Abstract
Frataxin is a mitochondrial protein with a central role in iron homeostasis. Defects in frataxin function lead to Friedreich’s ataxia, a progressive neurodegenerative disease with childhood onset. The function of frataxin has been shown to be closely associated with its ability to form oligomeric species; however, the factors controlling oligomerization and the types of oligomers present in solution is a matter of debate. Using small-angle X-ray scattering (SAXS), we found that Co2+, glycerol and a single amino acid substitution at the N-terminus, Y73A, facilitate oligomerization of yeast frataxin, resulting in a dynamic equilibrium between monomers, dimers, trimers, hexamers, and higher order oligomers. Using X-ray crystallography, we found that Co2+ binds inside the channel at the 3-fold axis of the trimer, which suggests that the metal has an oligomer-stabilizing role. The results reveal the types of oligomers present in solution and support our earlier suggestions that the trimer is the main building block of yeast frataxin oligomers. They also indicate that different mechanisms may control oligomer stability and oligomerization in vivo.
Keywords: Protein oligomerization, Protein flexibility, metal chaperone, Friedreich's ataxia, Neurodegenerative diseases
Introduction
Iron is one of the most abundant elements on earth and it is essential for most living organisms, despite the toxic effects associated with it. Thus, in an O2-containing environment and at physiological pH, Fe2+ (6 H20) is readily oxidized to the acidic Fe3+ form, which rapidly produces insoluble Fe(OH)3. In addition, the one-electron oxidation of iron by oxygen produces superoxide that can in turn react to form hydrogen peroxide. Reaction of hydrogen peroxide with Fe2+ produces the highly reactive and toxic hydroxyl radical through the Fenton reaction. Thus, unligated iron carries a potential danger for cells and may have implication in a number of human diseases, which explains the need for a tight control of iron chemistry in organisms 1,2. Two proteins, ferritin and frataxin, are known to be central to the storage and detoxification of iron in cells. The importance of frataxin was first shown by the discovery that the autosomal recessive disease Friedreich’s ataxia (FRDA) is linked to the presence of expanded GAA repeats in the first intron of the frataxin-encoding gene, and also to mutations in the gene sequence coding for the protein 3. FRDA affects approximately one in 40,000 individuals, with early onset—often before the age of 15. Frataxin deficiency results in aberrations of cellular iron homeostasis and high production of reactive oxygen species (ROS), leading to a progressive neurodegenerative disease that is also characterized by cardiomyopathy and diabetes mellitus 4; 5; 6.
Both ferritin and frataxin and their homologs have been implicated in the control of iron detoxification, which these proteins achieve by catalyzing the oxidation of ferrous iron to the ferric form and by storing it as a ferrihydrite biomineral within oligomeric species 7; 8; 9; 10; 11. In addition, the role of frataxin in iron homeostasis also includes iron delivery for iron-sulfur cluster synthesis, heme biosynthesis, and aconitase repair 12; 13; 14. Frataxin species from humans and Saccharomyces cerevisiae (Yfh1) have been studied most extensively. They are expressed in the cytoplasm as 210-residue and 174-residue precursor polypeptides with N-terminal mitochondrial-targeting sequences. Proteolytic cleavage by mitochondrial processing peptidase results in the 52–174 mature form of yeast frataxin, while the human protein appears to exist as several variants with N-termini of different lengths (see below) 15; 16; 17; 18.
The function of ferritin and frataxin has been strongly linked to their ability to form oligomeric species. In the case of ferritin, these oligomers are spontaneously assembled in cells, while for frataxin and its homologs there appears to be variation in the propensity of the proteins from different sources to form oligomers and in the type of oligomers formed 19; 20; 21. In the presence of ferrous iron and oxygen, yeast frataxin has been shown to undergo stepwise assembly from monomers to larger oligomers that can contain up to 24 subunits, and even larger complexes 19. Higher-order oligomers can store up to ~50–75 iron atoms per subunit in 1–2-nm cores 11; 19; 22. The oligomeric species of yeast frataxin can be easily dissolved into monomers, for example by the addition of reducing agents 19, suggesting that iron plays an active role in oligomer stabilization. In humans, at least 4 variants of frataxin have been isolated. The variants with the longer N-termini (56–210 and 42–210) can often assemble into larger structures in vivo and during expression in E. coli 16; 18. The large oligomers can be disassembled irreversibly into stable monomers by the addition of SDS 20. However, in contrast to the yeast Yfh1 protein, purified monomers of human frataxin do not appear to form oligomers in vitro, even in the presence of iron 16. This may indicate that assembly of the human protein requires additional assistance of some kind. Indeed, it has been shown that the Hsp70-type protein SSQ1 is required for Yfh1 to function in S. cerevisiae 23; 24. The two other known variants of human frataxin, 81–210 and 78–210, have been found to be unable to adopt higher-order oligomeric states 18; 25, indicating that residues 56–77 at the N-terminus of the protein are essential for oligomerization.
X-ray crystallographic studies of a variant of yeast frataxin in which amino acid Y73 was replaced by an alanine (Y73A) showed that this protein crystallized as a trimer, apparently stabilized by extensive interactions between the monomers around the threefold axis as well as by the N-terminus, which bridged the monomers by forming additional interactions between them 26. Size exclusion chromatography also showed that in contrast to wild-type Yfh1, which requires iron for assembly, the Y73A variant spontaneously assembled into larger oligomeric species that contained up to 24 monomers 26. In addition, single-particle reconstruction studies of both iron-loaded and iron-free 24-meric particles clearly demonstrated that they were built up of trimeric units 22; 26, suggesting that the trimer might be the main building block of larger frataxin oligomers. Interestingly, the arrangement of the functional features found in this structure, such as the position of the ferroxidation and iron mineralization sites and charge distribution inside and around the channel at the threefold axis, has striking similarities to the arrangement of the corresponding features in ferritin, despite the absence of any evolutionary relationships at the amino acid sequence level or at the three-dimensional structural level 26.
Given these data, the type of frataxin oligomers that exist in solution and the role of the different factors that control oligomer stability, is poorly understood. In the current work, using a combination of small-angle X-ray scattering, dynamic light scattering and X-ray crystallography, we studied the oligomeric states of yeast frataxin in solution and the effects of glycerol, Co2+ and amino acid substitution at the N-termini on these states. X-ray crystallography was used to reveal the cobalt binding site and its potential role in oligomer stabilization.
Results
Structure of yeast frataxin in solution
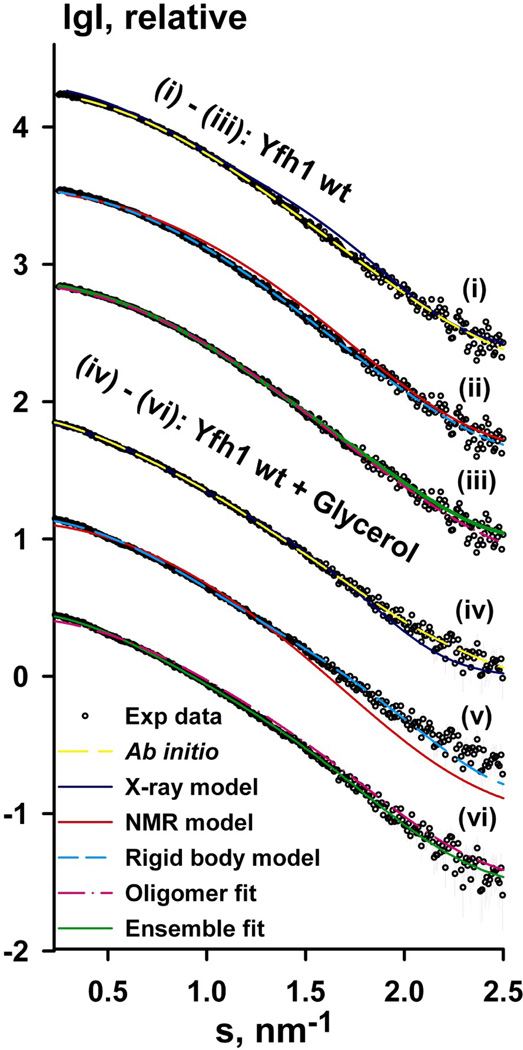
To gain some insight into the solution structure of wild type yeast frataxin (Yfh1wt; residues 52–174) under different conditions, we used SAXS. The advantages of using SAXS lie in the ability of this method to provide structural information for samples directly in solution. The method tolerates a wide variety range of protein or protein complex sizes 27 and has successfully been used for modeling of proteins with rather short flexible parts, e.g. hinge regions in multidomain proteins 28, as well as natively unfolded proteins 29; 30. The results of the SAXS experimental measurements, solution structure modeling (using the available X-ray and NMR model coordinates of yeast frataxin) and model fitting to the experimental data are summarized in Figure 1a and Table 2. Since no concentration effects were observed in the experiments with glycerol, data with the highest concentration were used for analysis and structural modeling (Figure 1a, iv–vi). The radius of gyration (Rg), estimated using Guinier approximation, was 1.80 ± 0.1 and 2.25 ± 0.1 nm for Yfh1wt and Yfh1wt + glycerol, respectively. Given that the ratio Vp/MW (Vp is the Porod volume, MW is the molecular weight) is expected to be approximately 1:1.7, the volume in the absence of glycerol is consistent with the molecular weight of the frataxin monomer (13.7 kDa). In the presence of glycerol, the volume is significantly greater, indicating the presence of higher-order oligomers. Theoretical scattering calculated for the X-ray structure of the Y73A variant of yeast frataxin yielded an Rg value of 2.07 nm (the new X-ray model, see below), while for the NMR structure of Yfh1wt (2GA5.PDB) the different conformers yielded Rg values in the range 1.54–1.72 nm. Although these values are close to the Rg estimated from the SAXS data for the protein without glycerol, as seen in Figure 1a (i–ii, Yfh1wt), and (iv–v, Yfh1wt + glycerol) neither the X-ray model nor any of the NMR models provided satisfactory fit to the experimental SAXS data (the corresponding discrepancies χ are shown in Table 2). We also estimated the maximum dimensions (Dmax, see Table 2) and the interatomic distance distribution function p(r) of Yfh1wt in the presence and absence of glycerol using GNOM 31. The p(r) for Yfh1wt + glycerol showed shoulders at larger distances and increase in Dmax value (Table 2), as compared to Yfh1wt. These features are typical of dynamic systems 32.
Figure 1. SAXS measurements of yeast frataxin.
(a) Experimental SAXS profiles (experimental data shown as black circles) for wild-type Yfh1wt (i–iii) and Yfh1wt + glycerol (iv–vi) were appropriately displaced along the logarithmic axis for better visualization and overlaid on the corresponding fits of the X-ray structure (i, iv), ab initio model (ii, v), lowest-energy NMR conformer (denoted NMR model on the figure, PDB ID: 2GA5), rigid body model from OLIGOMER (denoted oligomer fit) and the EOM ensemble (denoted Ensemble fit, iii, vi). Experimental SAXS data are shown to a maximal momentum transfer of s = 2.5 nm−1. See also Figure S1b for plots of the Guinier region.
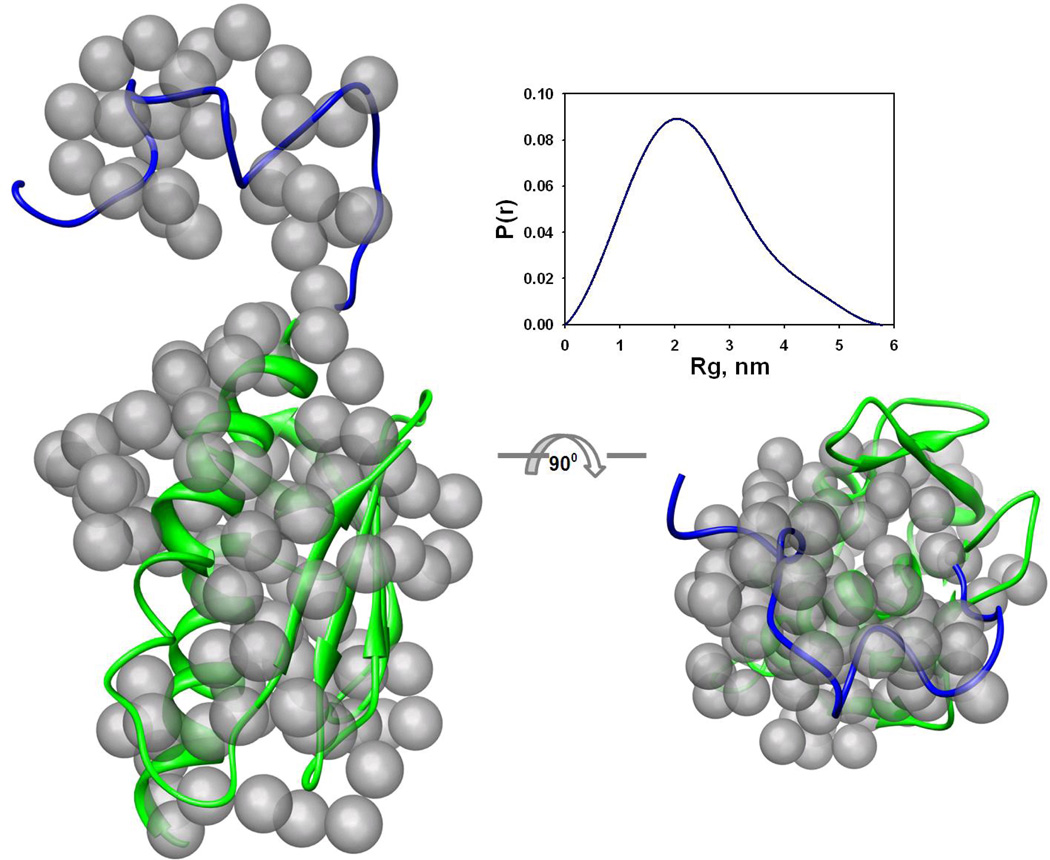
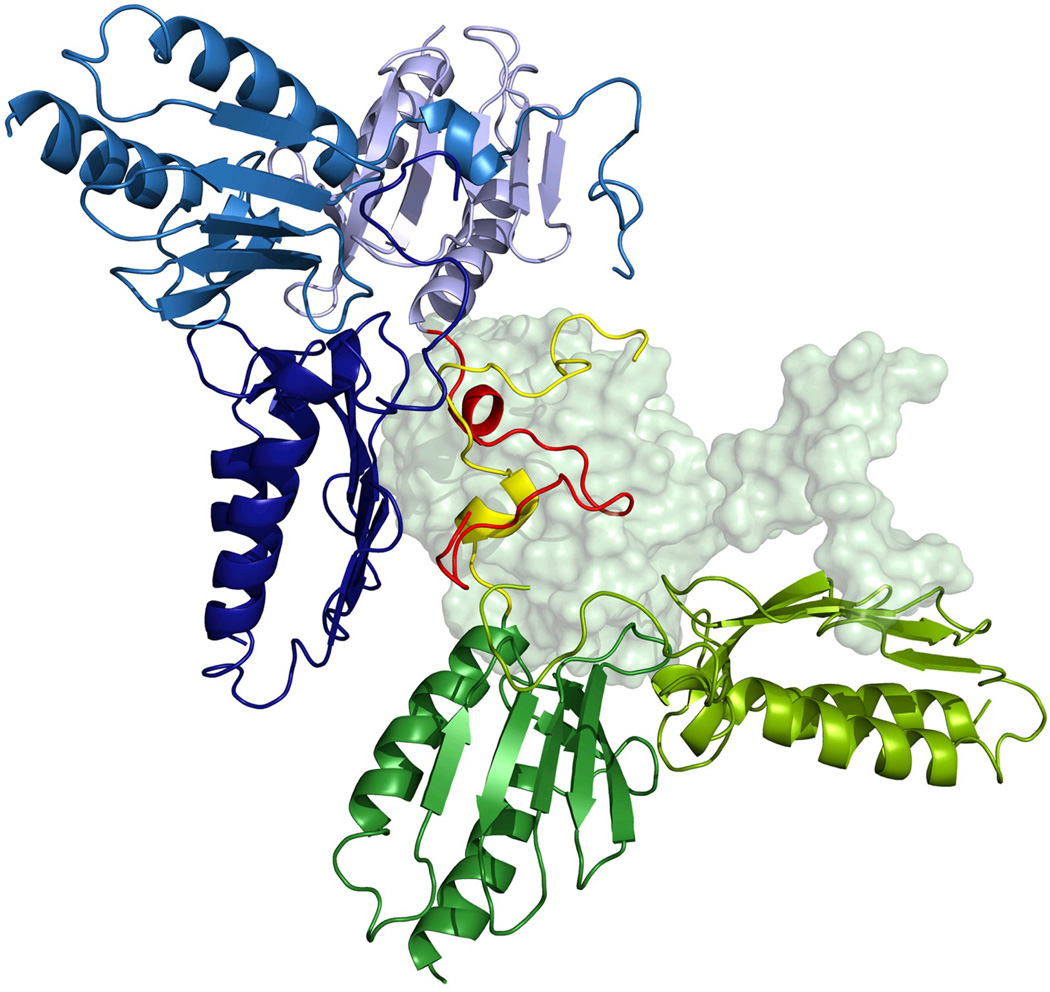
(b) Superposition of the reference ab initio model on the rigid body model. Transparent gray beads represent the reference ab initio model generated by GASBOR. The view on the right has been rotated by 90° about the horizontal axis. The cartoon representation is used for the BUNCH model of frataxin (see text). The blue part corresponds to the modeled N-terminus, and green to the available high-resolution structure used for modeling. The figures were prepared using the CHIMERA molecular graphics system 70. The insert shows the distance distribution function p(r) used for ab initio modeling.
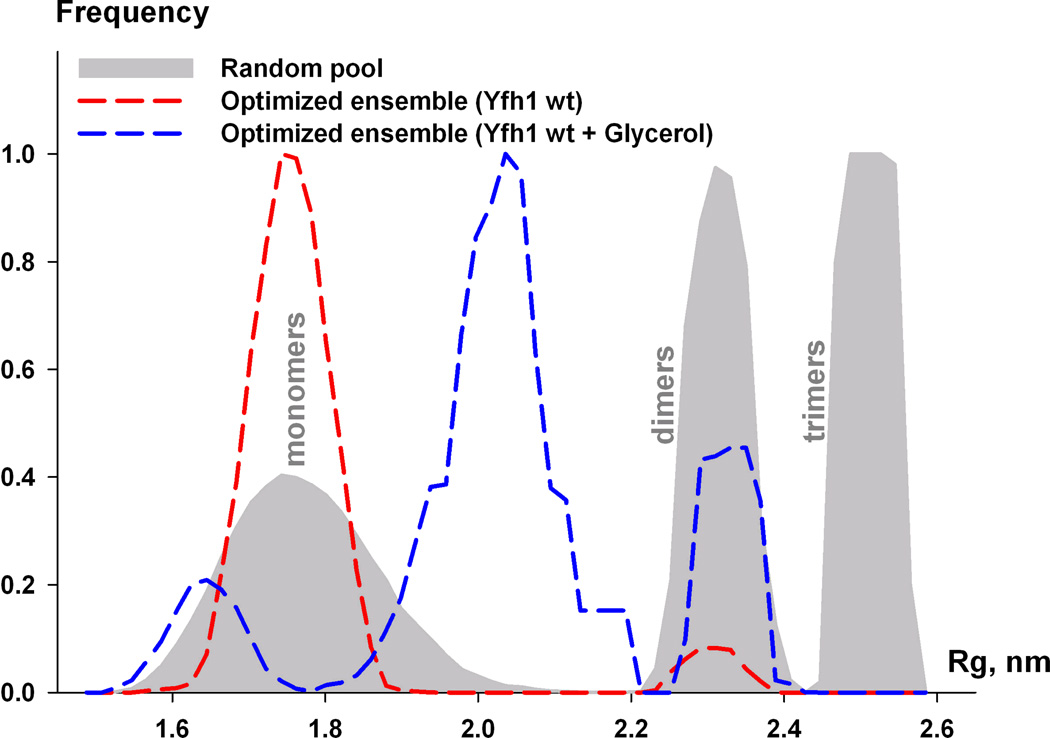
(c) Flexibility of frataxin in solution. Rg distribution for the random pool (6,000 models) containing equal fractions of monomers, dimers, and trimers is shown by the area filled with gray. Rg distribution of optimized ensemble corresponding to wild-type Yfh1 (red dashed line) and wild-type Yfh1 + glycerol (blue dashed line).
Table 2.
The results of SAXS measurements on the monomeric yeast frataxin in the absence and presence of glycerol
| Sample | Rg(nm) | Dmax(nm) | Ve(nm3) | Vp (nm3) | χg | χx | χn | χo | χb | χf |
|---|---|---|---|---|---|---|---|---|---|---|
| Yfh1wt Yfh1wt + glycerol |
1.8 2.25 |
6.8 7.5 |
28 32.2 |
24 44 |
1.16 1.36 |
1.19 1.2 |
1.16 3.26 |
1.03 2.53 |
1.04 1.4 |
1.02 1.01 |
Rg(nm), Dmax, Ve(nm3) and Vp(nm3) are radius of gyration, maximum size, excluded volume, and Porod volume, respectively. χg, discrepancy of GASBOR ab initio models; χx, discrepancy to new X-ray structure; χn, discrepancy to lowest-energy NMR conformer (model #1 in 2GA5.PDB); χb, discrepancy to the BUNCH model; χo, OLIGOMER discrepancy to twenty NMR conformers and X-ray model; χf, discrepancy to EOM ensemble model.
For the interpretation of the SAXS data we subsequently performed ab initio modelling using the DAMMIN software. Ten ab initio models independently generated using DAMMIN had a mean normalized spatial discrepancy (NSD), as calculated by DAMAVER, in the range 0.54–0.59 and 0.52–0.58 for Yfh1wt data and the Yfh1wt + glycerol data, respectively. The best ab initio model had a fit to the experimental scattering of χ = 1.16 (Table 2). At the next step higher resolution ab initio models were generated using GASBOR. Twenty GASBOR models had NSD in the range 0.9–0.96, which illustrates that all shapes were essentially similar and that a unique solution had been identified. The best GASBOR model was selected for rigid body superposition with the model of the molecule (Figure 1b).
The NMR model and the X-ray structure suggest that yeast frataxin monomer has a highly flexible N-terminus, which may explain the above mentioned poor agreement of the Xray and NMR models (PDB entry 2GA5) with the experimental scattering data. To fit the NMR and X-ray models to the SAXS data, a combination of ab initio and rigid body modeling approaches was employed to re-model the N-terminus, using the program BUNCH. During this process, the interatomic distances and angles between ‘dummy’ atoms of the ab initio model were constrained to mimic the Cα peptide chain. Ten BUNCH models were generated for the different data sets. The models showed good agreement with the Yfh1wt scattering data (s range from 0.020 to 2.50 nm−1) with χ in the range 1.04–1.40, but rather poor agreement with the Yfh1wt + glycerol data, with χ in the range 1.4–2.4 (compare Figure 1a (ii) and (v), red curve). Furthermore, assuming that different frataxin conformations exist in solution in equilibrium with each other, we used OLIGOMER to calculate the volume fractions of the separate NMR conformers contributing to the scattering profile. The results showed that only two conformers, # 8 and 16 (PDB entry 2GA5), contributed 32% and 68% volume fractions, respectively, to the scattering. This further improved the discrepancy of the fitting to the Yfh1wt data, as compared to the lowest-energy NMR conformer or the X-ray structure, but not for the case of the data in the presence of glycerol. This can be seen from the comparison of the discrepancy values in Table 2 and is also shown in Figure 1a for the data in the absence and presence of glycerol (pink curves, Figure 1a (iii) and (vi), respectively). In the latter case the poor fit of the model to the data is clearly visible in the low-resolution range. We also compared the reference ab initio model generated by GASBOR to the rigid body model with the lowest discrepancy generated by BUNCH (see Figure 1b). The superposition showed very good agreement, indicating that both reconstructions converged to a similar solution. It is noteworthy, that both ab initio and rigid body models are likely to represent an average over a set of conformations in solution and that the good agreement between them suggests validity of such models.
To improve the fit for the case of Yfh1wt + glycerol and quantitatively characterize the ensemble properties of Yfhwt and Yfh1wt + glycerol in solution, the ensemble optimization method (EOM) was applied to the measured data sets. The EOM analysis allowed us to further improve the fit to the experimental data sets (see Figure 1a (iii, vi), green curves, and Table 2 (χf)). As seen from Figure 1c, Rg distributions corresponding to Yfh1wt and Yfh1wt + glycerol ensembles show considerable differences. Thus, in the case of Yfh1wt, only a negligible fraction of dimers was selected. If a random pool only contained monomers, the discrepancy did not change as compared to the mixed full pool. On the other hand, the Yfh1wt + glycerol data could only be fitted with more extended monomers and a larger fraction of dimers, as compared to Yfh1wt. The data could not be fitted if a random pool only contained monomers. On the other hand, if only three curves per ensemble were allowed to be selected, dimers were always present in the ensemble and the discrepancy was in the range 1.06–1.07. The average radius of gyration of the EOM ensemble, as seen from Figure 1c, was found to be close to the Rg calculated by Guinier approximation (1.80 ± 0.1 and 2.25 ± 0.1 nm for Yfh1wt and Yfh1wt + glycerol, respectively, see above).
The results clearly show that Yfh1wt in solution exists in different conformations, and that the primary source of the conformational heterogeneity is the high flexibility of the N-terminus. Furthermore, while experimental SAXS data for Yfh1wt can be described with an ensemble model, mainly consisting of monomers, a dynamic monomer-dimer equilibrium exists in the case of Yfh1wt + glycerol. Hence, the addition of glycerol apparently results in the formation of dimers, which presumable could be considered as the first stage in the oligomerization process of frataxin.
Characterization of the Yfh1 Y73A variant oligomers using SAXS
Gel filtration studies have shown that the Yfh1 Y73A variant is eluted as two major fractions, one of lower molecular weight, suggested to represent a trimer, and the second of higher molecular weight, suggested to be a 24-mer. As noted above, the low molecular weight fraction was crystallized and revealed the trimeric frataxin structure, while the high molecular weight fraction was studied by single-particle EM reconstruction and revealed a 24-meric particle built up of 8 trimers 22; 26. For a better understanding of the role of the Y73A replacement on the oligomerization propensity of the protein, we studied the low molecular weight fraction using DLS (Figure 2a) and SAXS (Figure 2b and Table 3). The DLS measurements, analysis of the SAXS data and structural models generated to fit the data (see methods section) indeed reveal the presence of several oligomeric species in solution. As seen from Table 3, while the major part of the protein in solution is in monomeric form (around 60%), there is also a proportion of trimers, hexamers, and dodecamers (9%, 20%, and 11%, respectively). Two different models (based on NMR conformers and kinematic loop modeling) were needed to model the monomers, which confirm the high flexible nature of the monomeric species. With this pool of monomers and oligomers, the generated models could fit the SAXS data with χ2 = 0.44, as estimated by the program OLIGOMER. There were no signs of protein aggregation, as shown by the analysis of the Guinier region of the data (Figure S1a). These results clearly showed that Y73A modification of yeast frataxin results in a higher propensity to form higher-order oligomers, independently of the presence of metals, perhaps as a result of reduced flexibility of the N-terminal region of the protein. During crystallization, the trimeric form is presumably selected as the most stable in the formation of the crystal lattice.
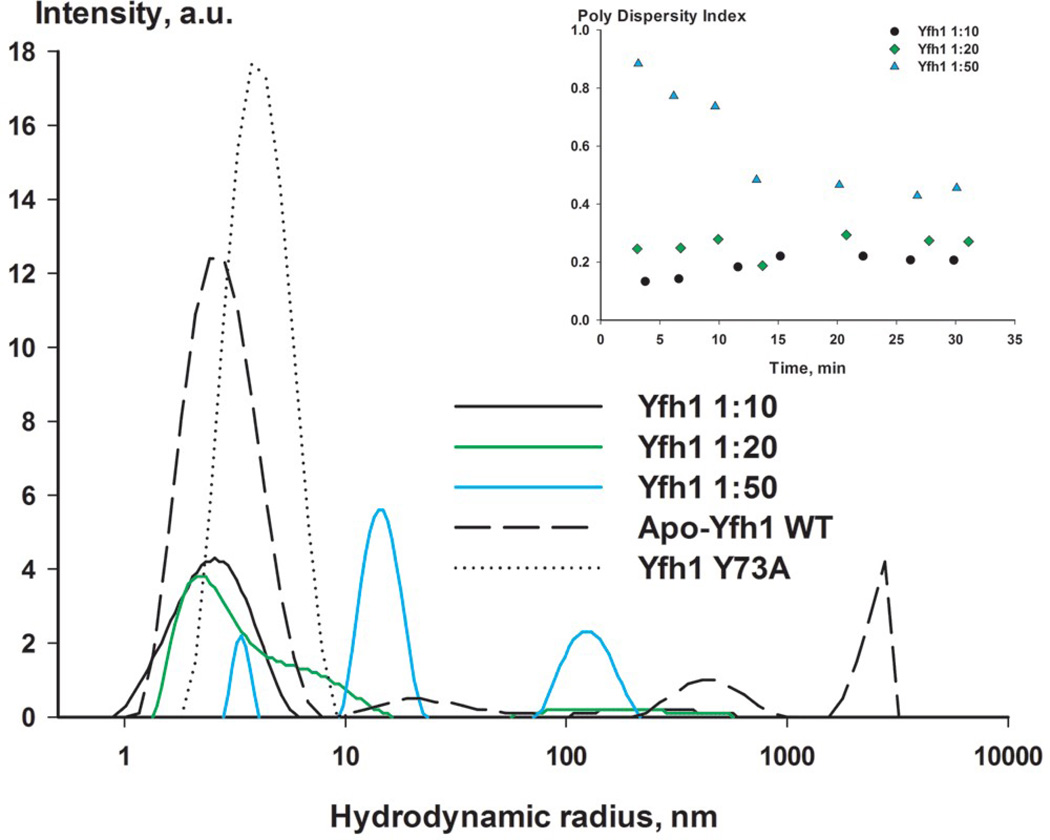
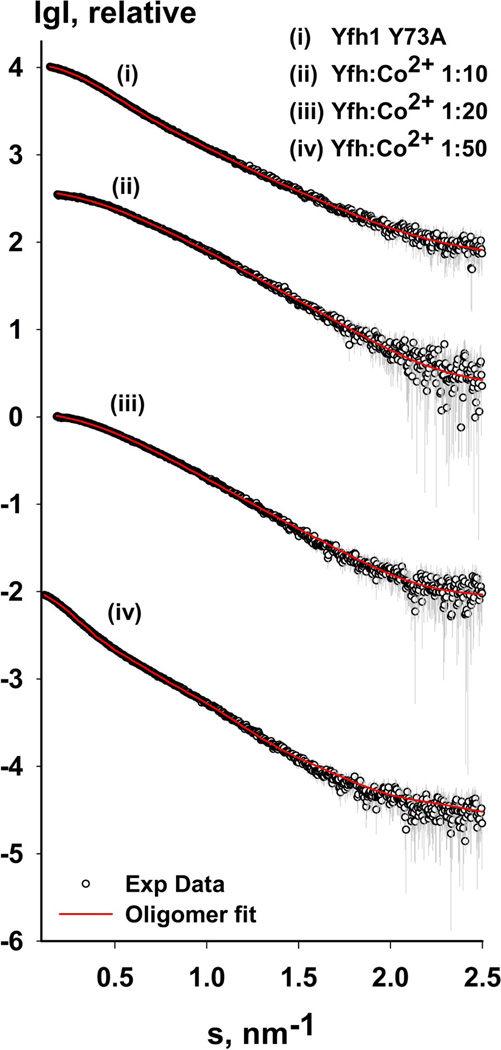
Figure 2.
Oligomerization of yeast frataxin homolog. (a) Sample size distribution (hydrodynamic radius) observed using DLS. Wild-type Yfh1 and the Y73A variant are shown in gray; Yfh1:CoCl2 ratios (1:10, 1:20, and 1:50) are shown in black, green, and blue, respectively. The insert shows the time-dependence of the polydispersity index after addition of CoCl2. (b) Experimental SAXS profiles (with experimental data shown as black circles) for the Y73A variant of Yfh1 and wild-type Yfh1 in the presence of different amounts of metal, as shown in the figure. The corresponding fit curve is overlaid on each profile (red) (see Table 3 for details). The data are shown to a maximum momentum transfer of s = 2.5 nm−1. See also Figure S1a for plots of the Guinier region.
Table 3.
The combinations of the models with different oligomeric states used for fitting to the experimental data (Figure 2b) are shown together with their respective distributions in the mixture and the discrepancy (χ2). Rg and Dmax are radius of gyration and maximum size, respectively
| Sample | Rg (nm) | Dmax (nm) | Oligomer Model | Distribution (%) | χ2 |
|---|---|---|---|---|---|
| Y73A Yfh1 | 3.2±0.1 | 11.5±0.5 | 1a/1b/3c/6e/12f | 36/24/9/20/11 | 0.44 |
| 1:10 Yfh1:Co[II] | 2.3±0.1 | 8±0.5 | 1a/3d/6e | 67/24/9 | 0.55 |
| 1:20 Yfh1:Co[II] | 2.5±0.1 | 9±0.5 | 1a/3d/6e | 54/33/13 | 0.55 |
| 1:50 Yfh1:Co[II] | 5.4±0.1 | 19±0.5 | 1a/3d/6e/42g | 26/57/6/11 | 0.67 |
NMR conformer(s),
Kinematic loop model(s),
Tilted trimer from Rosetta docking,
Trimer with rebuilt N-terminus using Rosetta,
Hexamer from EM density docking,
Dodecamer from EM density docking,
42-mer from combining two EM-docked 24-mers and removing two overlapping trimers.
Dynamic light scattering studies of cobalt-induced oligomerization of yeast frataxin
To further investigate the influence of various factors on the oligomerization of yeast frataxin, we used DLS to study the effects of Mg2+, Zn2+ and Co2+. The choice of divalent metals was dictated by the available biochemical data, which indicated that ferrous iron induces formation of Yfh1 oligomers. While Mg2+ did not show any visible effects in our experiments, the addition of Zn2+ resulted in aggregation and precipitation of the sample (data not shown). On the other hand, Co2+ appeared to be the most useful metal and induced the formation of higher order oligomers in a concentration-dependent fashion. It appears that at least in this respect, Yfh1 and its bacterial homologue CyaY behave similarly in response to metal binding 33. Other advantages of using cobalt include its stability, which allows avoiding the formation of metal oxidation products, and an ionic radius, which is similar to that of Fe2+.
The protein-to-metal ratios (Yfh1:CoCl2) in the experiments were optimized to 1:10, 1:20, and 1:50, and the protein concentration was 3 mg/ml. As controls, we also used the Y73A Yfh1 variant, which, as shown above, is prone to the formation of higher-order oligomers, and the wild-type Yfh1wt without metal. For each concentration of CoCl2, light scattering and the respective polydispersity indices were measured several times over a period of 30 min. After approximately 30 min, the polydispersity index was stable, indicating that equilibrium had been reached (Figure 2a, inset). This incubation time was also used later in our SAXS measurements.
As shown in Figure 2a, at a Yfh1wt:CoCl2 ratio of 1:10, no substantial changes in the DLS profile could be observed, as compared to the protein solution without added metal. Increasing the ratio to 1:20 resulted in tailing of the peak, with a particle size distribution around 2.3 nm. At a Yfh1wt:CoCl2 ratio of 1:50, three separate peaks with particle size distributions of around 3.5 nm, 15 nm, and 125 nm could be observed. This clearly demonstrates that in a concentration-dependent fashion, Co2+ induces oligomerization of yeast frataxin into larger particles. In the experiments we found the presence of 100 mM NaCl, which was required for optimal protein stabilization and for preventing aggregation. The presence of salt has previously been reported as monomer-stabilizing 34. However, in our experiments this did not prevent oligomer formation in the presence of Co2+.
Characterization of cobalt-induced yeast frataxin oligomers using SAXS
To further determine the type of oligomers induced by the addition of Co2+, we used SAXS to examine the solutions with the same protein-to-metal ratios, as in the DLS experiments (1:10, 1:20, and 1:50). Analysis of the Guinier region (Figure S1a) confirmed that addition of Co2+ to the samples at these concentrations did not induce aggregation, although the sample with 1:50 ratio was showing signs of polymerization, given the high average Rg value of 5.45 nm (around 1.8 nm for the monomeric protein, as described above). A comparison of the calculated average molecular weights, using excluded volume and lysozyme as a standard, revealed an increase with higher concentrations of Co2+. Figure 2b and Table 3 show that the data for the 1:10, 1:20, and 1:50 ratios could be fitted with χ2 values of 0.55, 0.55, and 0.67, respectively, given the input we provided from our pool of oligomers, and using the program OLIGOMER. Analysis of the volume distributions of oligomer fitting to the respective SAXS profiles showed that as Co2+ concentration increased, so did the volume fraction of frataxin trimer. In other words, Co2+ strongly supports the formation of trimers.
Analysis of the Co2+-induced oligomeric states (Table 3) clearly showed a gradual increase in the percentage of trimers and hexamers and a concomitant decrease in the percentage of monomers, from 67% at a protein-to-metal ratio of 1:10 to 26% at 1:50. It was also evident that at the 1:50 ratio, oligomers of considerably higher order were formed. Interestingly, while the percentage of trimers increases from 24% to 57% when the protein-to-metal ratio changes from 1:10 to 1:50, the number of hexameric species only reaches 9%. This is different from the oligomerization behavior of the Y73A variant, which shows higher percentage of hexameirc species, compared to trimers (20% and 9%, respectively, (Table 3)). These results show that the oligomerization behavior of frataxin can be changed relatively easily and that the presence of Co2+ specifically stabilizes the trimers, supporting earlier biochemical data on metal-induced oligomerization of yeast frataxin 35.
X-ray crystallographic characterization of cobalt binding to yeast frataxin
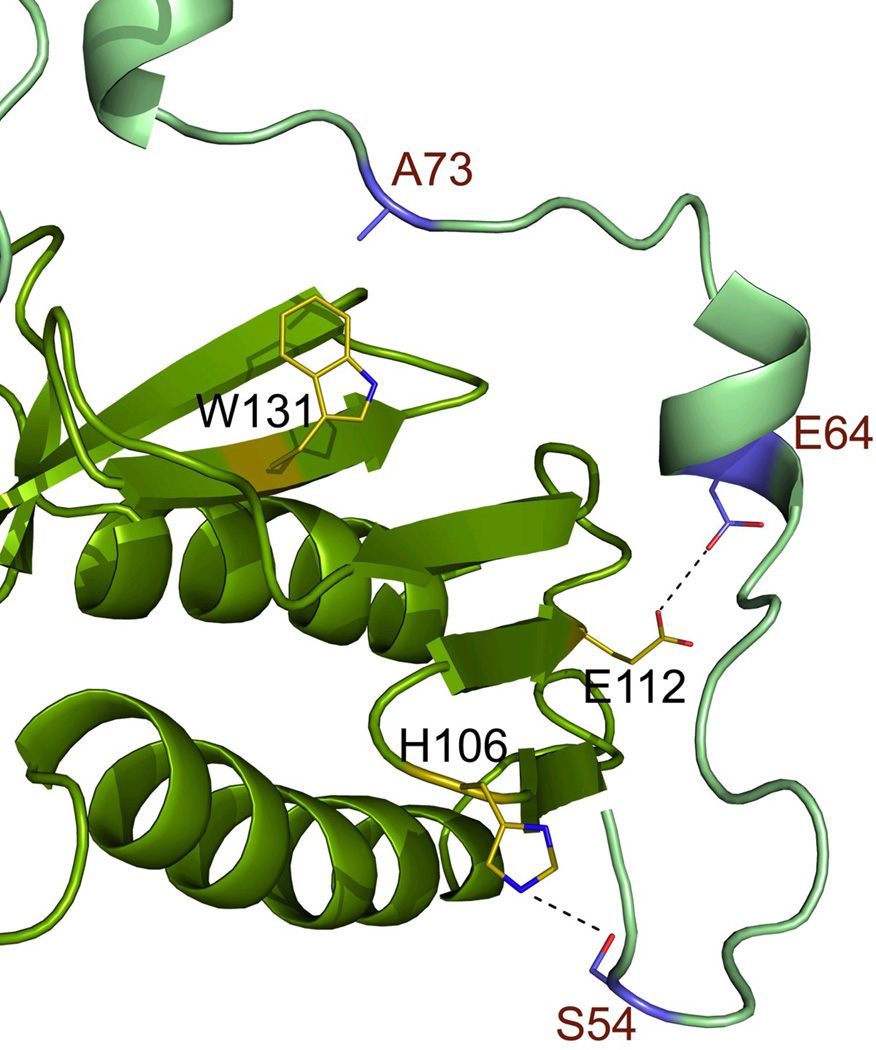
The crystals of the Y73A variant of Yfh1, which were produced from the modified crystallization conditions, belonged to space group I213 with one monomer in the asymmetric subunit and diffracted to 2.9 Å resolution (Table 1). As discussed previously 26, the monomers are involved in extensive interactions around the channel at the 3-fold axis. Their N-termini form a bridge between the monomers, which adds to the stability of the trimeric structure. The new data allowed the electron density for the complete N-terminal part of the structure to be traced, adding nine residues (V52–V60) to the new model. In the present conformation of the N-terminus, the side chains of E64 and E112 are within hydrogen bonding distance (3.1 Å) from each-other, while the newly added part of the N-terminus seems to be stabilized primarily by van der Waals contacts between side chains within the region V52-S54 and residues from the loop between β -strands β4- β5 (H106 is shown on Figure 3a).
Table 1.
X-ray data collection and refinement statistics for the crystal structure of the Y73A variant of Yfh1 with and without Co2+.
| Data collection | Y73A | Co(II)-soak |
|---|---|---|
| Beamline | I911-2 | I911-3 |
| Space group | I213 | I213 |
| Wavelength (Å) | 1.038 | 1.605 |
| Cell dimensions, a = b = c (Å) | 121.36 | 121.20 |
| Resolution range (Å) | 2.96 (3.03–2.96) | 3.2 (3.28–3.2) |
| Completeness (%) | 99.0 (99.1) | 99.8 (99.9) |
| I/σ(I) (%) | 22.6 (2.5) | 25.4 (3.1) |
| No. of unique reflections | 12,005 (929) | 9,507 (705) |
| Ramerge | 5.6 (65.3) | 4.6 (49.3) |
| Refinement | ||
| Rbcryst (Rfree) | 0.266 (0.292) | 0.204 (0.256) |
| R.m.s.d. bond (Å) | 0.014 | 0.014 |
| R.m.s.d. angles (°) | 1.71 | 1.83 |
Rmerge = Σ|Ii - <I>|/ΣI, where Ii is an individual intensity measurement and <I> is the average intensity for this reflection.
RCryst = Σ|Fobs - Fcalc|/ΣFobs, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rfree is the same as Rcryst but calculated on 5% of the data excluded from refinement.
Figure 3.
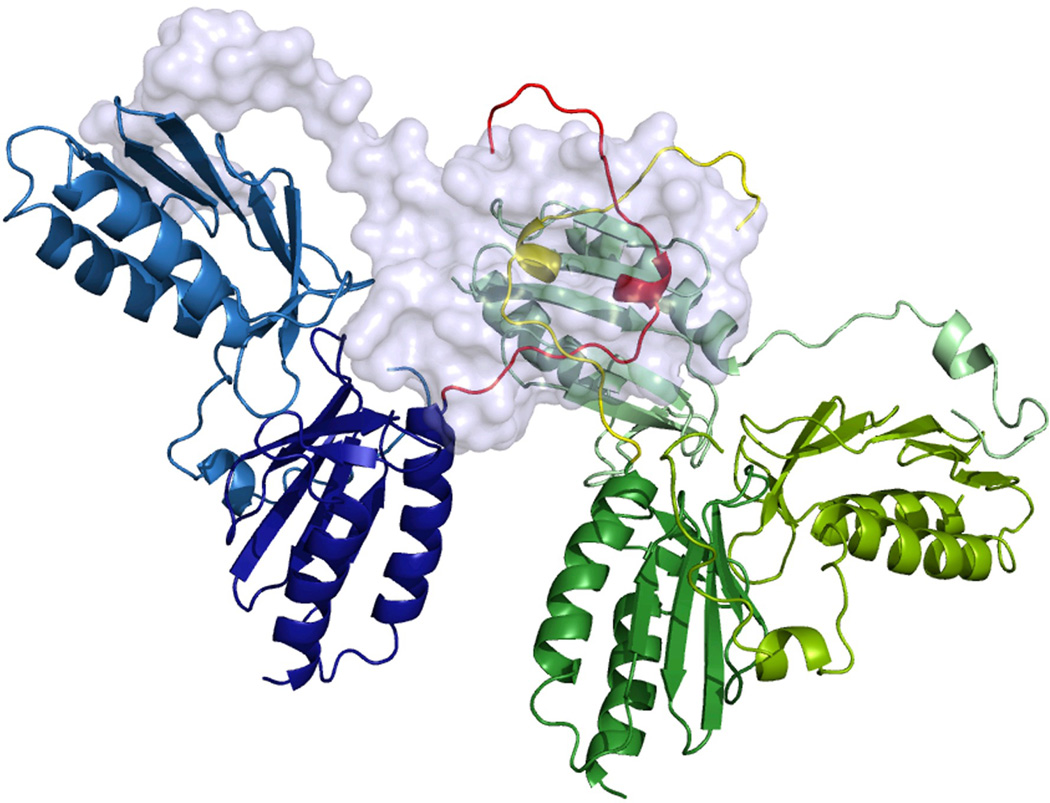
The X-ray structure of the Y73A frataxin variant. (a) The interaction of the N-terminal region of one of the subunits with the neighboring subunit. Amino acid side chains are shown as sticks and a possible hydrogen bond between E64-E112 as a dashed line. (b) The intersection of the N-termini of two monomers belonging to two different trimers within the crystal lattice. (c) Packing of Y73A frataxin trimers, with each trimer colored uniquely. One of the monomers is shown in surface representation for clarity. The N-terminal residues 52–60 are colored orange and brown. (d) Packing of the Y73A variant into hexamers, as suggested by the models of the hexamers used to fit the SAXS scattering data. (see Methods section).
Analysis of the packing and interactions within the crystal lattice is of interest for our understanding of the interactions that stabilize high-order oligomers. The previous model of frataxin (2FQL.PDB) showed that the trimers within the crystal lattice were packed against each other with the N-termini from neighboring trimers building an intersection (involving amino acid residues P69, L70, E71, and K72). In the new model, due to the extension of the N-terminal part, we could observe an additional intersection involving amino acid residues Q59, V60, and V61 (Figure 3b; only V60 is shown). This part of the N-terminus contributes to the stabilization of the interactions between trimers within the crystal lattice (Figure 3c) and could also stabilize transient monomer-monomer interactions in solution, thus driving the oligomerization towards higher-order oligomers. Upon hexamer formation, as suggested by the models used to fit the SAXS data, this type of interaction between the N-termini is preserved (Figure 3d). However, in this case the interacting N-termini have to be in a slightly different conformation in order to avoid a steric clash with neighboring trimers (further discussed in the Discussion section).
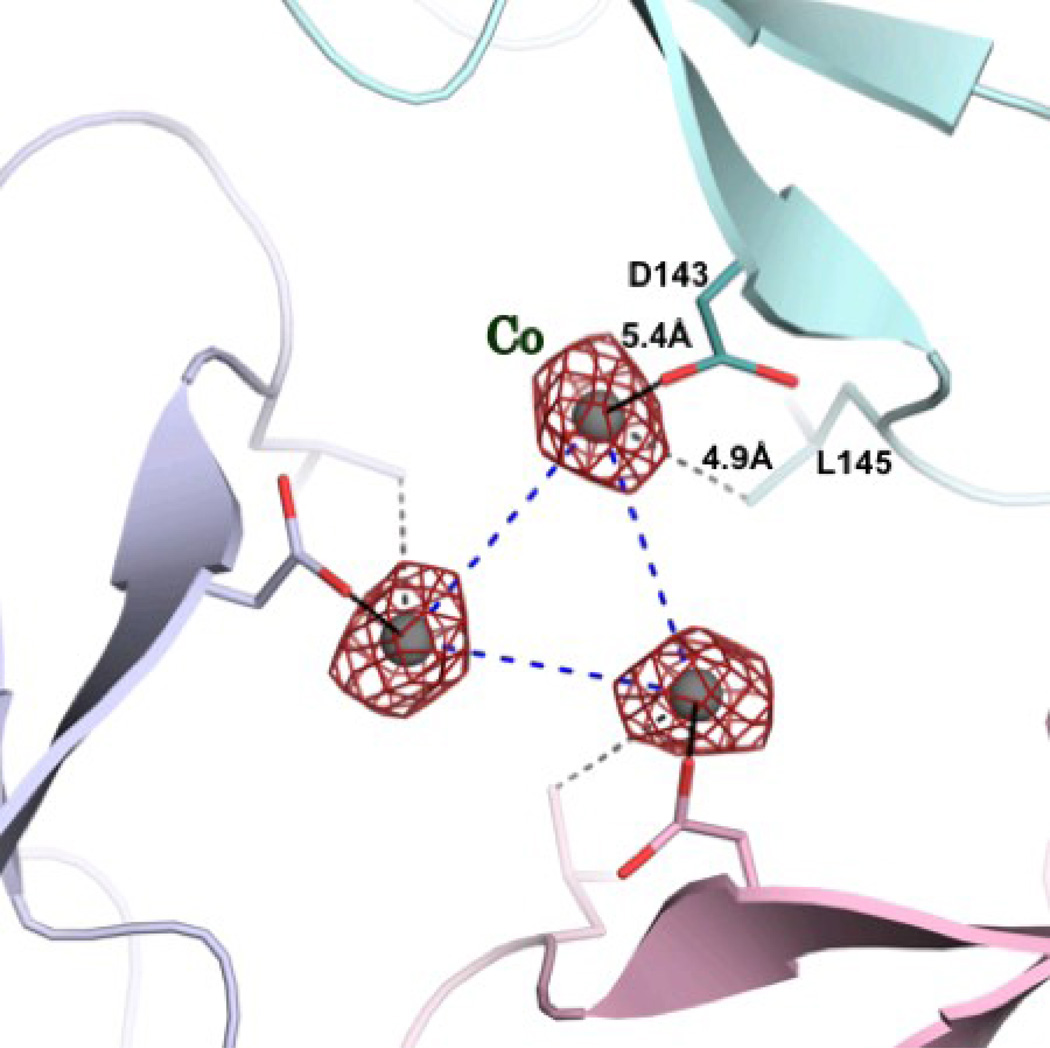
To assess the ability of cobalt to bind to frataxin, we first soaked the crystals of frataxin trimers with CoCl2. X-ray data were collected and the presence of bound metal in the structure was validated by the examination of the Fo-Fc and the anomalous Fourier difference maps. Our previous crystallographic work showed that the three-dimensional structure of frataxin trimers pre-loaded with iron contained a metal bound in the channel at the threefold axis of the trimer 26, although the resolution of that structure was only 3.5 Å. With the current higher resolution data, soaking with cobalt clearly showed a peak both in the Fo-Fc and anomalous difference density maps at levels of up to 6 σ and 5 σ, respectively. At the occupancy of 0.33 there was no residual density in the Fo-Fc maps and the B-factor of the metal refined to a value similar to that of the surrounding protein groups. Figure 4a shows three metal ions bound within the channel between the monomers. This mode of binding is different from that which was observed for iron binding in iron-preloaded trimers and suggests that one cobalt ion is shared by the three monomers in the trimer. This also suggests that the affinity of Co2+ to the binding site is lower than that of iron. While the electron density around the bound iron clearly suggested that the metal was hydrated 26, the low occupancy of the cobalt and the resolution of the structure would prevent the density of the solvent to be observed. However, the position of the metal at about 5 Å from the invariant D143 close to the wider opening of the channel suggests that some solvent molecules may be bound to the metal. It should be noted that even in the case of the iron-loaded trimer structure the metal did not show direct interactions with the invariant D143 26.
Figure 4.
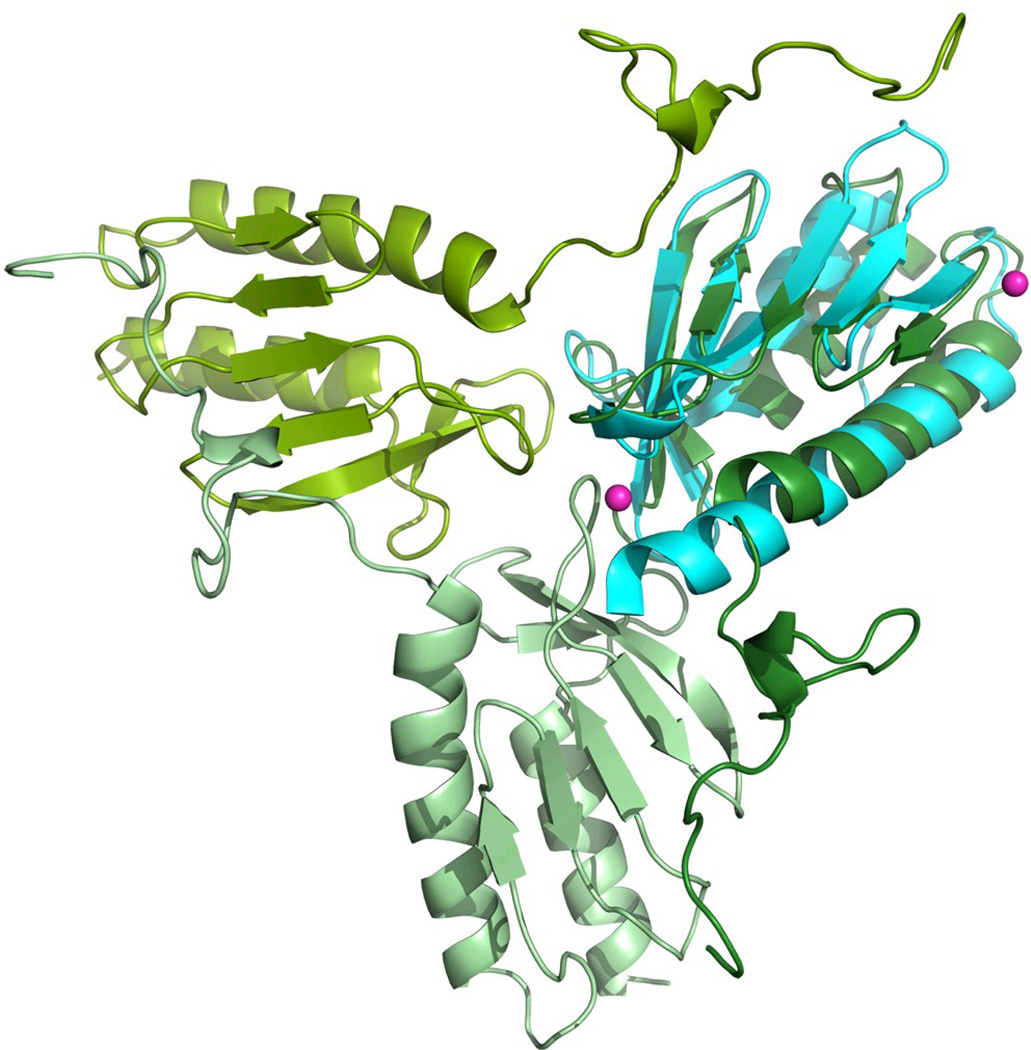
a) The 2Fo-Fc electron density map contoured at 1.0 σ for the cobalt. Three cobalt atoms are located around the threefold axis, with 30% occupancy each. b) Superposition of the structures of yeast frataxin (green) and its bacterial homologue CyaY (cyan, PDB code 2EFF) showing the positions of the cobalt ions (sphere) in CyaY.
Discussion
In this work, we have studied the effect of three different factors on the oligomerization properties of yeast frataxin Yfh1, and have shown that, depending on the conditions, it may exist in different oligomeric states in solution. Thus, the presence of glycerol facilitated the dimerization of the protein, resulting in dynamic monomer-dimer equilibrium. Glycerol is generally known to contribute to protein stability, by inducing protein compaction and by reducing flexibility 36. Taking into account the much higher viscosity of the native environment of proteins, and due to various crowding effects 37; 38, it would be logical to suggest that the monomeric form of yeast frataxin is rare in vivo. It may also be that in solution the dimer structure serves as a seed for the growth of higher-order oligomers, since the intertwined interaction between the monomers may be already formed at this stage.
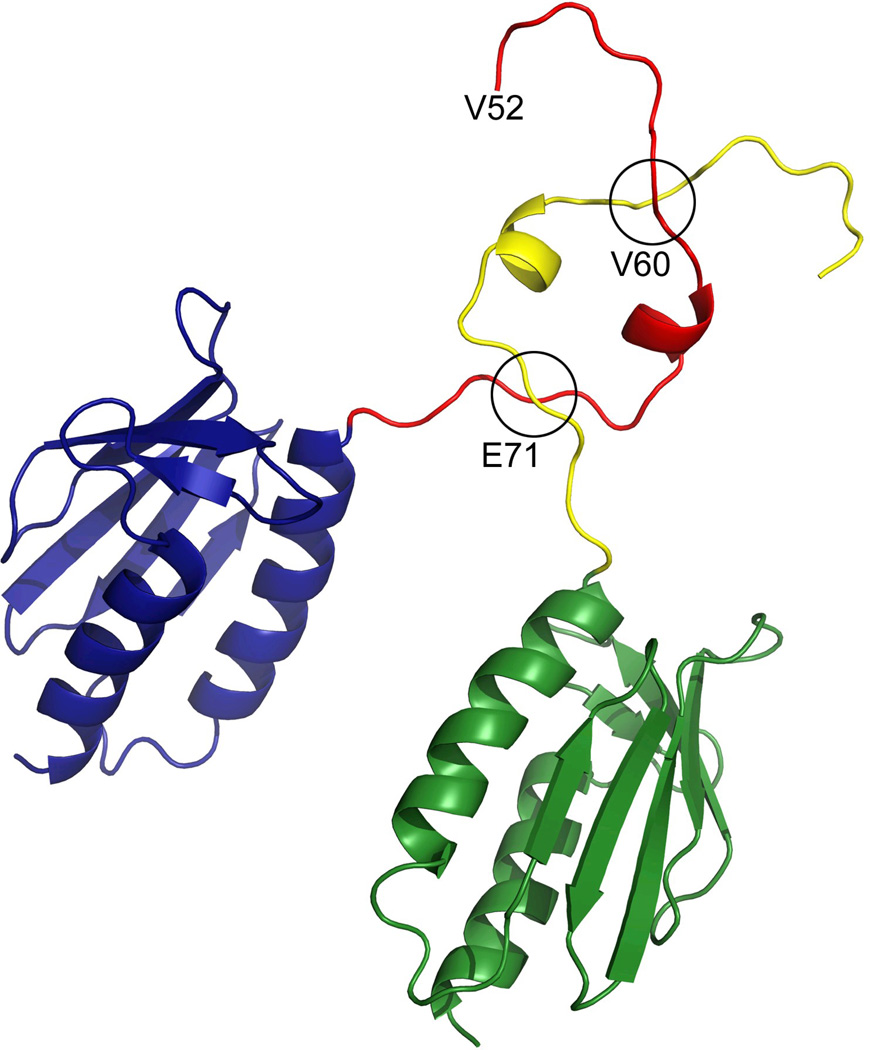
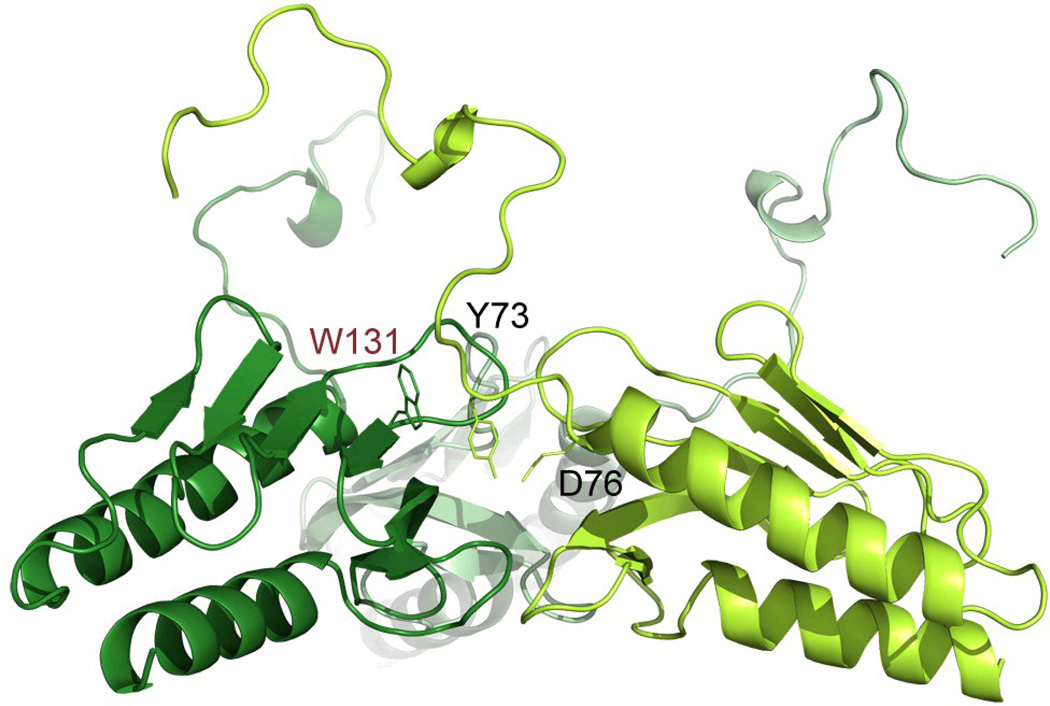
The data obtained from both the Yfh1WT and Y73A variant show that the N-terminus is highly flexible and that the conformation seen in the X-ray structure of the Y73A trimer is presumably a result of stabilization of the N-terminus by interactions with neighboring monomers as well as by crystal packing interactions. This is in contrast to solution data, which show smaller relative percentage of Y73A trimers in solution, as compared to hexamers (9% and 20%, respectively, Table 3). This suggests that in solution the interactions between trimers within higher order oligomers may contribute to trimer-, as well as higher order oligomer stabilization. The modeling of the hexamer model to fit the SAXS data required some modifications in the conformation of the N-terminal part of the protein, as compared to the conformation in the X-ray structure. One possible hydrogen bond involving the side chains of Y73 and D76 may contribute to the stabilization of this conformation (Figure 5). Interestingly, in this conformation W131 appears to be more exposed, than in the X-ray structure of the trimer (compare the conformation of the N-terminus in Figure 3a and Figure 5). This residue has been implicated in interactions with Isu1, the scaffold protein on which iron-sulfur clusters are assembled 39, suggesting that higher order oligomers may be required for the assembly of the Fe-S cluster synthesis machinery 40.
Figure 5.
The conformation of the N-terminal part within the trimer.
The trimer was generated as described in the methods section, to fit the SAXS data has a different conformation of the N-terminal part, as compared to the X-ray structure. In this conformation W131 is exposed to solvent, which could be used in the interaction with Isu1.
In the presence of Co2+ the trimeric form appears to be dominating. Higher metal concentrations lead to the formation of hexamers and higher-order oligomers. In contrast to the Y73A variant, which preferentially exists in hexameric and higher order oligomeric forms in solution, the presence of metal appears to be sufficient for the stabilization of the trimeric structure. However, from the present data it would be difficult to determine whether the higher-order oligomers are directly stabilized by the metal or whether their formation is merely a result of the increasing number of trimers, which may spontaneously assemble into higher-order structures. It is however known, that at least in the case of iron, the metal stabilizes higher-order oligomers, while the addition of reducing agents dissolves these complexes 19. In the case of the frataxin homolog CyaY from E. coli, it is also known that Fe2+, when added anaerobically, stimulates the formation of tetramers, while larger oligomers are formed in the presence of atmospheric oxygen 21; 41. It should be noted that the part of the N-terminus, which is involved in trimer stabilization in yeast frataxin, is absent in CyaY. This suggests that a different type of interactions may be involved in the stabilization of the oligomers in this case. Indeed, when co-crystallized with Co2+, no CyaY trimers can be observed in the crystals (space group P3221), although one of the metal ions in the structure was coordinated to H58, which belongs to the loop between strands 3 and 4 of the β-sheet 33. As shown in Figure 4b, the structurally homologous loop in the crystal structure of the Y73A trimer is located at the channel around the 3-fold axis 26. This position is at approximately 14 Å from the position of the cobalt atom observed in the current study. Earlier reports have indicated that iron binds to the acidic patch at helix 1 and strand 1 of the β-sheet of frataxin 42; 43; 44; 45. However, in the present study no metal density was found within this area.
The present data also demonstrate that the oligomerization behavior of frataxin depends on the amino acid composition of the N-terminus. Thus, the higher propensity of the Y73A variant to form oligomers shows that by a single amino acid replacement in this part of the protein, its properties shift to becoming more like human frataxin. This result, together with the known poor conservation of the amino acid sequence and the length of the N-terminus within the frataxin family, suggests that the oligomerization properties of the protein are different in different species. This may in turn contribute to the variations found in the function of the protein, which could occur even within a single organism. This may have interesting implications for the human protein, which is known to exist in at least four different forms, 78–210, 81–210, 56–210, and 42–210. It cannot be excluded that these forms may oligomerize differently depending on iron and external conditions. It should be noted that when we refer to “oligomerization” we distinguish it from the term “aggregation”, which is often used in the literature as a synonymous of “oligomerization”. In structural terms an oligomer (homo-oligomer or hetero-oligomer) is a structurally defined complex between two or more polypeptide units, with defined (and often conserved) interaction areas 46. On the other hand, a protein aggregate usually cannot be easily defined in structural terms since different aggregates do not necessarily have the same structure or number of subunits. Also the interactions between the subunits within the aggregate are not necessarily the same and may be difficult to describe in detail.
Materials and methods
Protein expression and purification
The Y73A frataxin variant from S. cerevisiae (residues 52–174 of the yeast frataxin sequence, corresponding to the mature form of the protein 47) was recombinantly expressed in E. coli and purified as described previsously 26. The wild-type protein was purified as described previsously 10. Protein concentration was determined from 280 nm light absorbance with the extinction coefficient, ε280 nm, of 20 000 M−1 cm−1.
Dynamic light scattering
Dynamic light scattering measurements were made on a Zetasizer Nano S (Malvern Instruments Ltd.) using a scattering angle of 173° and a laser working at 633 nm. A ZEN2112 quartz cuvette was used, with 20 µl reaction mixture, for each measurement. For the study of metal-induced oligomerization, wild-type yeast frataxin at a concentration of 3 mg/ml was mixed with a buffer containing CoCl2 to give the final protein:metal molar ratios 1:10, 1:20, and 1:50. In the absence of metal Yfh1WT and the Y73A variant were measured at 2 mg/ml. All solutions were centrifuged at 14,000 rpm for 10 min prior to the measurements.
SAXS measurements and modeling
Data collection and reduction
SAXS data were collected at beamline I711, MAX-Lab synchrotron and at the EMBL X33 beamline on the storage ring DORIS III (DESY) 48. Protein buffer consisted of 10 mM HEPES-NaOH and 100 mM NaCl. In separate measurements, 5% glycerol or 2.2–22 mM CoCl2 was added to the buffer. Yeast frataxin without glycerol and CoCl2 was measured at a concentration of 10 mg/ml. The data were recorded using a Titan CCD (Oxford Diffraction) detector at a sample-detector distance of 1.5 m and a wavelength of 1.1 Å, covering the range of momentum transfer 0.007 < s < 0.3 Å −1. Yeast frataxin samples in glycerol buffer were measured at 15 °C at a concentration range of 1.3–9.9 mg/ml. The data were recorded using a 1 M PILATUS detector (DECTRIS) at a sample-detector distance of 2.7 m and a wavelength of 1.5 Å, covering the range of momentum transfer 0.012 < s < 0.6 Å−1. No measurable radiation damage was detected on comparison of four successive time frames with 30 s exposures. The data were averaged after normalization to the intensity of the transmitted beam and the scattering of the buffer was subtracted using PRIMUS 49.
Forward scattering I(0) and the radius of gyration Rg were evaluated using the Guinier approximation 50. These parameters were also computed from the entire scattering patterns using the program GNOM 31, which provides the distance distribution functions p(r) and the maximum particle dimensions Dmax. Molecular weight estimates were made using lysozyme as a standard, or from the excluded volume of the hydrated particle (the Porod volume Vp), computed as reported by Porod 51.
Evaluation of the theoretical scattering curves from high-resolution X-ray and NMR structures of frataxin (PDB ID: 2FQL and 2GA5, respectively) and fitting to the experimental scattering data was performed using CRYSOL 52. Form factors corresponding to individual NMR conformers and X-ray structure were calculated using the “/nmr” option in the FFMAKER tool from the Atsas package 53. For fitting of the observed scattering curves with weighted combinations of form factors, the program OLIGOMER 49 was used.
Ab initio modeling
Low-resolution ab initio models were generated with DAMMIN 54, which represents a protein by an assembly of densely packed beads. Simulated annealing was used to build a compact interconnected configuration of beads that fitted the experimental data I(s) to minimize discrepancy:
where N is the number of experimental points; c is a scaling factor; and Iexp(s), Icalc(s), and σ(sj) are the experimental intensity, the calculated intensity, and the experimental error at the momentum transfer Sj, respectively. An alternative higher-resolution ab initio model, which models the particle in solution as a protein-like assembly of dummy residues, was constructed using GASBOR 55. This program represents the internal structure of the molecule more accurately than DAMMIN. Multiple DAMMIN and GASBOR calculations were performed to assess the stability of resulting solutions. Ten to 20 independent reconstructions were performed and the models were averaged with the program DAMAVER 56. The mean normalized spatial discrepancy (NSD) between models was in the range 0.537–0.590 and 0.897–0.962 for the DAMMIN and GASBOR models, respectively. Low NSD values for the DAMMIN and GASBOR models illustrated that all independently generated shapes were very similar and that a unique solution had been identified.
Combined ab initio and rigid body modeling
The N-terminus of frataxin would be expected to be flexible in solution. The X-ray structure lacks the first 9 residues of the N-terminus. NMR structure determination has shown different conformations for approximately the first 20 amino acids and for the C-proximal loop (residues 93–106). Thus, these regions were remodeled using the program BUNCH 57. The starting model generated with PRE_BUNCH consisted of two rigid bodies taken from the lowest-energy NMR conformer (model #1 in 2GA5.PDB), residues 21–92 and 107–123. The positions of rigid bodies were fixed as in the NMR structure. Ten BUNCH models were generated and the model with the lowest discrepancy was superposed with the ab initio model using SUPCOMB13 58.
Flexibility assessment
Independently, the SAXS data were analyzed using the ensemble optimization method (EOM)59 consisting of two separate programs: Random Chain (RANCH) and Genetic Algorithm Judging Optimization of Ensembles (GAJOE). The EOM assumes coexistence of a number of conformations in solution for a given construct in order to fit the experimental SAXS data, providing a quantitative characterization of flexibility and analysis of the size distribution of possible conformers. The X-ray model of frataxin (see below) showed trimers within the crystal lattice, and in addition, the structure of 24-meric oligomers, which was determined from EM clearly demonstrated, that the particles were built up by trimers 22. The presented in this work data (see below) also suggest that in the presence of glycerol, a large pool of dimmers exists in solution. Thus, it would be expected that the smallest oligomerization unit would consist of interacting dimmers, which might serve as a nucleus for the formation of trimers, which in turn will form higher-order oligomers. In order to create a pool of random models, we used independently generated BUNCH models. Three starting models were created with PRE_BUNCH, which consisted of one rigid body from the lowest- energy NMR conformer in 2GA5.PDB, residues 21–123. Each starting model corresponded to one of the monomers in the trimer structure of frataxin and was used to independently generate (with different random seed number) 13 BUNCH models. These monomers were then combined with each other model into trimers, using an in-house script in the Perl programming language, resulting in 2,197 trimer models with asymmetric N-termini. A similar procedure was used to generate 2,000 dimers. In addition, 2,000 monomer models were generated using RANCH with one rigid body (model #1 in 2GA5.PDB, residues 21–123). The final random pool consisted of 6,000 models, containing equal fractions of monomers, dimers, and trimers. Theoretical scattering curves were then calculated for each model in the pool using CRYSOL. An intensities master file was generated using the program ONEFILE2 from the EOM package. A Size_list file was created using GAJOE. After generation of the pool and additional files required for EOM, GAJOE was used to select subsets of protein models (~20). The average experimental scattering was calculated for each subset and fitted to experimental SAXS data. Subsets were selected many times in order to minimize the discrepancy. Multiple runs of GAJOE were performed (10 independent runs; for each GAJOE run, the genetic algorithm process was repeated 50 times using the default parameters of the genetic algorithm, i.e. 1,000 generations, 50 ensembles of theoretical curves, 20 curves per ensemble, 10 mutations per ensemble, and 20 crossings per generation).
Co2+-dependent oligomerization
For the study of metal-dependent oligomerization of yeast frataxin, three different Yfh1:CoCl2 ratios (1:10, 1:20, and 1:50) were prepared in a buffer consisting of 10 mM HEPES-NaOH and 100 mM NaCl, using a protein concentration of 3 mg/ml. The SAXS data were recorded at beamline I711 60 at the MAX-Lab synchrotron, using the same set-up and procedure as described above. For fitting of the coordinates of the frataxin model to the SAXS data, a pool of yeast frataxin oligomers was created. It consisted of (1) all monomeric NMR conformers (20 models) from 2GA5.PDB; (2) 10 variants of the Y73A asymmetric unit (a monomer) where residues 52–73 were remodeled using the kinematic loop modeling protocol 61 of the Rosetta 3.2 software suite 62 to account for its flexibility; and (3) eight Y73A crystallographic trimers that were first docked into the single-particle EM reconstruction of the 24-mer 26. Subsequently, the resulting 24-mer particle was stripped of trimers in stepwise fashion, creating 21-, 18-, 15-, 12-, 9-, 6-, and 3- subunit oligomers. In addition, the flexible N-terminus was remodeled, using the conformers from kinematic loop modeling above (2) and all steric clashes with the protein core of neighboring subunits were eliminated. We also created 42–78 subunit oligomers by combining 18- and 21-subunit oligomers as building blocks. Particles of this type have been observed on negatively stained EM micrographs of the Yfh1 Y73A variant (unpublished data), human frataxin, and horse spleen ferritin 16. Subsequently, chains A and B of the crystallographic trimer were used as input to the symmetric docking protocol 63 of Rosetta 3.2 in the “local refine” mode, allowing a maximum of 3 Å translation and 8° rotation in the creation of 2,000 dimer models. Similarly, chains A, B, and C of the crystallographic trimer were used for the creation of 2,000 trimer models. CRYSOL was used to calculate all the scattering patterns and OLIGOMER was used to calculate the volume fractions of the components that best fitted the observed scattering curves.
Oligomerization of yeast frataxin Y73A variant
For the study of oligomerization induced by replacement of tyrosine residue 73 with an alanine, we used the same methods as described for metal-dependent oligomerization above, at a concentration of 4 mg/ml for the low molecular weight fraction.
X-ray crystallography
Initial crystallization conditions were found at the MAX-Lab protein crystallization facility with the JCSG-plus screen (Molecular Dimensions Ltd.) set-up in Greiner low-profile 96-well plates using the Mosquito robot (TTP Labtech, UK). Well and drop solution volumes of 80 µl and 200 nl (100 nl protein and 100 nl well solution), respectively, were used at 20 °C. Crystals were found in the screen solution containing 1.26 M (NH4)2SO4, 0.2 M Li2SO4, and 0.1 M Tris, pH 8.5. Optimization of these conditions yielded crystals between 0.2 and 0.5 mm in size, which grew after two weeks at 15 °C. The crystallization drops were prepared by mixing 2 µl well solution containing 1.7 M (NH4)2 SO4, 0.2 M Li2SO4, 4% γ-butyrolactone, 0.1 M Tris HCl, pH 8.5, and 2 µl of protein solution containing 9.2 mg/ml Y73A Yfh1p in 10 mM HEPES, pH 7.3. These conditions are different from the earlier published crystallization conditions (0.1 M Bis-Tris, pH 5.5, 2.0 M (NH4)2 SO4 and 4% (v/v) γ-butyrolactone).
X-ray data to 2.9 Å resolution were collected at the MAX-Lab synchrotron, beamlines I911-2 and I911-3. Prior to data collection, the crystals were flash-frozen in a cryo-solution containing 2 M (NH4)2 SO4, 4% γ-butyrolactone, 20% glycerol, and 0.1 M Tris HCl, pH 8.5. Due to low-resolution diffraction overloads, data collection was divided into long and short exposure sets, with exposure times per image of 30 s and 10 s, respectively. Both sessions covered 90° rotation with 0.5° per image. For metal ion soaking, prior to flash-freezing, 4 mM CoCl2 was added to the cryo-solution and the crystals were incubated in this solution for 30 min. XDS software 64 was used for indexing, integration, scaling, and merging. PHASER software 65 was used for molecular replacement using the previously determined crystal structure of the Y73A variant of Yfh1 26 (PDB: 2FQL). Refinement was carried out using alternate runs of the program REFMAC5 66 (CCP4 6.0.2 suite 67) each followed by inspection and manual rebuilding of the model using COOT 0.4.1 68. For TLS refinement, the whole monomeric unit (residues 52–172) was considered as one TLS group. Splitting the structure into two (52–60 & 61–172) or three groups (52–60, 61–75 & 76–172) did not result in any improvement of map quality or model geometry. Omit maps 69 were used to localize the missing N-terminal residues (52–60), which were subsequently manually built into the density. The mean value for the B-factors for the cobalt free structure refined to 73 Å2 and for the Co2+ complex to 96 Å2. For the localization of metal positions, anomalous Fourier maps were calculated using the Bijvoet differences (F+ - F−) as coefficients. The metal density peaks were clearly visible both in the Fo-Fc and anomalous difference density maps at levels of up to 6 σ and 5 σ, respectively. The occupancy of the metal was determined by assigning different values to the occupancy followed by inspection of the difference electron density. At the occupancy value of 0.33 no residual electron density was present. At this occupancy the B-factor of the metal refined to a value similar to that of the neighboring protein groups (79 Å2+), which indicates the correctness of the chosen occupancy value. A summary of data collection and refinement statistics is presented in Table 1.
Highlights.
Co2+ and glycerol facilitate dynamic equilibrium between oligomeric states of frataxin
Co2+ binds inside the channel at the 3-fold axis of the trimer
Tentative models of yeast frataxin hexamers and dodecamers are presented
Flexibility of the yeast frataxin N-terminus is essential for oligomer formation
Supplementary Material
Acknowledgments
This project was supported by the Swedish Research Council (Vetenskapsradet), a MAX IV/ESS grant from the Natural Science Faculty of Lund University, the Crafoord Foundation, and the Carl Trygger Foundation to SAK and by grant AG15709 from the National Institute on Aging, National Institutes of Health, to GI. Alexander Shkumatov and Dmitri Svergun would like to further acknowledge BMBF Research Grant SYNC-LIFE (contract number 05K10YEA) and the EU FP7 e-Infrastructure grant WeNMR (contract number 261572). We also thank the EMBL-Hamburg and Lund university for synchrotron beam time allocation at DESY (Hamburg, Germany) and Max-Lab, respectively. Clement Blanchet and Tomas Plivelic are acknowledged for technical support at beam line X33 (DESY) and I711 (MAX-Lab), respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
PDB ID: 3OEQ; 3OER
References
- 1.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 2.Kell DB. Iron behaving badly: inappropriate iron chelation as a major contributor to the aetiology of vascular and other progressive inflammatory and degenerative diseases. BMC Med Genomics. 2009;2:2. doi: 10.1186/1755-8794-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 4.Geoffroy G, Barbeau A, Breton G, Lemieux B, Aube M, Leger C, Bouchard JP. Clinical description and roentgenologic evaluation of patients with Friedreich's ataxia. Canadian Journal of Neurological Sciences. 1976;3:279–286. doi: 10.1017/s0317167100025464. [DOI] [PubMed] [Google Scholar]
- 5.Harding AE, Hewer RL. The heart disease of Friedreich's ataxia: a clinical and electrocardiographic study of 115 patients, with an analysis of serial electrocardiographic changes in 30 cases. Q J Med. 1983;52:489–502. [PubMed] [Google Scholar]
- 6.Finocchiaro G, Baio G, Micossi P, Pozza G, di Donato S. Glucose metabolism alterations in Friedreich's ataxia. Neurology. 1988;38:1292–1296. doi: 10.1212/wnl.38.8.1292. [DOI] [PubMed] [Google Scholar]
- 7.Babcock M, deSilva D, Oaks R, DavisKaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 8.Chasteen ND, Harrison PM. Mineralization in ferritin: an efficient means of iron storage. J Struct Biol. 1999;126:182–194. doi: 10.1006/jsbi.1999.4118. [DOI] [PubMed] [Google Scholar]
- 9.Gakh O, Adamec J, Gacy AM, Twesten RD, Owen WG, Isaya G. Physical evidence that yeast frataxin is an iron storage protein. Biochemistry. 2002;41:6798–6804. doi: 10.1021/bi025566+. [DOI] [PubMed] [Google Scholar]
- 10.Gakh O, Park S, Liu G, Macomber L, Imlay JA, Ferreira GC, Isaya G. Mitochondrial iron detoxification is a primary function of frataxin that limits oxidative damage and preserves cell longevity. Human Molecular Genetics. 2006;15:467–479. doi: 10.1093/hmg/ddi461. [DOI] [PubMed] [Google Scholar]
- 11.Nichol H, Gakh O, O'Neill HA, Pickering IJ, Isaya G, George GN. Structure of frataxin iron cores: an X-ray absorption spectroscopic study. Biochemistry. 2003;42:5971–5976. doi: 10.1021/bi027021l. [DOI] [PubMed] [Google Scholar]
- 12.Yoon T, Cowan JA. Iron-sulfur cluster biosynthesis. Characterization of frataxin as an iron donor for assembly of [2Fe-2S] clusters in ISU-type proteins. Journal of the American Chemical Society. 2003;125:6078–6084. doi: 10.1021/ja027967i. [DOI] [PubMed] [Google Scholar]
- 13.Lesuisse E, Santos R, Matzanke BF, Knight SAB, Camadro JM, Dancis A. Iron use for haeme synthesis is under control of the yeast frataxin homologue (Yfh1) Human Molecular Genetics. 2003;12:879–889. doi: 10.1093/hmg/ddg096. [DOI] [PubMed] [Google Scholar]
- 14.Bulteau AL, O'Neill HA, Kennedy MC, Ikeda-Saito M, Isaya G, Szweda LI. Frataxin acts as an iron chaperone protein to modulate mitochondrial aconitase activity. Science. 2004;305:242–245. doi: 10.1126/science.1098991. [DOI] [PubMed] [Google Scholar]
- 15.Koutnikova H, Campuzano V, Koenig M. Maturation of wild-type and mutated frataxin by the mitochondrial processing peptidase. Human Molecular Genetics. 1998;7:1485–1489. doi: 10.1093/hmg/7.9.1485. [DOI] [PubMed] [Google Scholar]
- 16.Cavadini P, O'Neill HA, Benada O, Isaya G. Assembly and iron-binding properties of human frataxin, the protein deficient in Friedreich ataxia. Human Molecular Genetics. 2002;11:217–227. doi: 10.1093/hmg/11.3.217. [DOI] [PubMed] [Google Scholar]
- 17.Schmucker S, Argentini M, Carelle-Calmels N, Martelli A, Puccio H. The in vivo mitochondrial two-step maturation of human frataxin. Human Molecular Genetics. 2008;17:3521–3531. doi: 10.1093/hmg/ddn244. [DOI] [PubMed] [Google Scholar]
- 18.Gakh O, Bedekovics T, Duncan SF, Smith DYt, Berkholz DS, Isaya G. Normal and Friedreich ataxia cells express different isoforms of frataxin with complementary roles in iron-sulfur cluster assembly. Journal of Biological Chemistry. 2010;285:38486–38501. doi: 10.1074/jbc.M110.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park S, Gakh O, O'Neill HA, Mangravita A, Nichol H, Ferreira GC, Isaya G. Yeast frataxin sequentially chaperones and stores iron by coupling protein assembly with iron oxidation. Journal of Biological Chemistry. 2003;278:31340–31351. doi: 10.1074/jbc.M303158200. [DOI] [PubMed] [Google Scholar]
- 20.O'Neill HA, Gakh O, Park S, Jin C, Mooney SM, Sampson M, Ferreira GC, Isaya G. Assembly of human frataxin is a mechanism for detoxifying redox-active iron. Biochemistry. 2005;44:537–545. doi: 10.1021/bi048459j. [DOI] [PubMed] [Google Scholar]
- 21.Bou-Abdallah F, Adinolfi S, Pastore A, Laue TM, Chasteen ND. Iron binding and oxidation kinetics in frataxin CyaY of Escherichia coli. Journal of Molecular Biology. 2004;341:605–615. doi: 10.1016/j.jmb.2004.05.072. [DOI] [PubMed] [Google Scholar]
- 22.Schagerlof U, Elmlund H, Gakh O, Nordlund G, Hebert H, Lindahl M, Isaya G, Al-Karadaghi S. Structural basis of the iron storage function of frataxin from single-particle reconstruction of the iron-loaded oligomer. Biochemistry. 2008;47:4948–4954. doi: 10.1021/bi800052m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knight SAB, Sepuri NBV, Pain D, Dancis A. Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. Journal of Biological Chemistry. 1998;273:18389–18393. doi: 10.1074/jbc.273.29.18389. [DOI] [PubMed] [Google Scholar]
- 24.Voisine C, Schilke B, Ohlson M, Beinert H, Marszalek J, Craig EA. Role of the mitochondrial Hsp70s, Ssc1 and Ssq1, in the maturation of Yfh1. Mol Cell Biol. 2000;20:3677–3684. doi: 10.1128/mcb.20.10.3677-3684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Neill HA, Gakh O, Isaya G. Supramolecular assemblies of human frataxin are formed via subunit-subunit interactions mediated by a non-conserved amino-terminal region. Journal of Molecular Biology. 2005;345:433–439. doi: 10.1016/j.jmb.2004.10.074. [DOI] [PubMed] [Google Scholar]
- 26.Karlberg T, Schagerlof U, Gakh O, Park S, Ryde U, Lindahl M, Leath K, Garman E, Isaya G, Al-Karadaghi S. The structures of frataxin oligomers reveal the mechanism for the delivery and detoxification of iron. Structure. 2006;14:1535–1546. doi: 10.1016/j.str.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Svergun DI, Mertens HDT. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. Journal of Structural Biology. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 28.Verstraete K, Vandriessche G, Januar M, Elegheert J, Shkumatov AV, Desfosses A, Van Craenenbroeck K, Svergun DI, Gutsche I, Vergauwen B, Savvides SN. Structural insights into the extracellular assembly of the hematopoietic Flt3 signaling complex. Blood. 2011 doi: 10.1182/blood-2011-01-329532. [DOI] [PubMed] [Google Scholar]
- 29.Svergun DI, Mylonas E, Hascher A, Bernado P, Blackledge M, Mandelkow E. Domain conformation of tau protein studied by solution small-angle X-ray scattering. Biochemistry. 2008;47:10345–10353. doi: 10.1021/bi800900d. [DOI] [PubMed] [Google Scholar]
- 30.Shkumatov AV, Chinnathambi S, Mandelkow E, Svergun DI. Structural memory of natively unfolded tau protein detected by small-angle X-ray scattering. Proteins-Structure Function and Bioinformatics. 2011;79:2122–2131. doi: 10.1002/prot.23033. [DOI] [PubMed] [Google Scholar]
- 31.Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. Journal of Applied Crystallography. 1992;25:495–503. [Google Scholar]
- 32.Bernado P. Effect of interdomain dynamics on the structure determination of modular proteins by small-angle scattering. Eur Biophys J. 2010;39:769–780. doi: 10.1007/s00249-009-0549-3. [DOI] [PubMed] [Google Scholar]
- 33.Pastore C, Franzese M, Sica F, Temussi P, Pastore A. Understanding the binding properties of an unusual metal-binding protein - a study of bacterial frataxin. Febs Journal. 2007;274:4199–4210. doi: 10.1111/j.1742-4658.2007.05946.x. [DOI] [PubMed] [Google Scholar]
- 34.Adinolfi S, Trifuoggi M, Politou AS, Martin S, Pastore A. A structural approach to understanding the iron-binding properties of phylogenetically different frataxins. Human Molecular Genetics. 2002;11:1865–1877. doi: 10.1093/hmg/11.16.1865. [DOI] [PubMed] [Google Scholar]
- 35.Adamec J, Rusnak F, Owen WG, Naylor S, Benson LM, Gacy AM, Isaya G. Iron-dependent self assembly of recombinant yeast frataxin: Implications for Friedreich ataxia. American Journal of Human Genetics. 2000;67:549–562. doi: 10.1086/303056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vagenende V, Yap MG, Trout BL. Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry. 2009;48:11084–11096. doi: 10.1021/bi900649t. [DOI] [PubMed] [Google Scholar]
- 37.Ralston GB. Effects of Crowding in Protein Solutions. Journal of Chemical Education. 1990;67:857–860. [Google Scholar]
- 38.Minton AP. Implications of macromolecular crowding for protein assembly. Current Opinion in Structural Biology. 2000;10:34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 39.Foury F, Leidgens S, De Smet S. Frataxin interacts with Isu1 through a conserved tryptophan in its beta-sheet. Human Molecular Genetics. 2010;19:276–286. doi: 10.1093/hmg/ddp495. [DOI] [PubMed] [Google Scholar]
- 40.Li HQ, Gakh O, Smith DY, Isaya G. Oligomeric Yeast Frataxin Drives Assembly of Core Machinery for Mitochondrial Iron-Sulfur Cluster Synthesis. Journal of Biological Chemistry. 2009;284:21971–21980. doi: 10.1074/jbc.M109.011197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Layer G, Ollagnier-de Choudens S, Sanakis Y, Fontecave M. Iron-sulfur cluster biosynthesis - Characterization of Escherichia coli CyaY as an iron donor for the assembly of [2Fe-2S] clusters in the scaffold IscU. Journal of Biological Chemistry. 2006;281:16256–16263. doi: 10.1074/jbc.M513569200. [DOI] [PubMed] [Google Scholar]
- 42.Huang J, Dizin E, Cowan JA. Mapping iron binding sites on human frataxin: implications for cluster assembly on the ISUFe-S cluster scaffold protein. Journal of Biological Inorganic Chemistry. 2008;13:825–836. doi: 10.1007/s00775-008-0369-4. [DOI] [PubMed] [Google Scholar]
- 43.Nair M, Adinolfi S, Pastore C, Kelly G, Temussi P, Pastore A. Solution structure of the bacterial frataxin ortholog, CyaY: Mapping the iron binding sites. Structure. 2004;12:2037–2048. doi: 10.1016/j.str.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 44.He YN, Alam SL, Proteasa SV, Zhang Y, Lesuisse E, Dancis A, Stemmler TL. Yeast frataxin solution structure, iron binding, and ferrochelatase interaction. Biochemistry. 2004;43:16254–16262. doi: 10.1021/bi0488193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cook JD, Bencze KZ, Jankovic AD, Crater AK, Busch CN, Bradley PB, Stemmler AJ, Spaller MR, Stemmler TL. Monomeric yeast frataxin is an iron-binding protein. Biochemistry. 2006;45:7767–7777. doi: 10.1021/bi060424r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imperiali B, Ali MH. Protein oligomerization: How and why. Bioorganic & Medicinal Chemistry. 2005;13:5013–5020. doi: 10.1016/j.bmc.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 47.Branda SS, Cavadini P, Adamec J, Kalousek F, Taroni F, Isaya G. Yeast and human frataxin are processed to mature form in two sequential steps by the mitochondrial processing peptidase. Journal of Biological Chemistry. 1999;274:22763–22769. doi: 10.1074/jbc.274.32.22763. [DOI] [PubMed] [Google Scholar]
- 48.Roessle MW, Klaering R, Ristau U, Robrahn B, Jahn D, Gehrmann T, Konarev P, Round A, Fiedler S, Hermes C, Svergun D. Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. Journal of Applied Crystallography. 2007;40:S190–S194. [Google Scholar]
- 49.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- 50.Guinier A. La diffraction des rayons X aux tr`es petits angles: application `a l'étude de phénom`enes ultramicroscopiques. Th`ese, Masson et cie, Univ. de Paris; 1939. [Google Scholar]
- 51.Porod G. Small Angle X-Ray Scattering. In: Glatter O, K O, editors. Small Angle X-Ray Scattering. Academic Press; London: 1982. pp. 17–51. [Google Scholar]
- 52.Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- 53.Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI. ATSAS 2.1 - towards automated and web-supported small-angle scattering data analysis. Journal of Applied Crystallography. 2007;40:S223–S228. [Google Scholar]
- 54.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophysical Journal. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophysical Journal. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophysical Journal. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. Journal of Applied Crystallography. 2001;34:33–41. [Google Scholar]
- 59.Bernado P, Mylonas E, Petoukhov MV, Blackledge M, Svergun DI. Structural characterization of flexible proteins using small-angle X-ray scattering. Journal of the American Chemical Society. 2007;129:5656–5664. doi: 10.1021/ja069124n. [DOI] [PubMed] [Google Scholar]
- 60.Knaapila M, Svensson C, Barauskas J, Zackrisson M, Nielsen SS, Toft KN, Vestergaard B, Arleth L, Olsson U, Pedersen JS, Cerenius Y. A new small-angle X-ray scattering set-up on the crystallography beamline I711 at MAX-lab. Journal of Synchrotron Radiation. 2009;16:498–504. doi: 10.1107/S0909049509018986. [DOI] [PubMed] [Google Scholar]
- 61.Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nature Methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andre I, Bradley P, Wang C, Baker D. Prediction of the structure of symmetrical protein assemblies. Proc Natl Acad Sci U S A. 2007;104:17656–17661. doi: 10.1073/pnas.0702626104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 67.Dodson EJ, Winn M, Ralph A. Collaborative Computational Project, number 4: providing programs for protein crystallography. Methods Enzymol. 1997;277:620–633. doi: 10.1016/s0076-6879(97)77034-4. [DOI] [PubMed] [Google Scholar]
- 68.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bhat TN. Calculation of an Omit Map. Journal of Applied Crystallography. 1988;21:279–281. [Google Scholar]
- 70.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.