doi: 10.1016/j.cbpa.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 40.Gong P, Basu N, Perkins EJ. Neurochemical and electrophysiological diagnosis of RDX and CL-20 neurotoxicity in the earthworm Eisenia fetida. SETAC North America 27th Annual Meeting; SETAC; 2006. p. 363. [Google Scholar]
- 41.Garcia-Reyero N, Habib T, Pirooznia M, Gust KA, Gong P, Warner C, Wilbanks M, Perkins E. Conserved toxic responses across divergent phylogenetic lineages: a meta-analysis of the neurotoxic effects of RDX among multiple species using toxicogenomics. Ecotoxicology. 2011;20 (3):580–594. doi: 10.1007/s10646-011-0623-3. [DOI] [PubMed] [Google Scholar]
- 42.Abalis IM, Eldefrawi ME, Eldefrawi AT. Effects of insecticides on GABA-induced chloride influx into rat brain microsacs. J Toxicol Environ Health. 1986;18(1):13–23. doi: 10.1080/15287398609530844. [DOI] [PubMed] [Google Scholar]
- 43.Ono F. An emerging picture of synapse formation: a balance of two opposing pathways. Sci Signal. 2008;1(2):e3. doi: 10.1126/stke.12pe3. [DOI] [PubMed] [Google Scholar]
- 44.Volkov EM, Nurullin LF, Nikolsky E, Vyskocil F. Miniature excitatory synaptic ion currents in the earthworm Lumbricus terrestris body wall muscles. Physiol Res. 2007;56 (5):655–658. doi: 10.33549/physiolres.931269. [DOI] [PubMed] [Google Scholar]
- 45.Lewis JA, Berberich S. A detergent-solubilized nicotinic acetylcholine receptor of Caenorhabditis elegans. Brain Res Bull. 1992;29(5):667–674. doi: 10.1016/0361-9230(92)90136-l. [DOI] [PubMed] [Google Scholar]
- 46.Golan DE, Tashjian AH, Jr, Armstrong EJ, Armstrong AW. Principles of Pharmacology: The Pathophysiiologic Basis of Drug Therapy. Lippincott Williams & Wilkins; Baltimore, MD: 2008. [Google Scholar]
- 47.Shaw C, van HF, Cynader MS, Wilkinson M. A role for potassium channels in the regulation of cortical muscarinic acetylcholine receptors in an invitro slice preparation. Brain Res Mol Brain Res. 1989;5(1):71–83. doi: 10.1016/0169-328x(89)90019-3. [DOI] [PubMed] [Google Scholar]
- 48.Volkov EM, Sabirova AR, Nurullin LF, Grishin SN, Zefirov AL. Effect of GABAergic and adrenergic agents on activity of Na+/K+ pump and Cl(−)-cotransport in somatic muscle cells of earthworm Lumbricus Terrestris. Bull Exp Biol Med. 2006;141(5):633–635. doi: 10.1007/s10517-006-0239-4. [DOI] [PubMed] [Google Scholar]
- 49.Wang Y, Gao J, Mathias RT, Cohen IS, Sun X, Baldo GJ. alpha-Adrenergic effects on Na+-K+ pump current in guinea-pig ventricular myocytes. J Physiol. 1998;509 (Pt 1):117–128. doi: 10.1111/j.1469-7793.1998.117bo.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Olsen RW, DeLorey TM. GABA and Glycine. In: Siegel GJ, Agranoff BW, Albers RW, Fisher SK, Uhler MD, editors. Basic Neurochemistry: Molecular, Cellular and Medical Aspects. Lippincott Williams & Wilkins; Philadelphia, PA: 1999. pp. 335–346. [Google Scholar]
- 51.Miura M, Yoshioka M, Miyakawa H, Kato H, Ito KI. Properties of calcium spikes revealed during GABAA receptor antagonism in hippocampal CA1 neurons from guinea pigs. J Neurophysiol. 1997;78(5):2269–2279. doi: 10.1152/jn.1997.78.5.2269. [DOI] [PubMed] [Google Scholar]
- 52.Olivero-Verbel J, Guerrero-Castilla A, Ramos NR. Biochemical effects induced by the hexachlorocyclohexanes. Rev Environ Contam Toxicol. 2011;212:1–28. doi: 10.1007/978-1-4419-8453-1_1. [DOI] [PubMed] [Google Scholar]
- 53.Borodinsky LN, Spitzer NC. Activity-dependent neurotransmitter-receptor matching at the neuromuscular junction. Proc Natl Acad Sci U S A. 2007;104 (1):335–340. doi: 10.1073/pnas.0607450104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rowland AM, Richmond JE, Olsen JG, Hall DH, Bamber BA. Presynaptic terminals independently regulate synaptic clustering and autophagy of GABAA receptors in Caenorhabditis elegans. J Neurosci. 2006;26 (6):1711–1720. doi: 10.1523/JNEUROSCI.2279-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.You YJ, Kim J, Cobb M, Avery L. Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 2006;3 (4):237–245. doi: 10.1016/j.cmet.2006.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barna J, Csoknya M, Lazar Z, Bartho L, Hamori J, Elekes K. Distribution and action of some putative neurotransmitters in the stomatogastric nervous system of the earthworm, Eisenia fetida (Oligochaeta, Annelida) J Neurocytol. 2001;30(4):313–325. doi: 10.1023/a:1014456329814. [DOI] [PubMed] [Google Scholar]
- 57.Walker RJ, Holden-Dye L, Franks CJ. Physiological and pharmacological studies on annelid and nematode body wall muscle. Comp Biochem Physiol C. 1993;106(1):49–58. doi: 10.1016/0742-8413(93)90253-h. [DOI] [PubMed] [Google Scholar]
- 58.Sporhase-Eichmann U, Winkler M, Schurmann FW. Dopaminergic sensory cells in the epidermis of the earthworm. Naturwissenschaften. 1998;85:547–550. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.