Abstract
Cytoskeletal motors drive the transport of organelles and molecular cargoes within cells1, and have potential applications in molecular detection and diagnostic devices2,3. Engineering molecular motors with dynamically controllable properties will allow selective perturbation of mechanical processes in living cells, and yield optimized device components for complex tasks such as molecular sorting and directed assembly3. Biological motors have previously been modified by introducing activation/deactivation switches that respond to metal ions4,5 and other signals6. Here we show that myosin motors can be engineered to reversibly change their direction of motion in response to a calcium signal. Building on previous protein engineering studies7–11 and guided by a structural model12 for the redirected power stroke of myosin VI, we constructed bidirectional myosins through the rigid recombination of structural modules. The performance of the motors was confirmed using gliding filament assays and single fluorophore tracking. Our general strategy, in which external signals trigger changes in the geometry and mechanics of myosin lever arms, should enable spatiotemporal control over a range of motor properties including processivity, stride size13, and branchpoint turning14.
Controllable bidirectional motion is a basic feature of many man-made machines and is critical to the function of biological systems such as the bacterial flagellar motor, which reverses its direction of rotation in response to cellular signaling15. Bidirectional transport on cytoskeletal filaments is generally thought to rely on alternate engagement of distinct motors with differing fixed polarities, although context-dependent directionality has recently been observed in a mitotic kinesin16. No motors have been reported to move bidirectionally on actin: all characterized members of the myosin superfamily are specialized for motion toward either the (+) or the (−) end of the polarized filament. We endeavored to create new myosin motors that can be signaled to switch their direction of motion, challenging our understanding of mechanical adaptations in unconventional myosins.
In current formulations of the swinging crossbridge model17, conformational changes are propagated through the catalytic myosin head to drive a rotation of the converter domain. This rotation is amplified by a rigid lever arm extension, generating a directed power stroke toward the (+) end of the actin filament in most myosin classes. Artificial geometric redirection of the lever arm by insertion of a four-helix bundle generates a (−) end directed motor7. This effect mimics the proposed18 basis of natural (−) end directed motion in the specialized myosin VI motor, in which redirection is mediated by a unique insert between the converter domain and the lever arm19. Structural12 and functional8 studies of myosin VI converged on a refined model in which the unique insert conspires with a converter domain rearrangement12 to generate a large (−) end directed stroke. Engineered myosin VI motors10 with artificial lever arm structures derived from α-actinin20 showed that 17 residues of the unique insert are sufficient for lever arm redirection, and provided a starting point for the designs presented here.
We developed bidirectional transporters over the course of four successive design cycles: (1) minimization of the unique insert of myosin VI to generate a motor with an intermediate lever arm angle; (2) variation of lever arm length within this altered geometry, leading to length-dependent effects on directionality; (3) construction of a collapsible lever arm whose effective length is controlled by an external signal; and (4) assembly of monomeric motor units into processive dimers.
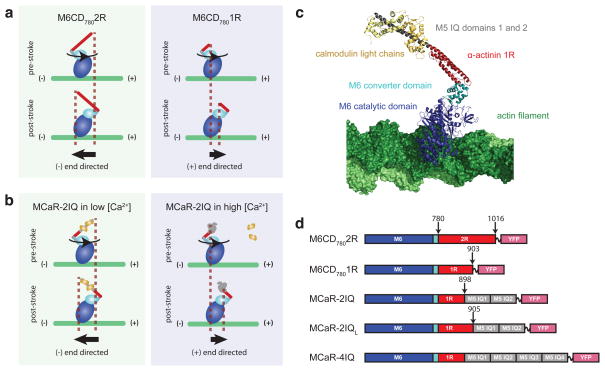
M6CD7802R (Fig. 1a) was generated by fusing two spectrin repeats of α-actinin10,20 following residue 780 of myosin VI, retaining only 7 residues of the unique insert. Gliding filament assays (Movie S1) show (−) end directed motion with lower velocities than M6CD7902R 10, as expected from a predicted lever arm angle that is intermediate between myosin VI and (+) end directed myosins. M6CD7801R was generated by shortening the lever arm of M6CD7802R to include only one spectrin repeat. M6CD7801R was found to be (+) end directed (Movie S1), implying that the lever arm was truncated beyond the neutral axis of the lever arm rotation (Figure 1a). In a panel of chimeric myosins with lever arms fused at five positions between 780 and 790, only fusions in the early portion of the unique insert (at 780 or 783) yielded directionalities that depended on lever arm length (Supplementary Figure 1).
Figure 1.
Engineered myosin designs. An artificial rigid lever arm20 composed of spectrin-like repeats (red) is fused to myosin VI after residue 780, retaining only 7 amino acids of the critical redirecting unique insert. This fusion is predicted to generate an exit angle that is intermediate between myosin VI and (+) end directed myosins. During the power stroke, rotation of the converter domain (light blue) leads to a net motion of the tip of the lever arm toward the (−) end of the actin filament for longer lever arms, and toward the (+) end for shorter lever arms. Fixed directionality constructs (a) with lever arms composed of differing numbers of spectrin repeats generate motion toward opposite ends of the actin filament. Calcium-controllable MCaR constructs (b) have chimeric lever arms composed of a single spectrin repeat fused to two IQ repeats (grey). Under low [Ca2+] conditions, the IQ domains are stabilized by binding to calmodulin (yellow), yielding a long lever arm and (−) end directed motion. High [Ca2+] favors dissociation of calmodulin, inducing a rigid to flexible transition that shortens the effective lever arm and reverses the direction of motion of the lever arm tip. An illustration (c) based on crystal structures shows the expected rigor conformation of MCaR-2IQ bound to the actin filament. In illustrations and block diagrams (d), structural modules are represented as follows: blue, myosin VI catalytic domain; light blue, myosin VI converter domain; red, one (1R) or two (2R) spectrin repeats from α-actinin; gray, IQ repeats from myosin V; ~, (GSG)4 flexible linker; pink, eYFP. Sequences of junctions between modules are shown in Supplementary Figure 4.
Dynamically controllable constructs (Figure 1b) were designed to transition between structures that mimic M6CD7801R and M6CD7802R. We chose calcium as the signal to control this transition, borrowing a signal that is widely used to control cytoskeletal motors in vivo, including distinct mechanisms used to activate skeletal21 and smooth22 muscle myosin. Calcium is used as both a natural23 and engineered5 regulator of kinesin function, and myosin V is activated by calcium in the Drosophila pupil24.
To achieve dynamic control over lever arm length (Fig. 1b), we incorporated IQ domains derived from the neck region of myosin V25,26. The helical structure of IQ domains is stabilized by binding to calmodulin. Elevated [Ca2+] can favor dissociation of calmodulin from IQ domains25,26, which is expected to destabilize helical structure. Thus, the IQ motif should function as a collapsible element whose effective length can be controlled by [Ca2+]. Dissociation of calmodulin causes inhibition of myosin V activity in vitro25, although it is not known whether this regulatory mechanism is used in vivo.
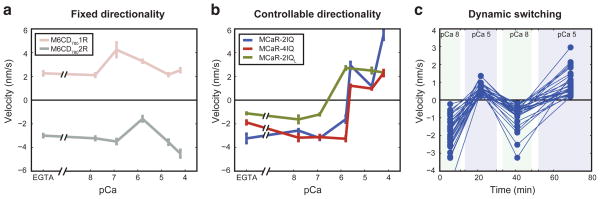
We constructed three monomeric myosins with calcium-controlled reversibility (MCaR-2IQ, MCaR-2IQL, and MCaR-4IQ) by fusing M6CD7801R to collapsible lever arm extensions composed of IQ domains (Figure 1b–d). In gliding filament assays (Movies S2–S4), all three MCaR constructs display calcium-sensitive directionality as expected, whereas the fixed directionality constructs are insensitive to calcium (Figure 2). We confirmed that gliding filament directionality could be switched by exchanging buffers in situ (Movies S5–S6), and we tested for dynamic and reversible directionality switching by cycling MCaR-2IQ repeatedly between low and high calcium concentrations (Figure 2c and Movie S6A,B).
Figure 2.
Gliding filament assays of engineered myosins. Filament velocities were determined in the presence of 2.5 μM calmodulin, 2 mM ATP, and the indicated concentrations of free calcium, reported as pCa = −log10([Ca2+]). Positive velocities indicate (+) end directed motion. Data points with error bars indicate mean ± SEM across sets of filaments (see Supplementary Table 1 and Supplementary Figure 2). For fixed directionality constructs (a), gliding direction is sensitive to engineered lever arm length but not to calcium (Movie S1). MCaR constructs (b) switch directionality near pCa 6, corresponding to ~μM concentrations of free calcium (Movies S2, S3, S4). A velocity time course (c) shows the results of a dynamic switching assay with MCaR-2IQ. pCa buffer was exchanged three times in the flow chamber while observing the same field of filaments (Movies S6A,B). Conditions alternated between pCa 8 (10 nM free calcium) and pCa 5 (10 μM free calcium). Out of 47 filaments analyzed, 30 filaments (shown here) switched directions three times; see Supplementary Figure 3 for complete tabulation of filament behaviors.
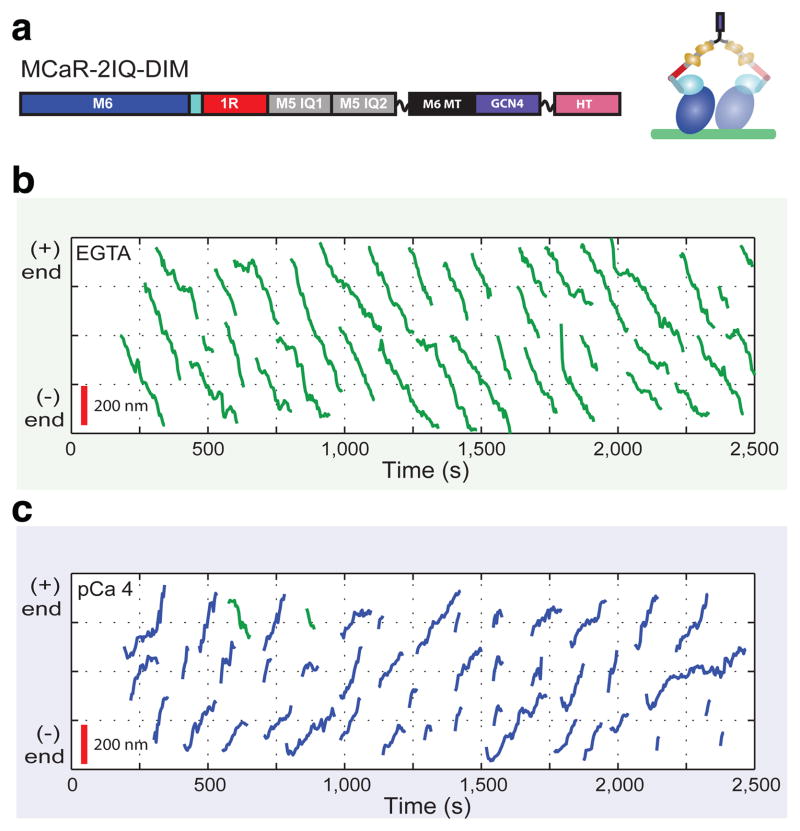
Molecular transport carried out by individual motor complexes depends on processive motion along cytoskeletal filaments, such as the hand-over-hand motion observed in dimeric myosin V and myosin VI. We constructed bidirectional dimers by fusing MCaR-2IQ to the medial tail region of myosin VI followed by a leucine zipper (Figure 3a) as described for previous engineered myosin VI variants10,11, and asked whether these constructs were capable of processive motion. Many models of processive dimer function emphasize the importance of coordination mediated by intramolecular tension, which may be missing or impaired in MCaR-2IQ-DIM, particularly in the high [Ca2+] state. However, recent studies of myosin VI constructs with artificial lever arms have found that processive motion may be achieved without optimizing lever arm geometry or mechanics10,11. Together with kinetic modeling11, these results argue that a high duty ratio may often be sufficient to confer processivity. Using single fluorophore tracking (Movies S7–S9), we found that MCaR-2IQ-DIM indeed supports processive motion toward both (−) and (+) ends of actin filaments (Figure 3b–c).
Figure 3.
Processive motility of a dimeric controllable motor. Dimers were constructed (a) by fusion of MCaR-2IQ to the medial tail region of myosin VI (black) followed by the GCN4 leucine zipper (green) and a HaloTag (pink). Assays of processive motion on immobilized actin filaments were performed in 50 μM ATP, 2 μM calmodulin, and EGTA (b) or pCa 4 (100 μM free calcium) buffer (c). Compiled traces from single fluorophore tracking (Movies S7–S9) are shown on a polarized axis representing distance traveled along the actin filament. Directionalities were scored by comparing to subsequent assays of control (−) end directed M6DI8162R-DIM motors10,11 on the same filaments (Movies S8, S9). For MCaR-2IQ-DIM, (−) end directed traces (green) are ubiquitous in EGTA but represent a small minority (6%) of traces in pCa 4. A total of 75 runs in EGTA and 36 runs in pCa 4 were analyzed for velocity and processivity, yielding average velocities of −2.9±1.0 nm/s in EGTA and +3.2±2.0 nm/s in pCa 4, and processive run lengths of 550 ± 50 nm in EGTA and 170 ± 30 nm in pCa 4.
The successful construction of controllable bidirectional motors broadly validates our design based on combining lever arm redirection with dynamic control of lever arm length. However, our results also contain unanticipated details that shed new light on structure/function relationships in myosin VI and suggest new structural hypotheses to guide future design cycles. Models of the power stroke (Supplementary Molecular Models) based on alignment to recently reported myosin VI structures12 do not correctly predict the location of the neutral axis in artificial lever arms fused at residues 780 or 783. The behavior of these constructs is more successfully predicted by a model that omits the reported converter domain rearrangement12, and our results suggest that the initial portion of the unique insert through V784 may be required for the rearrangement. In this region of the helical unique insert, residues L777, V780, V784, and N785 form a surface that makes differing contacts in the reported pre-stroke and rigor conformations of the converter12.
Simple geometric considerations are also insufficient to explain differences in behaviors between the three monomeric MCaR constructs (Figure 2b). We hypothesize that the low (−) end directed velocity of MCaR-2IQL relative to MCaR-2IQ may result from a decrease in lever arm rigidity due to the insertion of additional residues at the actinin/IQ junction. Accurate prediction of lever arm rigidities will be important for quantitative design of engineered myosins13,27. The local environment of the actinin/IQ junction may also influence the interaction of calmodulin with MCaR-2IQL, which requires lower calcium concentrations than MCaR-2IQ to switch to (+) end directed motility (Figure 2b). We should note that it is unknown whether calmodulin dissociates from both IQ domains in these constructs. In myosin V, calcium-induced dissociation occurs from IQ2 alone26, and our most successful structural model for MCaR-2IQ suggests that this could be sufficient for directionality reversal (Supplementary Molecular Models).
A future challenge will be to optimize the velocities and processivities of controllable bidirectional myosins. MCaR velocities are low in comparison to many natural myosins, which may be explained by short predicted stroke sizes (Supplementary Molecular Models), and construct-dependent kinetic perturbations that further extend the long-lived actin-bound phase of the myosin VI cycle, as seen earlier8. Velocity improvements may be achieved through alterations of the catalytic domain to accelerate nucleotide turnover, through mutagenesis or replacement of loops that act as kinetic determinants28, or construction of chimeras with faster-moving myosin classes. Higher processivities are also desirable since MCaR-2IQ-DIM is only robustly processive at subsaturating ATP concentrations (Figure 3), and might be achieved through mutagenesis of actin-binding loops29 or formation of higher-order complexes.
We have engineered a novel mechanical response to a natural cellular signal. Temporal and spatial modulation of calcium is used to regulate a diversity of cellular processes, and spatially localized changes in calcium levels may be artificially triggered using techniques such as laser-induced photolysis of caged compounds30. In a biological context, our designs allow rewiring of existing signaling mechanisms to control a new output31, but it would also be desirable to develop motors controlled by orthogonal signals such as small molecules or light32. Our conceptual designs based on rigid to flexible transitions are sufficiently general that they should be adaptable to a range of different control signals.
Transitions in lever arm structures such as those described here may be designed to cause changes in a wide range of motor functions controlled by lever arm geometry and mechanics, including step size selection13 and branchpoint turning14. Combined with the optimizations described above, these developments will expand the range of cellular mechanical functions engineered to respond to external signals31,32, and provide unprecedented levels of control over nanoscale motion in devices that harness the proven capabilities of biological molecular motors2,3.
METHODS
Proteins
DNA constructs were assembled from modules including codons 1–780 (encoding the catalytic domain) and codons 909–992 (encoding the medial tail) of porcine myosin VI, codons 266–502 of Dictyostelium α-actinin (truncated earlier when indicated), codons 766–813 (2IQ) or 766–861 (4IQ) of chicken myosin V, eYFP, and the GCN4 leucine zipper. Sequences of junctions between structural modules are shown in Supplementary Figure 4. All constructs were terminated in a C-terminal flag tag (GDYKDDDK), cloned into pBiEx-1 (Novagen), expressed by direct transfection of SF9 cells, and affinity purified as described earlier8,10,11. MCaR-2IQ-DIM protein was labeled with TMR-HaloTag ligand (Promega) before elution from anti-flag resin11.
Buffers for gliding filament assays and single fluorophore tracking
Buffers used for all incubation, wash, and assay steps were prepared at the indicated pCa values using 25 mM imidazole-HCl (pH. 7.45), 25 mM KCl, 10 mM DTT, 2 mM MgCl2, calmodulin as indicated, and Ca-EGTA buffer prepared as described33 with a total EGTA concentration of 1 mM. Free calcium concentrations were predicted using MaxChelator (http://maxchelator.stanford.edu) and measured using a calcium sensitive electrode (ThermoScientific). Gliding filament and single fluorophore tracking assays also included ATP as indicated, an ATP regeneration system (1 mM phosphocreatine, 100 μg/ml creatinephosphokinase), an oxygen scavenging system (0.4% glucose, 0.2 mg/mol glucose oxidase, and 36 μg/ml catalase), and 1.77 mM trolox as an antibleaching agent.
Gliding filament assays
Dual labeled gliding filament assays were performed as described earlier8,10 in flow cells coated with mouse monoclonal anti-GFP (Millipore) followed by incubation with myosin-YFP. Videomicroscopy data were collected using simultaneous two-channel imaging of Cy5-actin and TMR-phalloidin on an EMCCD camera (Andor) under objective-side TIRF illumination with a 532 nm DPSS laser (Newport) through a 1.49 NA 100X objective (Nikon). Filaments were tracked using custom software. Only polarity-marked filaments that traveled further than 100 nm were scored for directionality and velocity.
Single fluorophore tracking
Assays of dimeric myosin were performed as previously described11 using Alexa 633-phalloidin (Invitrogen) labeled actin filaments attached to the surface via N-ethyl maleimide inactivated full-length skeletal muscle myosin (a gift of Roger Cooke). Processive runs longer than 100 nm were scored for velocity and run length, and average run lengths were determined after correcting for this minimum threshold and accounting for premature termination events as described earlier11.
Supplementary Material
Acknowledgments
We thank M.W. Elting for valuable discussions and assistance, R. Cooke for providing full-length muscle myosin, and S. Sutton for providing purified actin. This work was supported by a Pew Scholars Award and NIH Grant DP2 OD004690 to Z.B., an NSF Graduate Research Fellowship to L.C., an AHA Predoctoral Fellowship to M.N., and a Stanford Graduate Fellowship to T.D.S.
Footnotes
AUTHOR CONTRIBUTIONS
L.C., M.N., and Z.B. designed the molecular motor constructs. L.C., M.N., and T.D.S. performed experiments on fixed directionality motors. L.C. and M.N. performed experiments on bidirectional motors. L.C. analyzed the experimental data. D.P., L.C., and M.N. performed structural modeling. Z.B. conceived and supervised the project. L.C. and Z.B. wrote the paper. All authors discussed the results and commented on the manuscript.
ADDITIONAL INFORMATION
Supplementary information accompanies this paper at www.nature.com/naturenanotechnology.
Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–80. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- 2.Korten T, Mansson A, Diez S. Towards the application of cytoskeletal motor proteins in molecular detection and diagnostic devices. Current opinion in biotechnology. 2010;21:477–88. doi: 10.1016/j.copbio.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Goel A, Vogel V. Harnessing biological motors to engineer systems for nanoscale transport and assembly. Nature Nanotechnology. 2008;3:465–75. doi: 10.1038/nnano.2008.190. [DOI] [PubMed] [Google Scholar]
- 4.Liu H, et al. Control of a biomolecular motor-powered nanodevice with an engineered chemical switch. Nat Mater. 2002;1:173–7. doi: 10.1038/nmat761. [DOI] [PubMed] [Google Scholar]
- 5.Konishi K, Uyeda TQ, Kubo T. Genetic engineering of a Ca(2+) dependent chemical switch into the linear biomotor kinesin. FEBS letters. 2006;580:3589–94. doi: 10.1016/j.febslet.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 6.Nomura A, Uyeda TQP, Yumoto N, Tatsu Y. Photo-control of kinesin-microtubule motility using caged peptides derived from the kinesin C-terminus domain. Chem Commun (Camb) 2006:3588–90. doi: 10.1039/b606538d. [DOI] [PubMed] [Google Scholar]
- 7.Tsiavaliaris G, Fujita-Becker S, Manstein DJ. Molecular engineering of a backwards-moving myosin motor. Nature. 2004;427:558–61. doi: 10.1038/nature02303. [DOI] [PubMed] [Google Scholar]
- 8.Bryant Z, Altman D, Spudich JA. The power stroke of myosin VI and the basis of reverse directionality. Proc Natl Acad Sci USA. 2007;104:772–7. doi: 10.1073/pnas.0610144104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park H, et al. The unique insert at the end of the myosin VI motor is the sole determinant of directionality. Proc Natl Acad Sci USA. 2007;104:778–83. doi: 10.1073/pnas.0610066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao JC, Elting MW, Delp SL, Spudich JA, Bryant Z. Engineered myosin VI motors reveal minimal structural determinants of directionality and processivity. Journal of Molecular Biology. 2009;392:862–7. doi: 10.1016/j.jmb.2009.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elting MW, Bryant Z, Liao JC, Spudich JA. Detailed tuning of structure and intramolecular communication are dispensable for processive motion of myosin VI. Biophysical Journal. 2011;100:430–9. doi: 10.1016/j.bpj.2010.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Menetrey J, Llinas P, Mukherjea M, Sweeney HL, Houdusse A. The structural basis for the large powerstroke of myosin VI. Cell. 2007;131:300–8. doi: 10.1016/j.cell.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 13.Vilfan A. Elastic lever-arm model for myosin V. Biophysical Journal. 2005;88:3792–805. doi: 10.1529/biophysj.104.046763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vilfan A. Myosin V passing over Arp2/3 junctions: branching ratio calculated from the elastic lever arm model. Biophysical Journal. 2008;94:3405–12. doi: 10.1529/biophysj.107.120568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee LK, Ginsburg MA, Crovace C, Donohoe M, Stock D. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature. 2010;466:996–1000. doi: 10.1038/nature09300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roostalu J, et al. Directional switching of the kinesin Cin8 through motor coupling. Science. 2011;332:94–9. doi: 10.1126/science.1199945. [DOI] [PubMed] [Google Scholar]
- 17.Holmes KC, Schröder RR, Sweeney HL, Houdusse A. The structure of the rigor complex and its implications for the power stroke. Philos Trans R Soc Lond, B, Biol Sci. 2004;359:1819–28. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells AL, et al. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–8. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 19.Menetrey J, et al. The structure of the myosin VI motor reveals the mechanism of directionality reversal. Nature. 2005;435:779–85. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anson M, Geeves MA, Kurzawa SE, Manstein DJ. Myosin motors with artificial lever arms. The EMBO journal. 1996;15:6069–74. [PMC free article] [PubMed] [Google Scholar]
- 21.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 22.Somlyo AP, Somlyo AV. Signal transduction and regulation in smooth muscle. Nature. 1994;372:231–6. doi: 10.1038/372231a0. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Schwarz TL. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 2009;136:163–74. doi: 10.1016/j.cell.2008.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satoh AK, Li BX, Xia H, Ready DF. Calcium-activated Myosin V closes the Drosophila pupil. Current biology : CB. 2008;18:951–5. doi: 10.1016/j.cub.2008.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krementsov DN, Krementsova EB, Trybus KM. Myosin V: regulation by calcium, calmodulin, and the tail domain. The Journal of Cell Biology. 2004;164:877–86. doi: 10.1083/jcb.200310065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trybus KM, et al. Effect of calcium on calmodulin bound to the IQ motifs of myosin V. J Biol Chem. 2007;282:23316–25. doi: 10.1074/jbc.M701636200. [DOI] [PubMed] [Google Scholar]
- 27.Parker D, Bryant Z, Delp SL. Coarse-Grained Structural Modeling of Molecular Motors Using Multibody Dynamics. Cellular and molecular bioengineering. 2009;2:366–374. doi: 10.1007/s12195-009-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sweeney HL, et al. Kinetic tuning of myosin via a flexible loop adjacent to the nucleotide binding pocket. The Journal of biological chemistry. 1998;273:6262–70. doi: 10.1074/jbc.273.11.6262. [DOI] [PubMed] [Google Scholar]
- 29.Krementsova EB, Hodges AR, Lu H, Trybus KM. Processivity of chimeric class V myosins. The Journal of biological chemistry. 2006;281:6079–86. doi: 10.1074/jbc.M510041200. [DOI] [PubMed] [Google Scholar]
- 30.Zheng JQ, Poo MM. Calcium signaling in neuronal motility. Annual review of cell and developmental biology. 2007;23:375–404. doi: 10.1146/annurev.cellbio.23.090506.123221. [DOI] [PubMed] [Google Scholar]
- 31.Yeh BJ, Rutigliano RJ, Deb A, Bar-Sagi D, Lim WA. Rewiring cellular morphology pathways with synthetic guanine nucleotide exchange factors. Nature. 2007;447:596–600. doi: 10.1038/nature05851. [DOI] [PubMed] [Google Scholar]
- 32.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsien R, Pozzan T. Measurement of cytosolic free Ca2+ with quin2. Meth Enzymol. 1989;172:230–62. doi: 10.1016/s0076-6879(89)72017-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.