Abstract
Focal adhesion kinase (FAK), a key regulator of cell adhesion and migration, is overexpressed in many types of cancer. The C-terminal focal adhesion targeting (FAT) domain of FAK is necessary for proper localization of FAK to focal adhesions and subsequent activation. Phosphorylation of Y926 in the FAT domain by the tyrosine kinase Src has been shown to promote metastasis and invasion in vivo by linking the FAT domain to the MAPK pathway via its interaction with Grb2. Several groups have reported that inherent conformational dynamics in the FAT domain likely regulate phosphorylation of Y926; however, what regulates these dynamics is unknown. In this paper, we demonstrate that there are two sites of in vitro Src-mediated phosphorylation in the FAT domain: Y926, which has been shown to affect FAK function in vivo, and Y1008, which has no known biological role. The phosphorylation of these two tyrosine residues is pH dependent, but this does not reflect the pH dependence of Src kinase activity. CD and NMR data indicate that the stability and conformational dynamics of the FAT domain are sensitive to changes in pH over a physiological pH range. In particular, regions of the FAT domain previously shown to regulate phosphorylation of Y926 as well as regions near Y1008 show pH-dependent dynamics on the μs-ms time scale.
Focal adhesion kinase (FAK) is a non-receptor tyrosine kinase. Upon activation of integrins, FAK is recruited to newly forming focal adhesions where it functions as both a signaling and scaffolding protein. It has been linked to the MAPK (1) and Rho and Ras GTPase (2–4) pathways and plays a role in cell adhesion (5, 6), migration (5), cell cycle progression (7), and cell survival (8–12). FAK consists of an N-terminal FERM (protein 4.1, ezrin, radixin, moesin) domain, a central kinase domain, and a C-terminal focal adhesion targeting (FAT) domain, which is responsible for localization of FAK to focal adhesions at least partially through its interaction with the protein paxillin (13). The FAK kinase domain is activated by autophosphorylation at Y397, creating a Src-homology (SH) 2 binding site that is recognized by Src kinase, among other proteins. Upon binding to Y397, Src phosphorylates several tyrosine residues in FAK including Y926 in the FAT domain (10, 14). Phosphorylation of Y926 creates a docking site for the SH2 domain of growth factor receptor bound protein 2 (Grb2) (10, 14, 15). The interaction between Grb2 and pY926 in the FAT domain has been shown to activate the MAPK pathway, resulting in increased angiogenesis via upregulation of vascular endothelial growth factor (VEGF) (16, 17), and promote microtubule-induced focal adhesion disassembly by linking FAK to dynamin (18). Moreover, melanoma cells expressing a nonphosphorylatable Y926F mutant of FAK showed decreased adhesion, migration, and invasion in vitro, which correlated with decreased metastasis in vivo. These results implicate the importance of phosphorylation of Y926 in cell migration and adhesion and subsequently in cancer progression (16).
There are several structures of the FAT domain including a solution structure at pH 6.0 (19) and two crystal structures at pH 7.0 (20, 21) in which the FAT domain adopts an anti-parallel four-helix bundle conformation. In addition, a crystal structure at pH 6.5 has been determined (21) in which the FAT domain exists as a domain-swapped dimer in which helix-1 partitions away from the bundle and packs against the other three helices of a second FAT molecule. The ability of helix-1 to adopt both intra- and inter-domain helix bundle conformations suggests that helix-1 possesses conformational mobility, and indeed several lines of evidence including NMR and molecular dynamics simulations have indicated that helix-1 undergoes a helix-coil conformational change in solution (22, 23).
The inherent conformational dynamics that regulate the partitioning of helix-1 in the FAT domain are thought to play a role in the phosphorylation of Y926 by Src and in binding of the phosphorylated FAT domain to the Grb2 SH2 domain (19, 21). Structural studies of kinases bound to substrate peptides suggest that the kinase recognizes an extended conformation for phosphorylation (24). Furthermore, structural evidence also suggests that Y926 must exist in a β-turn conformation to bind to Grb2 (25–28). However, because Y926 in the FAT domain is in a helix, it is not in a conformation that is conducive for phosphorylation by Src or for binding to Grb2. Therefore, it has been speculated that there must be a conformational change near Y926 to facilitate phosphorylation by Src and binding to the Grb2 SH2 domain (19, 21).
While it has been proposed that conformational dynamics in the FAT domain are important for regulating phosphorylation of Y926 and binding to Grb2, the mechanism(s) that regulate FAT domain dynamics in vivo is unclear. In this paper, we explore the effect of pH on the structure, dynamics, and Src-mediated phosphorylation of the FAT domain in vitro.
EXPERIMENTAL PROCEDURES
Expression and Purification of the FAT domain
The FAT domain was expressed as a GST fusion protein and purified as previously described (19).
Expression and Purification of the Src kinase domain
Plasmids containing the kinase domain of c-Src and the YopH phosphatase were provided by John Kuriyan’s lab at UC Berkeley. The expression and purification of the kinase domain of avian c-Src (251–533) have been described elsewhere (29). Purified protein was stored in 50% glycerol at −20°C until use.
In vitro Src-Mediated Phosphorylation of Full-Length FAT domain
Purified FAT domain was dialyzed into phosphorylation buffer (100 mM MES (pH 5.5, 6.0, or 6.5) or 100 mM HEPES (pH 7.0 or 7.5), 5.0 mM MgCl2, 1 mM DTT, 0.01% NaN3). Each reaction contained ~30 μM FAT domain, ~1 μM Src, and 1 mM ATP. Reactions were incubated at 37°C overnight in an eppendorf tube with a 2 kDa MW cutoff bottom floating in 100 mL of phosphorylation buffer to ensure that the pH remained constant throughout the course of the reaction. Samples were dialyzed into water before MS analysis.
In vitro Src-Mediated Phosphorylation of FAT Domain Synthetic Peptides
Two synthetic peptides containing either Y926 (SNDKV926YENVTGLVK-OH) or Y1008 (MKLAQQ1008YVMTSLQQEYK-OH) were synthesized at the UNC High-Throughput Peptide Synthesis and Arraying Facility. The peptides were dissolved in water and diluted with 10X phosphorylation buffer to a final concentration of 0.56 mg/mL in a final volume of 100 μL. Src (1 μL of 5 mg/mL) was added to each reaction, and the reactions were incubated at 37°C for one hour. In total, ten reactions were performed: phosphorylation of the Y926 peptide and the Y1008 peptide, each at pH 5.5, 6.0, 6.5, 7.0, and 7.5. Samples were kept at −20°C until analysis.
Mass Spectrometry-Based Phosphopeptide Identification
The sites of phosphorylation were identified by LC-MS/MS analysis of tryptic peptides derived from the phosphorylation reaction between FAT and Src. The experimental workflow is shown in Figure S1. The FAT domain was phosphorylated as described above, and the samples were digested with trypsin. The digestion reaction was monitored by SDS-PAGE to ensure that the reaction had reached completion. The digestion reactions were spiked with the phosphopeptide internal standards, re-suspended in an aqueous solution of 0.1% formic acid, and analyzed by reverse phase LC-MS/MS using a nano ultra LC system (Eksigent Inc, Dublin, CA) coupled to an LTQ-Orbitrap Velos system (Thermo Scientific, San Jose, CA). The LC-MS/MS data were analyzed as previously described (30). In brief, mass spectrometric data were acquired using the data-dependent mode to interrogate the top ten most abundant peptide ions via collision activated dissociation (CAD)-MS/MS. Mass spectra were processed, and peptide identification was performed using Mascot (Matrix Science Inc.). Peptides were identified using a target-decoy approach with a false discovery rate (FDR) of 1%. The Ascore algorithm was used to confidently localize the phosphorylation site on the peptide derived from the sample (31). In addition, the synthetic phosphopeptide standards were used to further verify the phospho-site assignment specificity using retention time and fragmentation patterns.
Peptide quantitation
Absolute quantitation of site-specific phosphorylation was performed on the peptides that contained the two identified phosphorylation sites in the FAT domain (pY926 and pY1008) with spiked-in heavy labeled internal standards. The following four phosphopeptide internal standards containing a 13C-labeled valine residue were synthesized at the UNC High-Throughput Peptide Synthesis and Arraying Facility: V(p926Y)ENVTGL(V13C5)K-OH, SNDKV(p926Y)ENVTGL(V13C5)K-OH, LAQQ(p1008Y)(V13C5)MTSLQQEYK-OH, and MKLAQQ(p1008Y)(V13C5)MTSLQQEYK-OH. In addition to the tryptic peptides, we also introduced an additional peptide standard for each site to compensate for possible variations in digestion efficiencies that would yield phosphopeptides containing a missed cleavage site. The internal standards were spiked-in, separately, at three-to-four standard concentrations (0.2, 0.4, 1, and 2 μg/mL) to the FAT trypsin digestion reactions. The raw peaks were processed, and extracted ion chromatograms (XIC) were generated from the full-scan MS using Xcalibur software (Thermo Scientific). Peptide peaks of interest were normalized by their stable isotope-labeled heavy internal standard counterpart using an in-house Perl script. Peak integration was performed using the Genesis algorithm (Thermo-Fisher Scientific, San Jose, CA). All deviations in retention times were corrected by peak alignment. The accuracy of the peak area ratios was validated by acquiring multiple chromatographic runs as described previously (30). Absolute quantitation was based on the relative peak area of the identified phosphopeptide and the corresponding heavy phosphopeptide internal standard. A standard curve was generated by plotting the log of the normalized peak area (area of analyte/area of standard) versus the standard concentration. The curves were fit by linear regression in Excel, and the x-intercept of the fit represents the analyte concentration. Both 3+ and 2+ charge states were quantified using these standard curves.
Relative changes in phosphorylation levels of the synthetic peptides were quantified as a function of pH using a single standard concentration (0.5 μg/mL of heavy internal standard). Phosphorylation reactions of the peptide containing Y926 were diluted 100 fold before analysis by MS whereas reactions of the peptide containing Y1008 were diluted 4 fold before analysis by MS so that the signal was in an appropriate linear dynamic range for quantitation.
Circular Dichroism
Purified FAT domain was dialyzed into CD buffer (10 mM K2HPO4, 0.01% NaN3) at the desired pH and concentrated to 10 μM. Far-UV CD (185–260 nm) spectra were collected at 25°C on an Applied Photophysics Pistar-180 spectropolarimeter. Thermal denaturation studies were conducted by monitoring the ellipticity at 221 nm over a temperature range of 25 to 95°C at increments of 0.5°C. Far-UV CD spectra (185 nm-260 nm) collected after the FAT domain had been heated to 95°C for 30 min showed that the FAT domain retained little or no helical character at 95°C. After each thermal denaturation, the temperature was reduced to 25°C for 15 min, and a far-UV spectrum was collected to verify that the FAT domain had regained its secondary structure.
Data analysis was conducted with the assumption that only populations of folded (F) and unfolded (U) species were being monitored. The denaturation curves were baseline corrected and fit to the following Boltzmann equation in SigmaPlot:
ΘObs, ΘF, and ΘU are the observed ellipticity at 221 nm, the ellipticity at 221 of the folded FAT domain, and the ellipticity at 221 of the unfolded FAT domain, respectively. Tm is the melting temperature or the temperature where half of the FAT domain is folded. Although the high temperature(s) required for most of the thermal denaturation studies limited the accuracy by which ΘU could be measured, the data were also fit to alternate equations (32, 33), and similar results were obtained.
NMR Spectroscopy
15N and 15N-13C-labeled FAT domain were expressed and purified as previously described (19). The purified FAT domain was exchanged into NMR buffer at the desired pH (25 mM Tris maleate, 150 mM NaCl, 10% D2O, 0.01% NaN3) and concentrated to ~0.2 mM. All NMR data were processed with NMRPipe (34) and analyzed with NMRView (35).
1H-5N HSQC spectra were collected as a function of pH, and a simultaneous 3D 13C/15N-NOESY (36, 37) with a 150 ms mixing time was collected at pH 6.0 and 7.5 on a Varian INOVA 800 MHz spectrometer at 37°C. While temperature changes in some buffered solutions can affect the pH, we used a tris-maleate buffer, which is relatively insensitive to temperature changes. The spectra were referenced to each other using peaks associated with the glycine linker.
Backbone assignments for the FAT domain at pH 6.0 had been obtained previously (19). Because several backbone resonances undergo chemical shift changes between pH 6.0 and 7.5, 3D NMR HNCACB (38) and CBCACONH (37) experiments were acquired at pH 7.5 on a sample of uniformly 13C-15N labeled FAT domain, and backbone assignments at pH 7.5 were determined as previously described (19) using the assignments at pH 6.0 as a starting point. Assignments at intermediate pH values were determined by tracking changes in chemical shift at pH 6.0 and pH 7.5.
Residual dipolar coupling (RDC) data were collected on an 15N-labeled sample of the FAT domain (0.4 mM). Pf1 phage (7.5 mg/mL ASLA Biotech Ltd.) was used as an alignment medium as previously described (19). 1H-15N-HSQC-IPAP (39) spectra were collected in NMR buffer alone and in the presence of phage.
Backbone Relaxation Analysis
15N-based 2D T1, T2, and heteronuclear NOE (HetNOE) spectra (40) were collected on uniformly 15N-enriched FAT domain samples at pH 6.0 and pH 7.5 on a Varian INOVA 600 MHz spectrometer at 37°C. Prior to data collection, the temperature was calibrated using a 100% MeOH standard sample. R1 and R2 relaxation rates were sampled with nine time points, three of which were collected in duplicate to estimate error. Delay values of 48, 136, 250, 385, 540, 708, 892, 1000, and 1300 ms were used for T1, and 7, 15, 23, 39, 62, 78, 93, 109, and 125 ms were used for T2 experiments. Two experiments comprising the 1H-15N NOE were acquired in an interleaved fashioned with a recycle delay ~ 5 sec. The T1 and T2 data were analyzed by fitting the intensities of resolved amide peaks to a single exponential decay, and the HetNOE data were analyzed by determining the difference in intensity between the irradiated and non-irradiated spectra using programs provided by the laboratory of Andrew Lee (UNC-CH).
T1, T2, and HetNOE data were used to evaluate backbone motions on the ps-ns timescale using the model-free formalism (41). The isotropic rotational correlation time (τm) was determined using the approach described by Dellwo and Wand (42). Rotational diffusion anisotropy was calculated using the local Di method (43) and the crystal structure of the FAT domain (PDB 1K40). Backbone relaxation data were fit using an anisotropic correction (Dpar/Dperp = 2.22 and 1.73 at pH 6.0 and pH 7.5, respectively) to minimize model selection error (44). Backbone relaxation rates were fit to one- or two-parameter model-free models (S2, S2 and τe, or S2 and Rex) using a program, relxn2.2, provided by the laboratory of Andrew Lee (UNC-CH) (45, 46).
RESULTS
Phosphorylation of the FAT Domain
Phosphorylation of Y926 has been implicated in excluding FAK from focal adhesions and promoting focal adhesion turnover (47), cell adhesion, migration, invasion in vitro and metastasis in vivo (16). Therefore, understanding how phosphorylation of Y926 affects FAK signaling on a structural level could provide key insights into how this site is involved in FAK-mediated cancer progression. We were interested in characterizing the phosphorylated species of the FAT domain because previous data from our lab and others have suggested that a structural rearrangement is necessary for recognition of the phosphorylated species by Grb2 (19, 21, 23). The FAT domain was phosphorylated in vitro using purified Src kinase domain. We varied several conditions, one of which was pH, to identify optimal conditions for phosphorylation. The site of phosphorylation was identified via microelectrospray Fourier transform ion cyclotron resonance mass spectrometry (μFT-ICR). The full MS of the intact protein revealed that the FAT domain was phosphorylated at a single site. Top-down electron capture dissociation tandem mass spectrometry (ECD-MS/MS) of the phosphorylated peak revealed the presence of two sites of phosphorylation in the FAT domain: Y926 and Y1008 at all pH values (pH 5.5, 6.0, 6.5, 7.0, and 7.5) (data not shown). Therefore, while we do not detect a doubly-phosphorylated species, it appears that there are two sites of tyrosine phosphorylation in the FAT domain.
To verify the full-length phosphorylation site assignments and to quantify the relative phosphorylation levels of each site, we subjected the phosphorylated samples to trypsin digestion and analyzed the peptides by reverse-phase LC-MS/MS. Figures S2 and S3 show the unambiguous assignment of the two peptides phosphorylated at Y926 and Y1008 (> 99% Mascot probability and Ascore corresponding to >99% site-localization probability). The phosphorylation site assignments were further verified by their heavy labeled spiked-in internal standard peptide counterparts allowing concurrent verification of the sequence ions in the MS/MS spectra. It is noteworthy that the mass spectra are identical in terms of their fragmentation patterns and differ only when the product ions contain the heavy valine substitution.
The bottom-up CAD-MS/MS data show that the two phosphorylation sites in the FAT domain, Y926 and Y1008, were phosphorylated at a range of biologically relevant pH conditions (pH 5.5, 6.0, 6.5, 7.0, and 7.5) (data not shown), in agreement with the top-down ECD-MS/MS data. Y926 is the only site of tyrosine phosphorylation in the FAT domain reported to modulate FAK function. Whereas phosphorylation of Y1008 has been previously observed by mass spectrometry in full-length FAK purified from Sf9 cells (48), it was not identified as a Src substrate. Furthermore, it is unclear whether phosphorylation of Y1008 is physiologically relevant, and no prior studies have established the relative levels of phosphorylation between these distinct sites. To quantify the relative and absolute levels of phosphorylation at Y926 and Y1008, we used a stable isotope dilution mass spectrometry-based method. We spiked the trypsin digestion reactions with various concentrations of an internal standard: a synthetic 13C-labeled phosphorylated peptide with a sequence identical to that of the tryptic peptide. The spiked-in internal standard allows both absolute quantitation and sequence verification of the peptide of interest.
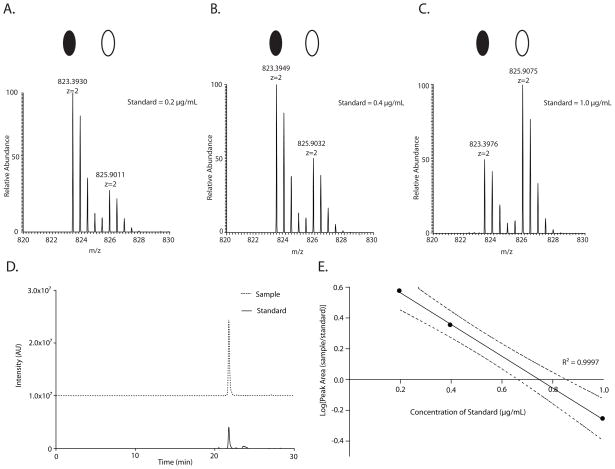
Figure 1 shows the stable isotope dilution mass spectrometry-based absolute quantification strategy that was used to determine the phosphorylation levels of Y926 at pH 5.5. The tryptic phosphopeptide that contains Y926 was quantified by the corresponding heavy-labeled spiked-in internal standard. The XIC peak areas of the phosphopeptide sample and internal standard (Figure 1D) were used to derive a linear relationship between the MS ion intensities and spiked-in standard. The linear regression of the standard curve, shown in Figure 1E, was then used to determine the absolute concentration of the phosphopeptide present in the sample at a given pH condition. We determined the concentration of both the 2+ and 3+ charge states of each peptide phosphorylated at Y926 and Y1008 at pH 5.5, 6.0, 6.5, 7.0, and 7.5.
FIGURE 1.
MS-based absolute quantification of the phosphopeptide containing Y926 (z=+2). MS spectra of the phosphopeptide containing Y926 with corresponding sample peptide (solid oval) and heavy internal standard peptide (empty oval) spiked in at A. 0.2 μg/mL, B. 0.4 μg/mL and C. 1.0 μg/mL. D. Extracted ion chromatogram (XIC) of sample and internal standard spiked in at 0.2 μg.mL−1. E. Linear regression fit of the normalized peak area of the sample as a function of spiked-in standard concentration. Dashed lines represent 95% confidence intervals.
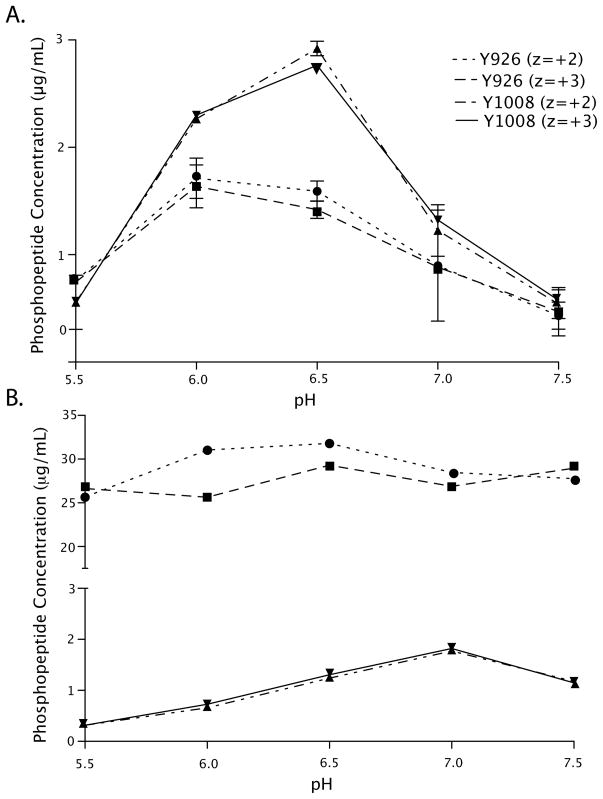
Figure 2A shows the levels of phosphorylation at Y926 and Y1008 phosphorylation as a function of pH. For Y926, phosphorylation levels are low at pH 5.5 (0.75 ± 0.04 μg/mL), reach a maximum at pH 6.0 (1.6 ± 0.2 μg/mL), and gently decrease to 0.4 ± 0.2 μg/mL at pH 7.5. The pH profile for phosphorylation of Y1008 is slightly different. As seen for phosphorylation of Y926, phosphorylation of Y1008 is low at pH 5.5 (0.5 ± 0.2 μg/mL) but reaches a maximum at pH 6.5 (2.85 ± 0.06 μg/mL) before sharply decreasing to 0.5 ± 0.1 μg/mL at pH 7.5.
FIGURE 2.
Site-specific quantitation of phosphorylation. A. Full-length FAT domain and B. FAT domain peptides were phosphorylated in vitro with Src kinase at pH 5.5, 6.0, 6.5, 7.0, and 7.5. After phosphorylation, the full-length FAT domain was trypsinized, and the levels of the Y926 phosphorylated peptide (● z=+2, ■ z=+3) and Y1008 (▲ z=+2, ▼ z=+3) were quantified as described in the text. Comparison of the phosphorylation profiles of the full-length FAT domain and of FAT domain peptides reveals that the specificity of phosphorylation is different in the peptides than in the full-length protein. Y1008 is preferred in the full-length FAT domain whereas Y926 is preferred in the peptides. Furthermore, the pH dependence of phosphorylation of the peptides is different than that of the full-length protein.
To determine whether the observed pH-dependent phosphorylation of the FAT domain is a result of the sensitivity of Src activity to pH or to pH-dependent changes in the FAT domain, we phosphorylated two synthetic peptides in vitro, one that contains Y926 and one that contains Y1008, (SNDKV926YENVTGLVK-OH and MKLAQQ1008YVMTSLQQEYK-OH) at pH 5.5, 6.0, 6.5, 7.0, and 7.5 and subjected them to the same analysis as the full-length protein. These peptides are expected to be unstructured, and therefore their structure should not be influenced by pH. Figure 2B shows the pH profile for phosphorylation of the synthetic peptides. Phosphorylation of both peptides is relatively insensitive to pH. Therefore, we conclude that the pH-dependent phosphorylation observed in the full-length FAT domain is a result of a pH-dependent change in the conformation or dynamics of the FAT domain and not Src activity.
Interestingly, in the context of the peptide, Y926 is phosphorylated to a greater extent than Y1008 (~25 fold). This result differs from the full-length protein in which phosphorylation levels at the two sites were either comparable or Y1008 was the major site, depending on pH. The fact that the Y926 peptide is phosphorylated to a greater extent than the Y1008 peptide is consistent with the fact that Y926 is the biologically relevant site of phosphorylation and that, in the absence of structure, i.e., in the context of a peptide, the sequence surrounding Y926 is a better substrate for Src than the sequence surrounding Y1008. Consistent with this finding, the sequence surrounding Y926 (DKVYENVT) is more similar to the Src consensus phosphorylation sequence (E/DEI/VYEGFF) (49) than that surrounding Y1008 (AQQYVMTS).
pH-Dependent Thermal Unfolding of the FAT domain
Based on the phosphorylation results, we believe that the pH-dependent phosphorylation observed in the full-length FAT domain is a result of a pH-dependent change in the conformation or dynamics of the FAT domain and not Src activity. We therefore investigated the effects of pH on the secondary structure of the FAT domain by CD. Far-UV CD spectra collected at pH 5.5, 6.0, 6.5, 7.0 and 7.5 show no change in secondary structure (data not shown). Therefore, pH does not have a major effect on the helical content of the FAT domain.
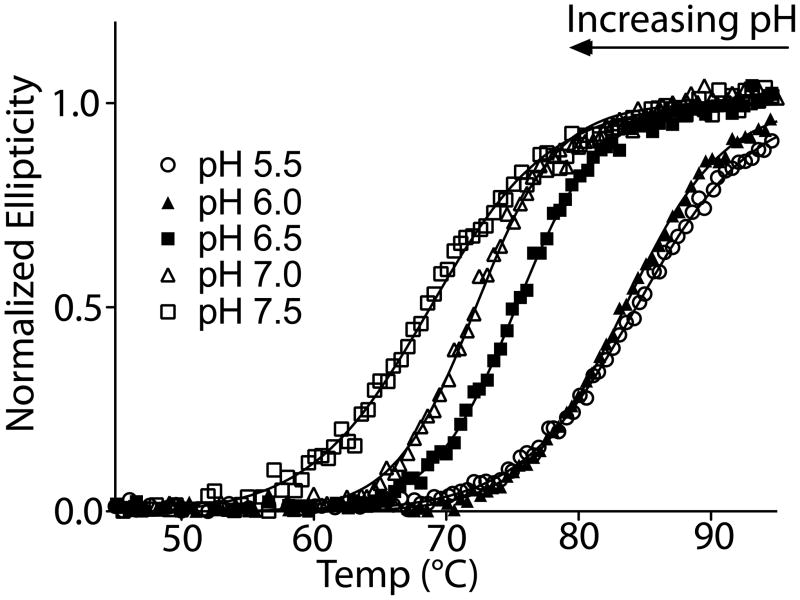
We also investigated the pH-dependence of thermal stability of the FAT domain by CD. Figure 3 shows the thermal denaturation curves at pH 5.5, 6.0, 6.5, 7.0, and 7.5. A sizable decrease in the Tm (approximately 16°C) is observed as the pH is increased from pH 5.5 to pH 7.5. Because the CD data indicate that the FAT domain retains helical content at higher pH, the decrease in stability at higher pH is likely a result of changes in tertiary packing interactions rather than loss of secondary structure.
FIGURE 3.
pH-dependent stability of the FAT domain. The thermal stability of the FAT domain was measured at pH 5.5, 6.0, 6.5, 7.0, and 7.5 by monitoring the CD signal at 221 nm from 25–95°C. The FAT domain is significantly more stable at pH 5.5 compared to pH 7.5, as indicated by the melting temperature.
1H-15N HSQC of the FAT Domain as a Function of pH
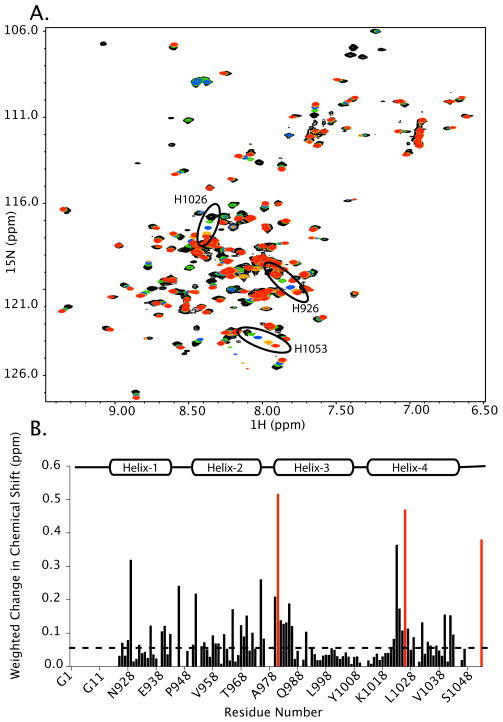
To evaluate possible pH-dependent perturbations on a per-residue basis, we collected high-resolution 1H-15N HSQC spectra of the FAT domain at various pH values. The 1H-15N HSQC spectrum provides the chemical shift for the amide nitrogen and proton atoms. The chemical shift value is sensitive to the electro-chemical environment around that atom, and therefore, changes in chemical shift indicate that the environment around the atom of interest is changing. Figure 4A shows an overlay of the 1H-15N spectra at pH 5.5, 6.0, 6.5, 7.0, and 7.5. While some of the NH resonances show significant pH-dependent chemical shift changes (δ > 0.05 ppm) over the 5.5–7.5 pH range, other NH resonances remain unchanged. Figure 4B shows the weighted changes in chemical shift mapped onto the sequence of the protein. Of interest is that several residues with significant changes in chemical shift are in helix-1, which contains one of the sites of phosphorylation, Y926. In fact, one of the residues with the largest change in chemical shift is V929, which is near Y926. Furthermore, significant chemical shift changes are observed in the C-terminal end of helix-1 and in the loop between helices-1 and 2. This region is of interest because dynamics in this region have been implicated in the regulation of Y926 phosphorylation (19, 21). Our findings that NH resonances corresponding to residues near Y926 and regions thought to regulate phosphorylation of Y926 (the loop between helices-1 and -2) show pH-dependent changes in chemical shift support our MS results that demonstrate that Y926 phosphorylation is affected by pH.
FIGURE 4.
1H-15N HSQC spectra as a function of pH. A. Overlay of 1H-15N HSQC spectra collected at pH 5.5 (black), 6.0 (green), 6.5 (blue), 7.0 (orange), and 7.5 (red). The chemical shifts of several peaks are affected by pH whereas others do not change. The chemical shifts of the three histidine residues in the FAT domain (H981, H1026, and H1053) are circled. B. The weighted changes in chemical shift from pH 5.5 to 7.5 were calculated. Changes greater than 0.05 ppm (dashed line) were considered significant. The histidine residues are shown in red.
The largest NH chemical shift changes were observed for the three histidine residues (H981, H1026, H1053) in the FAT domain, which is not surprising because histidine side chains typically have pKa values in the range of 6.0–6.5. We were able to track the chemical shift changes of the NH resonances of the histidine residues as a function of pH to estimate the pKa of each histidine within the FAT domain. Figure S4 shows the titration profiles used to estimate pKa values for each histidine. Estimated pKa values for H981, H1026, and H1053 are 5.82 ± 0.04, 6.21 ± 0.07, and 6.5 ± 0.1, respectively. Because these pKa values are distinct, we proceeded to calculate the relative pK values of all the residues that showed significant chemical shift changes. Chemical shift changes that resulted in a pK value ± 0.15 pH units to one of the histidine pKa values were considered correlated to that histidine. Figure S4d shows the residues whose pH-dependent changes in chemical shift correlate to specific histidine residues mapped onto the structure of the FAT domain. H981 and residues correlated to it are shown in blue whereas H1026 and correlated residues are shown in red. Titration of H981, located at the beginning of helix-3 correlates with the pH-dependent NH chemical shift changes of nearby residues, as expected. However, there are also several residues distal to H981 whose pH-dependent NH resonances correlate with this residue, including residues all along helix-2 and in the amino-terminus of helix-1, which contains Y926. Titration of H1026, located in the middle of helix-4, tracks with the pH-dependent NH chemical shift changes of surrounding residues. Interestingly, however, there are several residues in the C-terminus of helix-1 (V936, S940, S941, and S943) that titrate with H1026. Although the side chain of H1026 is mostly exposed to the solvent, the C-terminal region of helix-1 packs against helix-4 near H1026, and changes in helix-4 resulting from the titration of H1026 could affect residues in helix-1. These residues (V936, S940, S941, and S943) are near the loop between helices-1 and 2, which has been proposed to be involved in regulation of Y926 phosphorylation (19, 21). Histidine 1053, the last residue in FAK, does not appear to interact with other residues within the FAT domain, and titration of H1053 does not correlate with the pH-dependent changes in chemical shift of nearby residues. There were also a number of peaks, notably in the N-terminal region of helix-2, whose pH-dependent chemical shifts do not track with a particular histidine residue. pH-dependent NH chemical shift perturbations of these residues may be a result of de/protonation of both H1026 and H981 or of another titrating residue, possibly an aspartate or glutamate residue.
pH-Dependent Residual Dipolar Couplings
Because the 1H-15N HSQC spectra (Figure 4) show significant changes in chemical shift as a function of pH, we collected NH RDCs at pH 6.0 and pH 7.5 to further investigate possible pH-dependent structural changes in the FAT domain. RDCs provide long-range orientation information on the NH bond vector and are often used to assess structural perturbations upon mutation (50) and ligand binding (51). We chose pH 6.0 because the solution structure of the FAT domain had been solved at this pH (19). We were able to compare 47 out of a possible 138 NH resonances (146 residues minus 8 prolines), using Pf1 phage to induce weak alignment of the sample. Figure S5 shows the correlation between RDC values of two datasets collected at pH 6.0, two datasets collected at pH 7.5, and a comparison of a dataset collected at pH 6.0 and at pH 7.5. The correlation between the RDC values at pH 6.0 and pH 7.5 was 0.85, which is not significantly different from the correlation between two datasets collected at pH 6.0 (0.88) or two datasets collected at pH 7.5 (0.90). Therefore, the RDC data do not indicate a major structural change in the FAT domain from pH 6.0 to pH 7.5.
NOE Analysis Near Sites of Phosphorylation
To further assess possible structural alterations surrounding the two sites of phosphorylation as a function of pH, we collected a 3D 13C/15N-NOESY at pH 6.0 and pH 7.5. Figure S6 shows the NOESY strips for the Hδ and Hε of Y926 while Figure S7 shows the NOESY strips for the Hδ and Hε of Y1008 at pH 6.0 and pH 7.5. While there are minor changes in NOE intensity, the number of NOEs at the two pH values is similar, suggesting that the side chains of both of these tyrosines maintain short range (generally detected within 5 Å) NOE contacts as a function of pH. NOESY strips of other residues at the two pH values also show significant similarities.
1H-15N HSQC Analyses Suggest pH-Dependent Changes in FAT Domain Dynamics
While we were unable to detect any significant pH-dependent structural change, there are several residues in the 1H-15N HSQC spectra that show changes in NH peak intensity as a function of pH, suggesting a pH-dependent change in dynamics. In particular, as the pH is increased from pH 6.0 to pH 7.5, several residues show significant decreases in intensity resulting from peak broadening. Exchange broadening of NH resonances as the pH is increased can be difficult to interpret because a variety of mechanisms can contribute to this broadening, the most pertinent of which are pH-dependent differences in conformational dynamics (exchange between two or more states) and chemical exchange (an increase in exchange between the amide proton and the solvent). As expected, the N- and C-termini NH resonances are severely broadened at higher pH, likely due to an increase in base-catalyzed amide exchange. These residues are located in unstructured regions and consequently more exposed to the solvent. There are also several NH resonances associated with residues in the turn between helices 3 and 4 and in the N-terminus of helix 4 that broaden as the pH is increased. Figure 5A shows the residues that broaden as the pH is increased mapped onto the structure of the FAT domain. Interestingly, these residues are located near Y1008, one of the sites of phosphorylation. However, as stated above, it is difficult to distinguish whether broadening in this region is due conformational or chemical exchange. However, an increase in solvent exchange is likely, as these residues are in a turn and exposed to the solvent.
FIGURE 5.
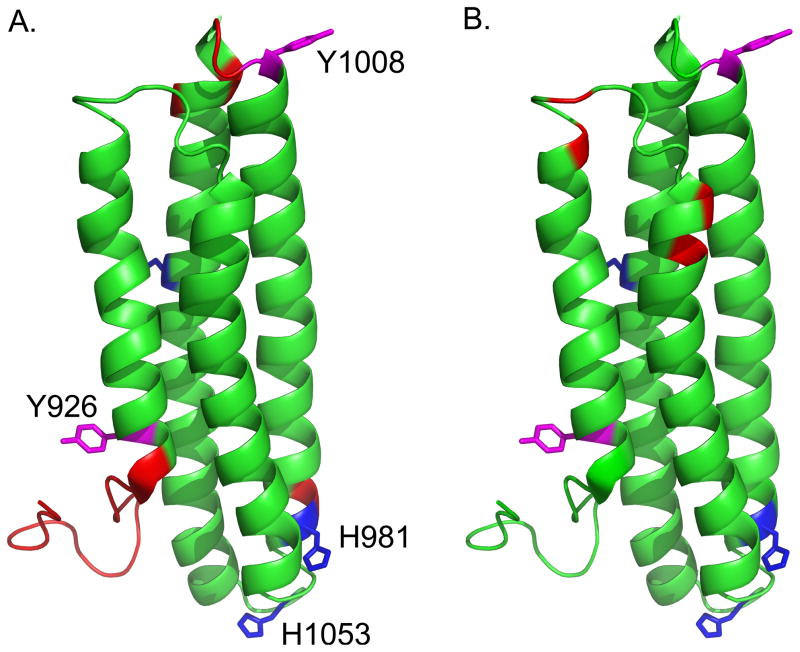
pH-dependent changes in intensity derived from 1H-15N HSQC spectra. A. Peaks that broaden in the 15N-1H HSQC spectra from pH 5.5 to pH 7.5 are shown in red. These residues localize to the N and C-termini, the turn between helices 3 and 4 and the N-terminus of helix-4. B. Peaks that narrow from pH 5.5 to pH 7.5 are shown in red. These residues localize to the “hinge region” between helices 1 and 2. The two sites of phosphorylation, Y926 and Y1008, are shown in magenta, and the histidine residues are shown in blue.
In addition to peaks that broaden as the pH is increased, there are also several peaks that increase in intensity upon increasing the pH from 6.0 to 7.5. Interestingly, several of these peaks are located at the end of helix-1, the beginning of helix-2, and the loop region in between these helices. Figure 5B shows the peaks that increase in intensity as the pH is increased mapped onto the structure of the FAT domain. Several of these residues were previously shown to be sensitive to temperature-dependent broadening (19), indicating that these residues experience dynamics on the intermediate to fast timescale. Conformational exchange in this region has previously been postulated to affect phosphorylation of Y926 in helix-1 (19, 21, 22). While it is possible that this region is becoming less dynamic, the residues that narrow as the pH is increased are the same residues that narrow as the temperature is increased. Hence, it is likely that increasing the pH enhances the kinetics of the process that contributes to the exchange-mediated dynamics in this region, most likely reflecting a transition from the intermediate timescale, where peaks are broader, to a faster timescale, where peaks become sharper.
Backbone Dynamics
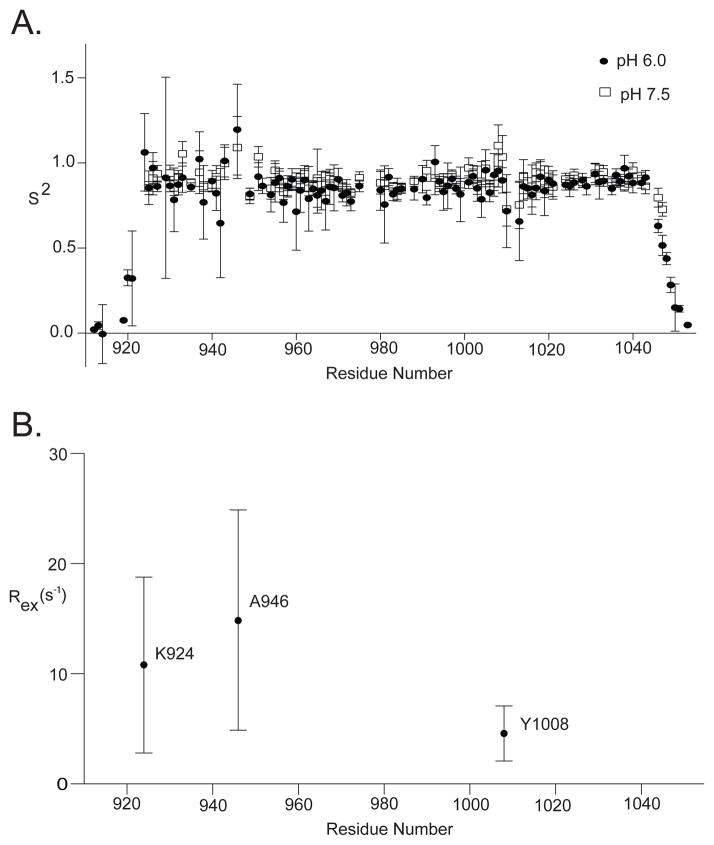
Because the pH-dependent 1H-15N HSQC data suggest changes in dynamics as a function of pH, we further investigated this effect by collecting 15N-backbone relaxation data (T1, T2, and HetNOE datasets) at pH 6.0 and pH 7.5. These measurements are sensitive to backbone dynamics on the ps-ns timescale as well as the ms timescale. The relaxation data were fit to a model-free formalism (41, 42) to obtain the overall correlation time (τm) and the order parameters S2 and τe, which measure the rigidity of the N—H bond and the internal correlation time, respectively. Changes in pH over this range did not affect the overall correlation time (τm) of the protein, which is generally dependent on the overall size and shape of the molecule. The isotropic τm was determined to be 11.3 ns at pH 6.0 and pH 7.5. This value is slightly higher than expected for a protein this size (~9.5 ns) likely because the FAT domain is not spherical (60 Å × 20 Å × 20 Å (20)). Therefore, model free fitting was performed using an anisotropic correction. Figure 6A shows the S2 values at pH 6.0 and pH 7.5 mapped onto the sequence of the FAT domain. No discernable difference was observed in the S2 order parameter as a function of pH. As expected, the S2 values are near 0.9 in the structured regions of the proteins and decrease to around 0.8 in the loops and turns. Residues in the N- and C-termini are not visible by NMR at pH 7.5 and have lower S2 values at pH 6.0 (~0–0.6), as these residues are less structured relative to those in the helix bundle, are more exposed to the solvent, and likely undergo enhanced solvent-mediated chemical exchange. Moreover, no difference in τe is observed as a function of pH.
FIGURE 6.
Backbone dynamics as a function of pH derived from 2D T1, T2, and HetNOE data as described in the Experimental Procedures. A. S2 order parameters at pH 6.0 and pH 7.5 mapped onto the sequence of the FAT domain. B. Rex values at pH 6.0.
It may not be surprising that we see little difference in S2 or τe as a function of pH because these values are sensitive to backbone motions on faster timescales (ps-ns) than the overall correlation time of the FAT domain. Thus, it is possible that the dynamic changes that occur as a function of pH occur on a slower timescale (μs-s) and may involve the side chains.
While the 15N-based T1, T2, and NOE relaxation measurements provide dynamic information on the ps-ns timescale, the measurements are also sensitive to motions on slower timescales (μs-ms). Indeed, several residues appear to be undergoing motion on these slower timescales, as indicated by contributions from Rex in the model-free fitting. At pH 6.0, there are three residues in helix-1, K924 (which is not visible at pH 7.5), A946, and Y1008, that experience slower μs-ms timescale dynamics (Figure 6B). The A946 NH resonance also narrows in the 1H-15N HSQC spectrum as the pH is increased, which provides further support that backbone dynamics in this region are susceptible to changes in pH. A946 is in a putative “hinge region”, which consists of the loop between helices 1 and 2. It has been postulated that dynamics in this region due to strain caused by a series of proline residues modulate a helix-coil transition in helix-1 that regulates phosphorylation of Y926 (19). It is also interesting that Y1008 displays pH-dependent μs-ms backbone dynamics as this is one of the sites of phosphorylation in the FAT domain, and our MS data indicate that phosphorylation at this site is sensitive to pH.
DISCUSSION
Phosphorylation of Y926 in the FAT domain has been shown to play a role in FAK signaling and localization. In particular, phosphorylation at this site has been linked to regulation of cell adhesion, migration, and invasion in vitro and metastasis in vivo (16). However, the mechanism by which phosphorylation of Y926 is regulated in vivo remains to be elucidated. While it has been speculated that a conformational change is necessary for phosphorylation of Y926 (19, 21), what regulates this change in vivo is unknown.
In this paper, we demonstrate that there are two sites of in vitro Src-mediated phosphorylation in the FAT domain: Y926 and Y1008. This is the first time to our knowledge that Y1008 has been identified as a Src phosphorylation site. Furthermore, phosphorylation of these sites is pH dependent. To distinguish whether the pH dependence of phosphorylation was due to pH-dependent changes in the FAT domain or in Src specificity, we phosphorylated two synthetic peptides corresponding to the tryptic fragments containing each of the FAT phosphorylation sites in vitro. Phosphorylation of the peptides does not show the same pH dependence as phosphorylation of the full-length FAT domain. We have interpreted the changes in phosphorylation of the peptides as too minor to be significant. However, even if the changes in phosphorylation of the peptides are considered, they do not show the same trend as phosphorylation of the full-length FAT domain. We therefore conclude that the pH-dependent phosphorylation observed in the FAT domain is due to changes in the FAT domain and not Src. We have previously shown that phosphorylation of Y926 is sensitive to conformation. In the context of a GST-helix-1 construct, in which helix-1 is expected to be unstructured, Y926 is phosphorylated by Src to a greater extent than in the full-length FAT domain construct in vitro (19). Therefore, it is not surprising that we observe higher phosphorylation levels of the synthetic peptides than of the full-length FAT domain. However, it is interesting that the preference for Y926 or Y1008 is different in the peptides versus the full-length protein. In the context of the peptide, Y926 is phosphorylated to a greater extent than Y1008 (~25 fold) whereas in the full-length protein, the levels of phosphorylation are either similar or Y1008 was the major site, depending on pH. While it is well known that proper sequence and conformation is necessary for recognition and subsequent phosphorylation by kinases, we think that this is an interesting example in which one site (Y926) appears to have a favorable sequence but poor conformation for phosphorylation by Src whereas a second site (Y1008) has a poor sequence but favorable conformation for phosphorylation. The fact that Y926 is the biological site of phosphorylation may suggest that, in vivo, sequence is more important than conformation for recognition by Src. However, the use of non-specific phosphorylation antibodies and peptide libraries to investigate sites of phosphorylation and determine kinase consensus sequences may have resulted in Y1008 being overlooked as a possible substrate for Src. Further work is necessary to determine whether Y1008 is phosphorylated in vivo.
Regardless of the biological relevance of phosphorylation of Y1008, the phosphorylation data indicate that either the structure or dynamics of the FAT domain are sensitive to pH. NMR-derived backbone RDCs and NOEs collected at pH 6.0 and pH 7.5 do not support a pH-dependent structural change near the two sites of phosphorylation. Furthermore, comparison of the solution structure solved at pH 6.0 (19) and the crystal structure solved at pH 7.0 (21) does not indicate a change in structure near Y926 or Y1008.
The second explanation for the observed pH-dependent phosphorylation is that a pH change over the 5.5 to 7.5 range affects the conformational dynamics of the FAT domain. Amide resonances associated with the turn between helices-3 and -4 and the N-terminus of helix-4, which is near Y1008, show pH-dependent changes in intensity in the 1H-15N HSQC, and the amide resonance of Y1008 shows μs-ms timescale motions. These data indicate that this region is in conformational exchange on this timescale and suggest that dynamics in this region may play a role in phosphorylation of Y1008.
A second region that experiences pH-dependent changes in dynamics is the loop between helices-1 and -2. Resonances in the 1H-15N HSQC corresponding to this region narrow as the pH is increased, and we also observe μs-ms timescale motions in this region, suggesting that the loop between helices-1 and -2 is in exchange between two or more conformations on the μs-ms timescale. Indeed, previous NMR and hydrogen exchange-directed molecular dynamics data have suggested that dynamics in this region, due to strain caused by a series of proline residues, modulate a helix-coil transition in helix-1 (19, 22). An open, intermediate state of the FAT domain in which helix-1 is partitioned away from the bundle was detected (22). We have attempted to investigate backbone dynamics as a function of pH via CPMG-based relaxation dispersion experiments. However, we were unable to detect Rex differences with this experiment likely because the population of the open, intermediate state is too low to be detected by these measurements. Previous work with a Y926E mutant of the FAT domain demonstrated that the intermediate form is populated at 0.1% (23). Hence, if a weakly populated pH dependent state exists it may be too lowly populated to be quantified by relaxation dispersion. Therefore, while we have evidence that dynamics in the hinge region and in the region near Y1008 are sensitive to pH on the μs-ms timescale, we were unable to quantify these dynamics.
Dynamics in the hinge region of the FAT domain have been proposed to control phosphorylation of Y926; however, what regulates these dynamics is unclear. Based on our data, it appears that pH affects dynamics in this region; however, we have not been able to quantify the rates and populations, possibly due to the presence of weakly populated states. Interestingly, the crystal structure of the domain-swapped dimer of the FAT domain was solved at pH 6.5 whereas the crystal structure of the monomeric four-helix bundle was solved at pH 7.0 (21). It is unclear whether formation of the domain-swapped dimer was induced by the lower pH; however, it is possible that lower pH favors partitioning of helix-1 and therefore the domain-swapped dimer. This is consistent with our observation that phosphorylation of Y926 is higher at pH 6.5 than at pH 7.0, suggesting that lower pH facilitates partitioning of helix-1, which would be expected to promote phosphorylation of Y926.
Because our results suggest that pH affects the dynamics of the FAT domain, we have attempted to determine the titrating residue that affects the dynamics. We have individually mutated the three histidine residues in the FAT domain as well as D994, D1040, and E949 to alanine. The histidines were mutated because the NMR data indicate that the histidine side chains titrate over this pH range. The aspartate and glutamate mutants were chosen based on their position in the structure of the FAT domain. D994 is in an area of negative charge, which might cause the side chain to be protonated; D1040 is in helix-4 and makes contacts with residues in helix-1 near Y926, and E949 is in the hinge region of the FAT domain. None of these mutants abrogate the pH-dependent stability of the FAT domain. It is possible that the pH-dependent effects that we observe are due to titration of a residue that we have not yet investigated or to titration of more than one residue.
This is the first study to quantitatively characterize the in vitro Src-mediated phosphorylation of the FAT domain of FAK. We have revealed that, in vitro, Src phosphorylates two tyrosine residues in the FAT domain, Y926 and Y1008. To our knowledge, phosphorylation of Y1008 by Src has not been shown. We also show that phosphorylation of Y926 and Y1008 varies as a function of pH and that, although the pH profile of these two sites differs, we were unable to find conditions that would exclusively favor phosphorylation of one site over the other. Therefore, studies that involve phosphorylation of the FAT domain, particulary in vitro, should be carefully conducted to identify the site of phosphorylation. Furthermore, based on our NMR, we believe that the pH depedence of phosphorylation of Y926 and Y1008 is due to changes in dynamics of the FAT domain, which could represent an interesting means of regulating phosphorylation in vivo.
Supplementary Material
Acknowledgments
Funding: This work was supported by National Institutes of Health grant 5R01GM080568 (to S.L.C.)
We thank Dr. Greg Young and Dr. Ashutosh Tripathy for technical support in collecting the NMR and CD data, respectively. We also thank Dr. Mary Carroll in the laboratory of Dr. Drew Lee for help in analyzing the NMR relaxation data and Min-Qi Lu for extensive help in sample preparation.
Abbreviations
- FAT
focal adhesion targeting
- FAK
focal adhesion kinase
- FERM
protein 4.1, ezrin, radixin, moesin
- Grb2
growth factor receptor bound protein 2
- SH
Src homology
- NHE1
sodium/proton exchanger 1
- CAD
collision activated dissociation
- FDR
false discovery rate
- XIC
extracted ion chromatograms
- RDC
residual dipolar coupling
- HetNOE
heteronuclear nuclear Overhauser effect
- μFT-ICR
microelectrospray fourier transform ion cyclotron resonance
- ECD
electron capture dissociation
Footnotes
SUPPORTING INFORMATION PARAGRAPH Determination of the sites of phosphorylation by mass spectrometry including quantitation strategy and MS/MS spectra of phosophopeptides, titiration profiles of histidine residues, pH-dependent RDC data, and pH-dependent NOE data for Y926 and Y1008. This material is available free of charge via the Internet at http://pubs.acs.org
References
- 1.Schlaepfer DD, Hunter T. Focal Adhesion Kinase Overexpression Enhances Ras-dependent Integrin Signaling to ERK2/Mitogen-activated Protein Kinase through Interactions with and Activation of c-Src. The Journal of Biological Chemistry. 1997;272:13189–13195. doi: 10.1074/jbc.272.20.13189. [DOI] [PubMed] [Google Scholar]
- 2.Ren X, Kiosses W, Sieg D, Otey C, Schlaepfer D, Schwartz M. Focal adhesion kinase suppresses Rho activity to promote focal adhesion turnover. J Cell Sci. 2000;113:3673–3678. doi: 10.1242/jcs.113.20.3673. [DOI] [PubMed] [Google Scholar]
- 3.Hildebrand J, Taylor J, Parsons J. An SH3 domain-containing GTPase-activating protein for Rho and Cdc42 associates with focal adhesion kinase. Mol Cell Biol. 1996;16:3169–3178. doi: 10.1128/mcb.16.6.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomar A, Lim S, Lim Y, Schlaepfer D. A FAK-p120RasGAP-p190RhoGAP complex regulates polarity in migrating cells. J Cell Sci. 2009;122:1852–1862. doi: 10.1242/jcs.046870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ilic D, Furuta Y, Kanazawa S, Takeda N, Sobue K, Nakatsuji N, Nomura S, Fujimoto J, Okada M, Yamamoto T, Aizawa S. Reduced cell motility and enhanced focal adhesion contact formation in cells from FAK-deficient mice. Nature. 1995;377:539–544. doi: 10.1038/377539a0. [DOI] [PubMed] [Google Scholar]
- 6.Webb DJ, Donais K, Whitmore LA, Thomas SM, Turner CE, Parsons JT, Horwitz AF. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nature Cell Biology. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 7.Zhao J, Reiske H, Guan J. Regulation of the cell cycle by focal adhesion kinase. J Cell Biol. 1998;143:1997–2008. doi: 10.1083/jcb.143.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ryu SJ, Cho KA, Oh YS, Park SC. Role of Src-specific phosphorylation site on focal adhesion kinase for senescence-associated apoptosis resistance. Apoptosis. 2006;11:303–313. doi: 10.1007/s10495-006-3978-9. [DOI] [PubMed] [Google Scholar]
- 9.Hungerford J, Compton M, Matter M, Hoffstrom B, Otey C. Inhibition of pp125FAK in cultured fibroblasts results in apoptosis. J Cell Biol. 1996;135:1383–1390. doi: 10.1083/jcb.135.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlaepfer DD, Mitra SK. Multiple Connections link FAK to Cell Motility and Invasion. Current Opinion in Genetics and Development. 2004;14:92–101. doi: 10.1016/j.gde.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Ilic D, Almeida EAC, Schlaepfer DD, Dazin P, Aizawa S, Damsky CH. Extracellular Matrix Survival Signals Transduced by Focal Adhesion Kinase Suppresses p53-mediated Apoptosis. J Cell Biol. 1998;143:547–560. doi: 10.1083/jcb.143.2.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu L, Owens L, Sturge G, Yang X, Liu E, Craven R, Cance W. Attenuation of the expression of the focal adhesion kinase induces apoptosis in tumor cells. Cell Growth Differ. 1996;7:413–418. [PubMed] [Google Scholar]
- 13.Tachibana K, Sato T, D’Avirro N, Morimoto C. Direct Assocation of pp125FAK with Paxillin, the Focal Adhesion-Targeting Mechanism of pp125FAK. Journal of Experimental Medicine. 1995;182:1089–1100. doi: 10.1084/jem.182.4.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlaepfer DD, Hunter T. Evidence for in vivo phosphorylation of the Grb2 SH2-domain binding site on focal adhesion kinase by Src-family protein-tyrosine kinases. Mol Cell Biol. 1996;16:5623–5633. doi: 10.1128/mcb.16.10.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katz BZ, Romer L, Miyamoto S, Volberg T, Matsumoto K, Cukierman E, Geiger B, Yamada KM. Targeting Membrane-localized Focal Adhesion Kinase to Focal Adhesions: Roles of Tyrosine Phosphorylation and Src Family Kinases. The Journal of Biological Chemistry. 2003;278:29115–29120. doi: 10.1074/jbc.M212396200. [DOI] [PubMed] [Google Scholar]
- 16.Kaneda T, Sonoda Y, Ando K, Suzuki T, Sasaki Y, Oshio T, Tago M, Kasahara T. Mutation of Y925F in focal adhesion kinase (FAK) suppresses melanoma cell proliferation and metastasis. Cancer Letters. 2008 doi: 10.1016/j.canlet.2008.05.042. [DOI] [PubMed] [Google Scholar]
- 17.Mitra S, Mikolon D, Molina J, Hsia D, Hanson D, Chi A, Lin S-T, Bernard-Trifilo J, Ilic D, Stupack D, Cheresh D, Schlaepfer D. Intrinsic FAK Activity and Y925 Phosphorylation Facilitate an Angiogenic Switch in Tumors. Oncogene. 2006:1–16. doi: 10.1038/sj.onc.1209588. [DOI] [PubMed] [Google Scholar]
- 18.Ezratty E, Partridge M, Gundersen G. Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat Cell Biol. 2005;7:581–590. doi: 10.1038/ncb1262. [DOI] [PubMed] [Google Scholar]
- 19.Prutzman KC, Gao G, King ML, Iyer VV, Mueller GA, Schaller MD, Campbell SL. The Focal Adhesion Targeting Domian of Focal Adhesion Kinase Contains a Hinge Region that Modulates Tyrosine 926 Phosphorylation. Structure. 2004;12:881–891. doi: 10.1016/j.str.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 20.Hayashi I, Vuori K, Liddington RC. The Focal Adhesion Targeting (FAT) Region of Focal Adhesion Kinase is a Four-Helix Bundle that Binds Paxillin. Nature Structural Biology. 2002;9:101–106. doi: 10.1038/nsb755. [DOI] [PubMed] [Google Scholar]
- 21.Arold ST, Hoellerer MK. The Structural Basis of Localization and Signaling by the Focal Adhesion Targeting Domain. Structure. 2002;10:319–327. doi: 10.1016/s0969-2126(02)00717-7. [DOI] [PubMed] [Google Scholar]
- 22.Dixon RDS, Chen Y, Ding F, Khare SD, Prutzman KC, Schaller MD, Campbell SL, Dokholyan NV. New Insights into FAK Signaling and Localization Based on Detection of a FAT Domain Folding Intermediate. Structure. 2004;12:2161–2171. doi: 10.1016/j.str.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Z, Feng H, Bai Y. Detection of a Hidden Folding Intermediate in the Focal Adhesion Targetin Domain: Implications for its Function and Folding. Proteins: Structure, Function, and Bioinformatics. 2006;65:259–265. doi: 10.1002/prot.21107. [DOI] [PubMed] [Google Scholar]
- 24.Hubbard SR. Crystal Structure of the Activated Insulin Receptor Tyrosine Kinase in Complex with Peptide Substrate and ATP Analog. The EMBO Journal. 1997;16:5573–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Songyang Z, Shoelson SE, McGlade J, Olivier P, Pawson T, Bustelo XR, Barbacid M, Sabe H, Hanafusa H, Yi T. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol Cell Biol. 1994;14:2777–2785. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahuel J, Gay B, Erdmann D, Strauss A, Garcia-Echeverria C, Furet P, Caravatti G, Fretz H, Schoepfer J, Grutter MG. Structural basis for specificity of GRB2-SH2 revealed by a novel ligand binding mode. Nature Structural and Molecular Biology. 1996;3:586–589. doi: 10.1038/nsb0796-586. [DOI] [PubMed] [Google Scholar]
- 27.Ogura K, Tsuchiya S, Terasawa H, Yuzawa S, Hatanaka H, Mandiyan V, Schlessinger J, Inagaki F. Solution Structure of the SH2 Domain of Grb2 Complexed with the Shc-derived Phosphotyrosine-containing Peptide. J Mol Biol. 1999;289:439–445. doi: 10.1006/jmbi.1999.2792. [DOI] [PubMed] [Google Scholar]
- 28.Nioche P, Liu WQ, Broutin I, Charbonnier F, Latreille MT, Vidal M, Roques B, Garbay C, Ducruix A. Crystal structures of the SH2 domain of grb2: highlight on the binding of a new high-affinity inhibitor. Journal of Molecular Biology. 2002;315:1167–1177. doi: 10.1006/jmbi.2001.5299. [DOI] [PubMed] [Google Scholar]
- 29.Seeliger MA, Young M, Henderson MN, Pellicena P, King DS, Falick AM, Kuriyan J. High Yield Bacterial Expression of Active c-Able and c-Src Tyrosine Kinases. Protein Science. 2006;14:3135–3139. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunawardena H, Huang Y, Kenjale R, Wang H, Xie L, Chen X. Unambiguous characterization of site-specific phosphorylation of leucine-rich repeat Fli-I-interacting protein 2 (LRRFIP2) in Toll-like receptor (TLR4)-mediated signaling. J Biol Chem. 2011;286:10897–10910. doi: 10.1074/jbc.M110.168179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beausoleil S, Villen J, Gerber S, Rush J, Gygi S. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 32.John DM, Weeks KM. van’t Hoff enthalpies without baselines. Protein Science. 2000;9:1416–1419. doi: 10.1110/ps.9.7.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen DL, Pielak GJ. Baseline length and automated fitting of denaturation data. Protein Science. 1998;7:1262–1263. doi: 10.1002/pro.5560070524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectal processing system based on UNIX pipes. J Biom NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 35.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 36.Pascal SM, Muhandiram DR, Yamazaki T, Forman-Kay JD, Kay LE. Simultaneous acquisition of 15N and 13C NOE spectra of proteins in H2O. J Magn Reson Series B. 1994;103:197–201. [Google Scholar]
- 37.Grzesiek S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance nmr. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 38.Wittekind M, Mueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha-and beta-resonances in proteins. J Magn Reson Series B. 1993;101:201–205. [Google Scholar]
- 39.Ottiger M, Delaglio F, Bax A. Measurement of J and dipolar couplings from simplified two-dimensional NMR specra. J Magn Reson. 1998;131:373–378. doi: 10.1006/jmre.1998.1361. [DOI] [PubMed] [Google Scholar]
- 40.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone Dynamics of a Free and a Phosphopeptide-Complexed Src Homology 2 Domain Studied by 15N NMR Relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 41.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. Journal of the American Chemical Society. 1982;104:4559. [Google Scholar]
- 42.Dellwo MJ, Wand AJ. Model-independent and model-dependent analysis of the global and internal dynamics of cyclosporin A. J Am Chem Soc. 1989;111:4571–4578. [Google Scholar]
- 43.Lee L, Rance M, Chazin W, Palmer A. Rotational diffusion anisotropy of proteins from simultaneous analysis of 15N and 13C alpha nuclear spin relaxation. J Biomol NMR. 1997;9:287–298. doi: 10.1023/a:1018631009583. [DOI] [PubMed] [Google Scholar]
- 44.Osborne M, Wright P. Anisotropic rotational diffusion in model-free analysis for a ternary DHFR complex. J Biomol NMR. 2001;19:209–230. doi: 10.1023/a:1011283809984. [DOI] [PubMed] [Google Scholar]
- 45.Clarkson MW, Gilmore SA, Edgell MH, Lee AL. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry. 2006;45:7693–7699. doi: 10.1021/bi060652l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee AL, Flynn PF, Wand AJ. Comparison of H-2 and C-13 NMR relaxation techniques for the study of protein methyl group dynamics in solution. J Am Chem Soc. 1999;121:2891–2902. [Google Scholar]
- 47.Katz BZ, Romer L, Miyamoto S, Volberg T, Matsumoto K, Cukierman E, Geiger B, Yamada KM. Targeting membrane-localized focal adhesion kinase to focal adhesions: roles of tyrosine phosphorylation and SRC family kinases. J Biol Chem. 2003;278:29115–29120. doi: 10.1074/jbc.M212396200. [DOI] [PubMed] [Google Scholar]
- 48.Ciccimaro E, Hevko J, Blair IA. Analysis of phosphorylation sites on focal adhesion kinase using nanospray liquid chromatography/multiple reaction monitoring mass spectrometry. Rapid Communications in Mass Spectrometry. 2006;20:3681–3692. doi: 10.1002/rcm.2783. [DOI] [PubMed] [Google Scholar]
- 49.Songyang Z, Carraway KL, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, Lorenzo MJ, Ponder BAJ, Mayer BJ, Cantley LC. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 50.Mauldin RV, Lee AL. Nuclear magnetic resonance study of the role of M42 in the solution dynamics of Escherichia coli dihydrofolate reductase. Biochemistry. 2010;49:1606–1615. doi: 10.1021/bi901798g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Amero CD, Byerly DW, McElroy CA, Simmons A, Foster MP. Ligand-Induced Changes in the Structure and Dynamics of Escherichia coli Peptide Deformylase. Biochemistry. 2009;48:7595–7607. doi: 10.1021/bi900600b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.