Studies of oncogenic retroviruses provided much of the intellectual foundation for our current understanding of the molecular mechanisms of carcinogenesis. The now well-established role of protein tyrosine kinases and Ras in growth stimulation and the myriad ways by which dysregulated expression of growth control genes can lead to tumor development are concepts deeply rooted in retrovirology. Indeed, more than 50 oncogenes now known to be involved in human cancers were first discovered and studied in retroviral models (1). Nonetheless, only one retrovirus, human T cell lymphotrophic virus (HTLV), is oncogenic in humans, and, as we enter the 21st century, retroviral oncogenesis models may seem to be of largely historical interest. However, analyses of jaagsiekte sheep retrovirus (JSRV) by Maeda et al. (2) and Rai et al. (3) in this issue of PNAS reveal that oncogenic retroviruses still hold important secrets that may be directly relevant to human cancer.
Analyses of jaagsiekte sheep retrovirus reveal that oncogenic retroviruses still hold important secrets that may be directly relevant to human cancer.
JSRV is the causative agent of ovine pulmonary carcinoma (OPC), a contagious tumor that afflicts sheep in the United Kingdom, South Africa, and several other countries, posing a significant veterinary problem in some flocks (4, 5). In addition, OPC shares clinical, and histologic features with human bronchioalveolar carcinoma (BAC), an important, non-smoking-related lung cancer that accounts for about 25% of all lung tumors in the United States (5, 6). Although the retroviral etiology of OPC has been suspected for more than 20 years, the absence of an in vitro culture system and the difficulties inherent in dealing with a large animal tumor model in which retroviruses had not been thoroughly studied slowed research. However, the cloning of JSRV from a primary lung tumor and subsequent demonstration that this agent was necessary and sufficient to induce OPC in new-born lambs (7) paved the way for experiments addressing the pathogenesis of the disease. The present studies imply a novel mechanism of transformation involving interaction of the viral Env protein with the cellular receptor used to mediate viral entry. The first of the papers used screening of Stanford G3 panel of whole human genome radiation hybrids to identify the cellular receptor as the sheep homolog of human HYAL2 (3), a protein encoded by a gene contained within a deletion found in a significant proportion of human lung cancer (8, 9). The second report used immortalized rodent cell lines to demonstrate that Env is the transforming gene of JSRV (2). Similar results have been obtained by using an immortalized chicken fibroblast cell line (J. C. DeMartini, personal communication).
The finding that the JSRV env gene product is capable of transforming cells is unexpected. OPC is a multifocal tumor that arises rapidly in experimentally infected lambs. This pattern of disease is almost always characteristic of retroviruses that carry oncogenes, viral versions of highly conserved cellular genes that play key roles in growth and development (1). However, JSRV lacks an oncogene, containing only gag, pol, and env, the three genes typically found in simple, replicating retroviruses (7). These genes encode virion structural proteins, enzymes involved in the replication cycle and the envelope protein, all of which normally do not alter cellular growth. Oncogenic viruses of this type are typically associated with tumors that arise after a long latent period. Invariably, such tumors contain an integrated provirus that alters the expression of cellular proto-oncogenes (1).
Retroviral Env proteins mediate viral entry by interacting with a cell surface receptor. In virtually all instances, this interaction does not affect cell growth, and most known retroviral receptors do not play an obvious role in growth signaling (10). Nonetheless, ligand–surface receptor interaction is a common way in which growth signals are transmitted to cells, and a recent report indicates that the env gene product of avian hemangioma virus, a retrovirus that induces vascular endothelial cell tumors, can also stimulate proliferation of monkey epithelial cells and the NIH 3T3 mouse cell line (11). Until these reports, the classic instance of Env-mediated cell proliferation involved spleen focus-forming virus (SFFV), an agent that induces rapid erythroleukemias in mice. This virus stimulates erythroid precursor cells through interactions between Env and the erythropoietin receptor (12). However, in contrast to JSRV and the hemangioma virus, the SFFV Env protein is a truncated recombinant molecule that no longer interacts with the receptor that mediates viral infection.
The JSRV receptor gene, HYAL2, is contained within a region on human chromosome 3p21.3 that is deleted in a substantial frequency of lung and breast tumors, suggesting that it may have tumor suppressor functions (9, 13). Although a possible role for a hyaluronidase in oncogenesis (perhaps through effects on angiogenesis or cell–cell interaction) can be envisioned, the protein encoded by HYAL2 appears atypical, in that it has very low hyaluronidase activity or lacks this function completely (3, 14). Nonetheless, HYAL2 is a cell surface GPI-anchored protein with the potential to participate in cell signaling interactions (3). However, analyses have so far failed to clearly assign the putative tumor suppressor function associated with 3p21.3 to a particular gene. Nineteen candidates map to this region, and HYAL2 is not among the small subset that display mutational patterns characteristic of the classical pattern of loss of tumor suppressor function (9). However, the possibility that haplo-insufficiency plays an important role in the tumor suppressor function cannot yet be excluded.
The way in which env expression stimulates cell growth will be an active topic of future studies. Rodent cells similar to those used for the transformation studies are not susceptible to JSRV infection (15), in large part because they do not express an Hyal2 protein that is competent to mediate entry (3, 16). The murine and rat Hyal2 genes have been cloned, but why these proteins fail to support viral entry is not known. In addition, neither the viral nor HYAL2 sequences required to mediate entry into human cells have been elucidated. This information, along with additional transformation experiments, should determine which of several possible mechanisms is responsible for growth stimulation. For example, if HYAL2 is a tumor suppressor and functions to limit cell growth, interaction with the JSRV Env protein may block this suppression, leading to a growth stimulatory signal (Fig. 1a). Alternatively, the JSRV Env protein may mediate transformation by interacting with HYAL2 and sending a growth stimulatory signal as part of the adsorption process (Fig. 1b). A third model predicts that the protein interacts with another cellular protein, either at the surface or on the inner face of the plasma membrane, and stimulates growth (Fig. 1c). Oncogenic strains of JSRV differ from highly related endogenous, nononcogenic sheep retroviruses within the transmembrane portion of the Env protein (17, 18), suggesting that this part of the protein, which does not interact with the cellular receptor, could be involved in growth signaling. Because cell division is required for simple retroviruses to gain entry into the nucleus and establish infection (19), stimulation of cell division during entry could allow the virus access to cell types that are less actively cycling. Interestingly, the type II pneumocysts and Clara cells that make up the tumors in OPC and BAC are not actively dividing cells under normal circumstances.
Figure 1.
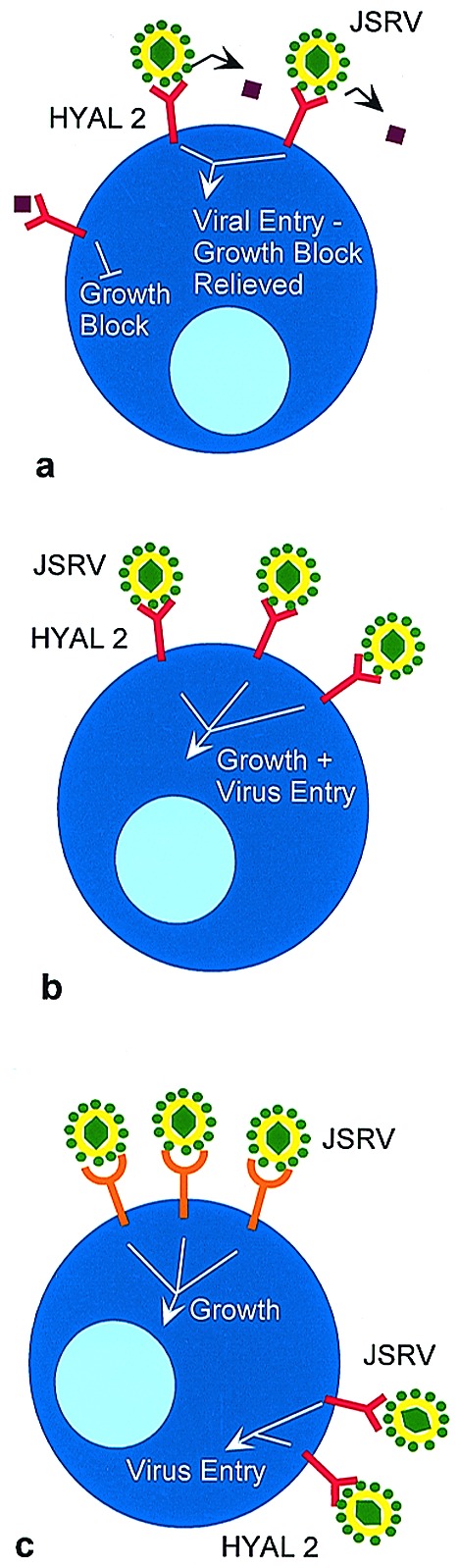
Possible mechanisms of transformation mediated by the JSRV Env protein. (a) HYAL2 normally functions to suppress cell growth. The mechanism by which suppression is mediated is not known, but one possibility is that Env-HYAL2 binding and virus entry blocks interaction with a ligand that could be soluble (as illustrated by the purple diamond) or cell-associated. (b) Env-HYAL2 interaction stimulates cell growth and mediates virus entry. (c) Env-HYAL2 interaction mediates virus entry, and Env interaction with another cellular protein stimulates cell growth. Although interaction with a cell surface molecule is illustrated, this interaction need not occur at the cell surface.
Despite the compelling in vitro experiments, and the demonstration that chicken cells transformed by JSRV env can form tumors in nude mice (J. C. DeMartini, personal communication), it is prudent to remember that tumor induction in normal lung cells may be more complex than transformation of immortalized cell lines in vitro. Unlike normal lung cells, immortalized cell lines have undergone genetic changes that make them acutely sensitive to transformation. Although JSRV is sufficient to induce OPC, newborn lambs inoculated with tumor homogenates develop tumors much faster than those injected with cloned virus produced in vitro (7). In addition, most naturally infected sheep display a much longer course of tumor development (4, 5). Some of these differences may relate to virus titers that are difficult to standardize, differences in strains of sheep, or differences among JSRV strains (18). All JSRV strains retain an ORF called orfx within the pol gene that is not required for transformation in vitro (2), but could play some part in tumor development in vivo. Based on sequence similarity, the orfx product may be related to the G-protein-coupled receptor family (18).
Although analyses of different env mutants is clearly an important next step, direct testing of mutants in vivo is complicated because sheep and goats are currently the only models for monitoring tumor induction. Transgenic mice expressing the HYAL2 receptor may allow infection of mice and analysis of JSRV mutants. The lung is the primary site of JSRV replication in infected sheep (20), suggesting that targeting the receptor to the appropriate lung cells may allow the disease process to be recapitulated. Targeting expression of the JSRV env gene to the lung by using an appropriate promoter may also facilitate the development of a more tractable animal model. However, in this instance, screening mutants will be somewhat more cumbersome. Future work will determine whether these approaches recapitulate events in the natural host.
A second important aspect of the analysis of JSRV that should not be overshadowed by the novel transformation aspect is that further understanding of this virus may lead to the development of important vectors to target cells within the lung. JSRV is unique among retroviruses in its target cell specificity in vivo, a feature that partly reflects the ability of the virus to maintain infectivity in the presence of lung fluids. In addition, the promoter and enhancer sequences contained within the viral long terminal repeat are highly active in at least some lung cell lines that resemble the in vivo target cell of the virus (21). Thus, depending on the sequences required to target these cells for infection and those required to stimulate transformation, vectors based on JSRV may be particularly useful for treating diseases such as cystic fibrosis.
The ability of the human HYAL2 protein to mediate JSRV entry raises the possibility that JSRV infection might be involved in some human cases of BAC. However, no evidence of human infection has been presented, and an epidemiologic connection between individuals with non-smoking-related lung cancer and exposure to infected sheep has never been made. Reliable reagents to screen for the presence of antibodies and JSRV-related proteins are still lacking. However, material that reacts with anti-JSRV Gag antisera was detected in some lung cancer specimens in one recent study (22). Certainly a careful PCR analyses of lung cancer and other specimens by using primers that detect sequences related to JSRV is merited. Irrespective of the outcome of these analyses, experience with other oncogenic retroviruses has taught us that the likelihood of shared pathways in OPC and BAC is extremely high, suggesting that the JSRV model may reveal significant insights into an important human malignancy.
Acknowledgments
I thank Dr. John Coffin for helpful discussions and Zohar Sachs for assistance with the manuscript. This paper was supported by grants CA33771 and CA24220 from the National Institutes of Health/National Cancer Institute.
Footnotes
References
- 1.Rosenberg N, Jolicoeur P. In: Retroviruses. Coffin J M, Hughes S E, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 475–586. [PubMed] [Google Scholar]
- 2.Maeda N, Palmarini M, Murgia C, Fan H. Proc Natl Acad Sci USA. 2001;98:4449–4454. doi: 10.1073/pnas.071547598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rai S K, Duh F-M, Vigdorovich V, Danilkovitch-Miagkova A, Lerman M I, Miller A D. Proc Natl Acad Sci USA. 2001;98:4443–4448. doi: 10.1073/pnas.071572898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmarini M, Fan H, Sharp J M. Trends Microbiol. 1997;5:478–483. doi: 10.1016/S0966-842X(97)01162-1. [DOI] [PubMed] [Google Scholar]
- 5.DeMartini J C, York D F. Vet Clin N Am Food Anim Pract. 1997;13:55–70. doi: 10.1016/s0749-0720(15)30364-9. [DOI] [PubMed] [Google Scholar]
- 6.Barsky S H, Cameron R, Osann K E, Tomita D, Holmes E C. Cancer. 1994;73:1163–1170. doi: 10.1002/1097-0142(19940215)73:4<1163::aid-cncr2820730407>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 7.Palmarini M, Sharp J M, de las Heras M, Fan H. J Virol. 1999;73:6964–6972. doi: 10.1128/jvi.73.8.6964-6972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wistuba I I, Behrens C, Virmani A K, Mele G, Milchgrub S, Girard L, Fondon J W, Garner H R, McKay B, Latif F, et al. Cancer Res. 2000;60:1949–1960. [PubMed] [Google Scholar]
- 9.Lerman M I, Minna J D. Cancer Res. 2000;60:6116–6133. [PubMed] [Google Scholar]
- 10.Hunter E. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 71–120. [Google Scholar]
- 11.Alian A, Sela-Donenfeld D, Panet A, Eldor A. Virology. 2000;276:161–168. doi: 10.1006/viro.2000.0550. [DOI] [PubMed] [Google Scholar]
- 12.Ruscetti S. Int J Biochem Cell Biol. 1999;31:1089–1109. doi: 10.1016/s1357-2725(99)00074-6. [DOI] [PubMed] [Google Scholar]
- 13.Sekido Y, Ahmadian M, Wistuba I I, Latif F, Bader S, Wei M H, Duh F M, Gazdar A F, Lerman M I, Minna J D. Oncogene. 1998;16:3151–3157. doi: 10.1038/sj.onc.1201858. [DOI] [PubMed] [Google Scholar]
- 14.Lepperdinger G, Strobel B, Kreil G. J Biol Chem. 1998;273:22466–22470. doi: 10.1074/jbc.273.35.22466. [DOI] [PubMed] [Google Scholar]
- 15.Palmarini M, Sharp J M, Lee C, Fan H. J Virol. 1999;73:10070–10078. doi: 10.1128/jvi.73.12.10070-10078.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rai S K, DeMartini J C, Miller A D. J Virol. 2000;74:4698–4704. doi: 10.1128/jvi.74.10.4698-4704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmarini M, Hallwirth C, York D, Murgia C, de Oliveira T, Spencer T, Fan H. J Virol. 2000;74:8065–8076. doi: 10.1128/jvi.74.17.8065-8076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai J, Bishop J V, Carlson J O, DeMartini J C. Virology. 1999;258:333–343. doi: 10.1006/viro.1999.9728. [DOI] [PubMed] [Google Scholar]
- 19.Brown P. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 161–203. [Google Scholar]
- 20.Palmarini M, Dewar P, de las Heras M, Inglis N F, Dalziel G, Sharp J M. J Gen Virol. 1995;76:2731–2737. doi: 10.1099/0022-1317-76-11-2731. [DOI] [PubMed] [Google Scholar]
- 21.Palmarini M, Datta S R, Omid R, Murgia C, Fan H. J Virol. 2000;74:5776–5787. doi: 10.1128/jvi.74.13.5776-5787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de las Heras M, Barsky S H, Hasleton P, Wagner M, Larson E, Egan J, Ortin A, Gimenez-Mas J A, Palmarini M, Sharp J M. Eur Resp J. 2000;15:330–332. doi: 10.1034/j.1399-3003.2000.16b23.x. [DOI] [PubMed] [Google Scholar]


