Abstract
AIM: To identify differentially expressed genes in quiescent and activated hepatic stellate cells (HSCs) and explore their functions.
METHODS: HSCs were isolated from the normal Sprague Dawley rats by in suit perfusion of collagenase and pronase and density Nycodenz gradient centrifugation. Total RNA and mRNA of quiescent HSCs, and culture-activated HSCs were extracted, quantified and reversely transcripted into cDNA. The global gene expression profile was analyzed by microarray with Affymetrix rat genechip. Differentially expressed genes were annotated with Gene Ontology (GO) and analyzed with Kyoto encyclopedia of genes and genomes (KEGG) pathway using the Database for Annotation, Visualization and Integrated Discovery. Microarray data were validated by quantitative real-time polymerase chain reaction (qRT-PCR). The function of Wnt5a on human HSCs line LX-2 was assessed with lentivirus-mediated Wnt5a RNAi. The expression of Wnt5a in fibrotic liver of a carbon tetrachloride (CCl4)-induced fibrosis rat model was also analyzed with Western blotting.
RESULTS: Of the 28 700 genes represented on this chip, 2566 genes displayed at least a 2-fold increase or decrease in expression at a P < 0.01 level with a false discovery rate. Of these, 1396 genes were upregulated, while 1170 genes were downregulated in culture-activated HSCs. These differentially expressed transcripts were grouped into 545 GO based on biological process GO terms. The most enriched GO terms included response to wounding, wound healing, regulation of cell growth, vasculature development and actin cytoskeleton organization. KEGG pathway analysis revealed that Wnt5a signaling pathway participated in the activation of HSCs. Wnt5a was significantly increased in culture-activated HSCs as compared with quiescent HSCs. qRT-PCR validated the microarray data. Lentivirus-mediated suppression of Wnt5a expression in activated LX-2 resulted in significantly impaired proliferation, downregulated expressions of type I collagen and transforming growth factor-β1. Wnt5a was upregulated in the fibrotic liver of a CCl4-induced fibrosis rat model.
CONCLUSION: Wnt5a is involved in the activation of HSCs, and it may serve as a novel therapeutic target in the treatment of liver fibrosis.
Keywords: Hepatic stellate cells, Wnt5a, Microarray, Bioinformatics analyses, Liver fibrosis
INTRODUCTION
Liver fibrosis is a common pathological change characterized by excessive deposition of extracellular matrix (ECM) that occurs in most types of chronic liver diseases. Hepatic stellate cells (HSCs) are a major source of ECM and activation of HSCs is one of the key steps in the development of liver fibrosis[1,2]. However, the molecular mechanisms underlying the activation of HSCs are not fully understood.
Previous studies have reported the differentially expressed gene profiles in quiescent and activated HSCs with gene chip. De Minicis et al[3] determined gene expression changes in three different models of HSC activation: culture-activated HSCs, HSCs isolated from CCl4-treated mice and mice underwent bile duct ligation (BDL). They found that the gene expression pattern of culture-activated HSCs was different from that of BDL- and CCl4-activated HSCs. Woo et al[4] performed a time-based study to investigate the differences in gene expression patterns during the activation of HSCs with microarrays and revealed a number of newly discovered genes involved in fibrogenesis during the activation of HSCs. Sancho-Bru et al[5] investigated the differences in phenotypic, genomic and functional characteristics between HSCs from human cirrhotic livers and HSCs from normal livers after prolonged culture. However, these three studies are lack of intense bioinformatics analyses.
In this study, we performed gene expression profile analyses using oligonucleotide microarrays to identify the genes involved in HSCs activation. These analyses revealed the altered expression of 2566 genes. On this basis, bioinformatics analyses were done to classify the function and explore the involved signaling pathway of these differentially expressed genes. Bioinformatics analyses revealed that these differentially expressed transcripts were grouped into 545 GO based on biological process GO terms. The most enriched GO terms included response to wounding, wound healing, regulation of cell growth, vasculature development, and actin cytoskeleton organization. It is reported that Wnt5a signaling takes part in wound healing, regulation of cell proliferation, vasculature development and the formation of cell polarity [6-9]. After intensive analyses of Kyoto encyclopedia of genes and genomes (KEGG) pathways, we found that Wnt5a signaling pathway participated in the HSCs activation, an observation verified by quantitative real-time polymerase chain reaction (qRT-PCR).
Because the role of Wnt5a in HSCs activation is unknown and the direct evidence has not been reported, we treated human HSCs line LX-2 with lentivirus-mediated Wnt5a ShRNA and found that knockdown of Wnt5a gene expression in LX-2 led to significantly impaired proliferation, downregulated expressions of type I collagen and transforming growth factor-β1 (TGF-β1). Wnt5a was upregulated in fibrotic liver of a carbon tetrachloride (CCl4)-induced fibrosis rat model. Collectively, our data suggest that Wnt5a may play a role in HSCs activation and liver fibrogenesis. We expect that our results will help illustrate the molecular mechanisms involved in liver fibrogenesis and provide preliminary experimental evidence for the feasibility of targeting Wnt5a in gene therapy of liver fibrogenesis.
MATERIALS AND METHODS
Cell isolation, identification and culture
HSCs were isolated from normal Sprague Dawley (SD) rats by collagenase-pronase perfusion and subsequent density centrifugation on Nycodenz gradients as described previously[10]. Purity was tested by retinoid autofluorescence and exceeded 95% in all isolations. HSCs were cultured in Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), glutamine, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer and antibiotics. After 1 (quiescent HSCs) and 7 (activated HSCs) d of culture, total RNA was extracted using an RNeasy mini-kit (Qiagen, Valencia, CA, United States). Human HSCs line LX-2 was cultured in DMEM (Invitrogen) supplemented with 10% FBS.
Microarray experiments and bioinformatics analysis
We used rat genome 430 2.0 array gene chips (Affymetrix, Santa Clara, CA). The arrays were hybridized, washed and scanned according to the standard Affymetrix protocol. The gene chip tests were performed by professional staffs of Shanghai Biochip Company. Chips were scanned with a Genechip Scanner 3000 7G (Affymetrix). Data analysis was performed using GCOS1.2 software. After background correction, we performed normalization for each array and gene. Data were filtered based on both signal intensity and detection call. Gene activity was considered to differ between quiescent HSCs and culture-activated HSCs (P < 0.01) when compared by the unpaired Student’s t test using multiple testing correction (Benjamini and Hochberg False Discovery Rate). Molecular function and classification of differentially expressed genes were analyzed with GO and KEGG pathway using the Database for Annotation, Visualization and Integrated Discovery Bioinformatics Resources 6.7[11] (http://david.abcc.ncifcrf.gov).
Quantitative real-time polymerase chain reaction
Five differentially expressed genes were selected for qRT-PCR analysis, with glyceraldehydes-3-phosphate dehydrogenase (GAPDH) as a control, to verify the reliability of the array data. The primer sequences of analyzed genes are shown in Table 1. Super Script III reverse transcriptase (Invitrogen) was used for reverse transcription reactions. PCR amplification was performed using TaqMan universal Master Mix (Applied Biosystems, Foster City, CA, United States). Amplified reactions were quantified using an ABI PRISM 7900 Sequence detection system (Applied Biosystems, Foster City, CA, United States). Experiments were performed in triplicate. Relative gene quantities were obtained using the comparative Ct method after normalization to GAPDH control genes.
Table 1.
Primer sequences of genes validating the microarray analysis by quantitative real-time polymerase chain reaction
| Gene name | Sense primer | Antisense primer |
| Wnt5a | AGGGCTCCTACGAGAGTGCT | GACACCCCATGGCACTTG |
| Frizzled2 | TCTCTGAGGACGGTTATCGCA | CAGAATCACCCACCAGATGGA |
| Calcium calmodulin mediated kinase IIdelta | TCGGCTCACACAGTACATGGA | CCCCGAACGATGAAAGTGAA |
| α-1 type I collagen | ACAGACTGGCAACCTCAAGAAG | AAGCGTGCTGTAGGTGAATCG |
| Connective tissue growth factor | GTTGGCGAACAAATGGCCTT | TGCCTCCCA AACCAGTCATAG |
| Glyceraldehydes-3-phosphate dehydrogenase | TCCTGCACCACCAACTGCTTAG | AGTGGCAGTGATGGCATGGACT |
Construction of lentivirus-mediated Wnt5a RNAi
Lentiviral vector pll3.7 expressing human Wnt5a ShRNA targeting 5’-GGACUUUCUCAAGGACAGA-3’ (538-556) was constructed by Shanghai Biyang Biotechology. Plasmids from correct clones were amplified by transforming into DH5α Escherichia coli cells. We used tronolab lentiviral vector system (pRsv-REV, pMDlg-pRRE and pMD2G) to perform lentivirus-mediated Wnt5a RNAi. An additional scrambled sequence was produced as a negative siRNA control. The scrambled sequence was not homologous with any known gene in mammalian cells as determined by a Basic Local Alignment Search Tool. 293T cells were transfected with polybrene. The crude virus lysates were amplified in 293T cells to generate higher titre viral stocks. Virus particles were purified and concentrated. The lentivirus titer was determined by a limited dilution plaque assay. LX-2 transfected with lentivirus-mediated pll3.7 Wnt5a ShRNA at concentrations of MOI 10 and MOI 20 was used as study cells; cells transfected with negative siRNA were negative controls, and cells without transfection were blank controls. The effect of Wnt5a knockdown was validated with Western blotting. Blots were developed with enhanced chemiluminescence detection reagents (Santa Cruz Biotechnology Inc., CA, United States), exposed on Kodak Xdmat blue XB-1 film and quantified by Bandscan 5.0 software using β-actin as internal control.
Effect of Wnt5a RNAi on cell proliferation
Cells were spread onto 96-well flat-bottom plates at a density of 5 × 104/mL, 100 μL per well, and cultured in DMEM including 5% fetal calf serum. Subgroups included: control group, negative siRNA control group and Wnt5a ShRNA group. Each group was repeated for 6 wells. The plates were incubated in a 5% CO2 humidified incubator at 37 °C for 24 h and 200 μL of 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) (5 mg/mL) was added into each well. The plates were then incubated for 4 h and the supernatants were removed. After 150 μL DMSO was added into each well, the plates were agitated for 20 min to dissolve the crystals. Finally, the A values were measured using the enzyme-linked immunoassay at a wave-length of 450 nm.
Effect of Wnt5a RNAi on the expression of collagen I and transforming growth factor-β1
Effects of Wnt5a RNAi on the expression of collagen I and TGF-β1 in LX-2 were analyzed with RT-PCR in vitro. Experiments were performed in triplicate. The primer of α-1 type I collagen (COL1A1) : sense primer: 5’-TGCCAAGGGAGATGCTGGTC-3’, antisense primer: 5’-TTCACCCTTAGCACCAACAGCA-3’, product size 238 bp; TGFβ1: sense primer: 5’-TGCAAGACTATCGACATGGAG-3’, antisense primer: 5’-ACTTGAGCCTCAGCAGACGCA-3’, product size 389 bp. The amplified products were applied to electrophoresis on 1% agarose gel analysis. The relative expression levels of COL1A1 and TGFβ1 were assessed based on the β-actin control by calculating the average ratios of light density using symmetry computerized gel imaging system.
Experimental fibrotic model
Liver fibrosis was induced by subcutaneous injection at hind leg of 40% CCl4-tea oil , 0.5 mL/100 g body weight for the first time and then 0.3 mL/100 g body weight twice a week for 8 wk. Control rats received only tea oil. The amount of CCl4 administered was adjusted for body weight each week. Liver tissue samples obtained from CCl4-treated and non-treated control rats were dehydrated, embedded in paraffin, sectioned and stained with hematoxylin and eosin and Masson staining. The expression of Wnt5a was analyzed with Western blot with antibodies to Wnt5a (sc-30224, Santa Cruz, United States). The bands were semi-quantitatively evaluated by densitometric analysis. Protein expression levels of Wnt5a were thereby normalized to those of the housekeeping gene β-actin.
Statistical analysis
Statistical analyses were performed using SPSS 13.0 software. Two-way analysis of variance was performed to detect the effects of Wnt5a knockdown on cell proliferation and Student t test was used to detect the difference between any two groups. The results were considered statistically significant at P < 0.05.
RESULTS
Microarray analysis and validation of microarray data
Culture-activated HSCs exhibited myofibroblast-like appearance. cRNAs, prepared from quiescent HSCs and culture-activated HSCs, were hybridized to a rat genome 430 2.0 array gene chips. Of the 28 700 genes represented therein, 2566 genes displayed at least a 2-fold increase or decrease in expression at the P < 0.01 level with a false discovery rate. Of these, 1396 genes were upregulated, while 1170 genes were downregulated in culture-activated HSCs. As expected, genes such as COL1A1 and connective tissue growth factor (CTGF) were significantly upregulated. We were particularly interested in Wnt proteins in view of their potential role in HSCs activation. Wnt5a was upregulated by approximately 6-fold in activated HSCs in comparison with quiescent HSCs , its downstream factors Frizzled2 was upregulated by 3.2-fold and calcium calmodulin mediated kinase II (CamKII) was upregulated by 1.7-fold. Changes in gene expression profiles observed in the microarrays were validated by qRT-PCR analysis using the same total RNAs analyzed in the microarrays. We verified the upregulation of Wnt5a, Frizzled2 and CamKII as well as COL1A1, CTGF (Figure 1). This verification confirmed that all the qRT-PCR results were in agreement with the microarray data.
Figure 1.
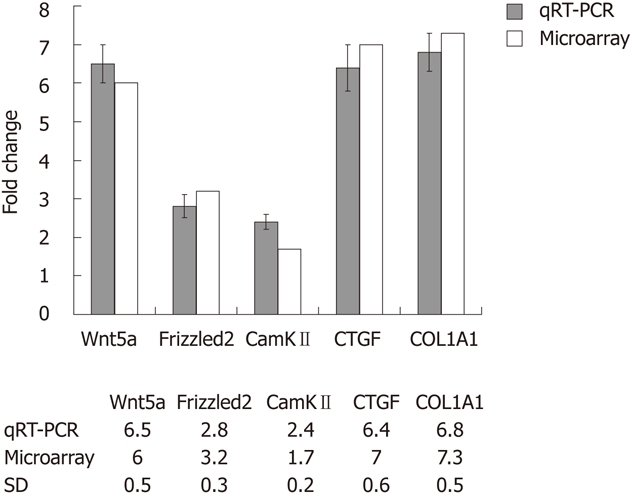
Validation of microarray data by quantitative real-time polymerase chain reaction. Comparison between expression values of five selected differently expressed genes estimated by quantitative real-time polymerase chain reaction (qRT-PCR) and gene expression data. The detected changes measured by qRT-PCR reflected different changes in gene expression between quiescent and activated hepatic stellate cells obtained by microarray analysis. SD: Sprague Dawley; CTGF: Connective tissue growth factor; CamKII: Calcium calmodulin mediated kinaseII.
Gene ontology analysis
To elucidate the relationship between gene differential expression patterns of quiescent HSCs and culture-activated HSCs, we examined the functional bias of 2566 differentially expressed transcripts according to Gene Ontology (GO) classifications. These differentially expressed transcripts were grouped into 545 GO based on biological process GO terms. The most enriched GO terms included response to wounding, wound healing and regulation of cell growth (Table 2). Analyses of GO also indicated that there were 102 GO terms identified by cellular component classification, and 90 GO terms identified by molecular function classification.
Table 2.
Top 20 enriched Gene Ontology terms based on Gene Ontology classifications
| Biological process | Count | Percent | Molecular function | Count | Percent | Cellular component | Count | Percent |
| Response to wounding | 78 | 7.24 | Cytoskeletal protein binding | 73 | 6.78 | Extracellular region part | 125 | 11.61 |
| Wound healing | 46 | 4.27 | Actin binding | 52 | 4.83 | Extracellular region | 180 | 16.71 |
| Regulation of cell growth | 43 | 3.99 | Growth factor binding | 24 | 2.23 | Extracellular matrix | 69 | 6.41 |
| Vasculature development | 50 | 4.64 | Glycosaminoglycan binding | 24 | 2.23 | Proteinaceous extracellular matrix | 61 | 5.66 |
| Actin cytoskeleton organization | 42 | 3.90 | Calcium ion binding | 76 | 7.06 | Extracellular matrix part | 34 | 3.16 |
| Actin filament-based process | 43 | 3.99 | Extracellular matrix structural constituent | 14 | 1.30 | Cytoskeleton | 125 | 11.61 |
| Blood vessel development | 48 | 4.46 | Heparin binding | 19 | 1.76 | Basement membrane | 25 | 2.32 |
| Cell migration | 51 | 4.73 | Polysaccharide binding | 24 | 2.23 | Extracellular space | 71 | 6.59 |
| Cell adhesion | 72 | 6.69 | Pattern binding | 24 | 2.23 | Cell projection | 92 | 8.54 |
| Biological adhesion | 72 | 6.69 | Insulin-like growth factor binding | 11 | 1.02 | Cytoskeletal part | 88 | 8.17 |
| Regulation of growth | 55 | 5.11 | Integrin binding | 12 | 1.11 | Cell surface | 53 | 4.92 |
| Regulation of cellular component size | 46 | 4.27 | Carbohydrate binding | 39 | 3.62 | Actin cytoskeleton | 39 | 3.62 |
| Extracellular matrix organization | 27 | 2.51 | Chemokine activity | 10 | 0.93 | Actin filament bundle | 14 | 1.30 |
| Regulation of cell proliferation | 87 | 8.08 | Chemokine receptor binding | 10 | 0.93 | Plasma membrane | 216 | 20.06 |
| Cytoskeleton organization | 54 | 5.01 | Protein complex binding | 29 | 2.69 | Collagen | 12 | 1.11 |
| Response to organic substance | 109 | 10.1 | Identical protein binding | 57 | 5.29 | Actomyosin | 14 | 1.30 |
| Response to hormone stimulus | 72 | 6.69 | Collagen binding | 8 | 0.74 | Stress fiber | 13 | 1.21 |
| Response to endogenous stimulus | 77 | 7.15 | Growth factor activity | 20 | 1.86 | Microtubule cytoskeleton | 53 | 4.92 |
| Cell motion | 62 | 5.76 | Platelet-derived growth factor binding | 5 | 0.46 | Cell leading edge | 26 | 2.41 |
| Blood vessel morphogenesis | 38 | 3.52 | Phospholipid binding | 20 | 1.86 | Vesicle lumen | 15 | 1.39 |
Kyoto encyclopedia of genes and genomes pathway analysis
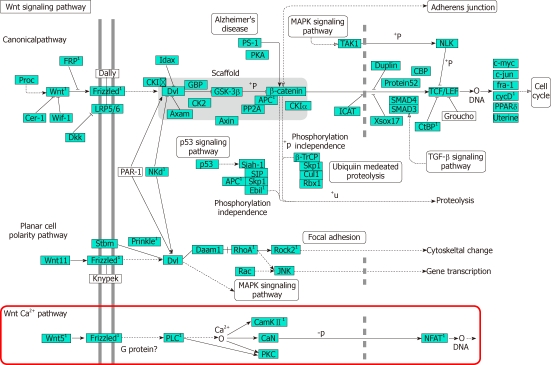
Analysis of KEGG pathways revealed many enrichment-related pathways including focal adhesion, ECM-receptor interaction, regulation of actin cytoskeleton, pathways in cancer, dilated cardiomyopathy, hypertrophic cardiomyopathy, TGF-β signaling pathway, chemokine signaling pathway, p53 signaling pathway, renal cell carcinoma, Wnt signaling pathway, leukocyte transendothelial migration, MAPK signaling pathway in HSCs activation. The top 20 significantly perturbed pathways are listed in Table 3. Of these, Wnt signaling pathway is shown in Figure 2. It revealed that Wnt5a/Frizzled2/PLC/CamKII pathway participated in the activation of HSCs.
Table 3.
Signaling pathway related to activation of hepatic stellate cells
| Terms | Count | Percent | P value | Fold enrichment | Benjamini |
| rno04510: Focal adhesion | 51 | 4.73 | 6.34E-19 | 4.16 | 9.90E-17 |
| rno04512: Extracellular matrix-receptor interaction | 27 | 2.50 | 9.93E-13 | 5.30 | 7.75E-11 |
| rno04810: Regulation of actin cytoskeleton | 36 | 3.34 | 5.18E-08 | 2.74 | 2.69E-06 |
| rno05200: Pathways in cancer | 40 | 3.71 | 2.74E-05 | 2.00 | 0.001066 |
| rno05414: Dilated cardiomyopathy | 18 | 1.67 | 3.27E-05 | 3.18 | 0.00102 |
| rno05410: Hypertrophic cardiomyopathy | 17 | 1.57 | 5.01E-05 | 3.22 | 0.001302 |
| rno04350: Transforming growth factor-beta signaling pathway | 17 | 1.57 | 6.76E-05 | 3.148 | 0.001505 |
| rno04540: Gap junction | 15 | 1.39 | 4.27E-04 | 2.94 | 0.008302 |
| rno04110: Cell cycle | 19 | 1.76 | 7.69E-04 | 2.40 | 0.013254 |
| rno04666: Fc gamma R-mediated phagocytosis | 15 | 1.39 | 0.001011 | 2.71 | 0.015653 |
| rno05222: Small cell lung cancer | 14 | 1.29 | 0.001758 | 2.68 | 0.024645 |
| rno04062: Chemokine signaling pathway | 22 | 2.04 | 0.002121 | 2.04 | 0.027229 |
| rno04115: p53 signaling pathway | 12 | 1.11 | 0.002359 | 2.89 | 0.027945 |
| rno05211: Renal cell carcinoma | 12 | 1.11 | 0.003385 | 2.76 | 0.03708 |
| rno04310: Wnt signaling pathway | 19 | 1.76 | 0.003846 | 2.08 | 0.039281 |
| rno04670: Leukocyte transendothelial migration | 16 | 1.48 | 0.005063 | 2.21 | 0.048281 |
| rno04010: MAPK signaling pathway | 28 | 2.59 | 0.008006 | 1.67 | 0.071107 |
| rno05219: Bladder cancer | 7 | 0.64 | 0.022754 | 3.09 | 0.172201 |
| rno04270: Vascular smooth muscle contraction | 14 | 1.29 | 0.0235 | 1.97 | 0.169299 |
| rno05212: Pancreatic cancer | 10 | 0.92 | 0.027028 | 2.30 | 0.184168 |
Figure 2.
Wnt signaling pathway participated in the activation of hepatic stellate cells. 1Differentially expressed genes revealed by microarray. TGF-β1: Transforming growth factor-β1; PKC: Protein kinase; CamKII: Calcium calmodulin mediated kinase II; LRP: Low density lipoprotein receptor-related protein; GBP: Paternally expressed 12; APC: Adenomatous polyposis coli; CBP: CREB binding protein; PKC: Protein kinase C; NFAT: Nuclear factor of activated T cells; PLC: Phospholipase c.
Effect of Wnt5a knockdown on LX-2 cells
LX-2 cells were infected with lentiviral vector encoding GFP at a multiplicity of infection (MOI) of 10, resulting in GFP expression in the majority of cultured cells. Western blot analysis showed that the protein expression levels of Wnt5a were lower in the Wnt5a ShRNA group than in the control and negative siRNA group (P < 0.01) (Figure 3). MTT assay indicated that the proliferation of LX-2 cells was inhibited significantly after transfection with Wnt5a ShRNA as compared with the control and negative siRNA group (P < 0.05) (Figure 4). RT-PCR showed that Wnt5a gene silencing remarkably decreased the levels of COL1A1 and TGF-β1 mRNA (P < 0.05, respectively) as shown in Figure 5.
Figure 3.
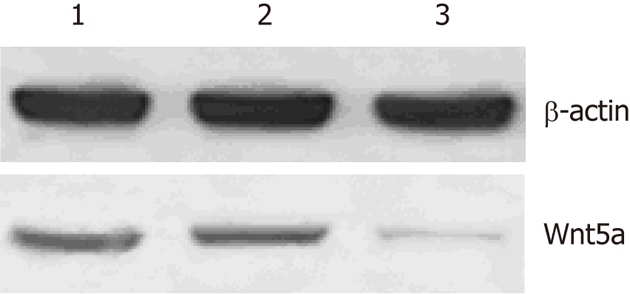
Expression levels of Wnt5a proteins in three groups detected by Western blotting. 1: Control; 2: Negative siRNA; 3: Wnt5a ShRNA.
Figure 4.
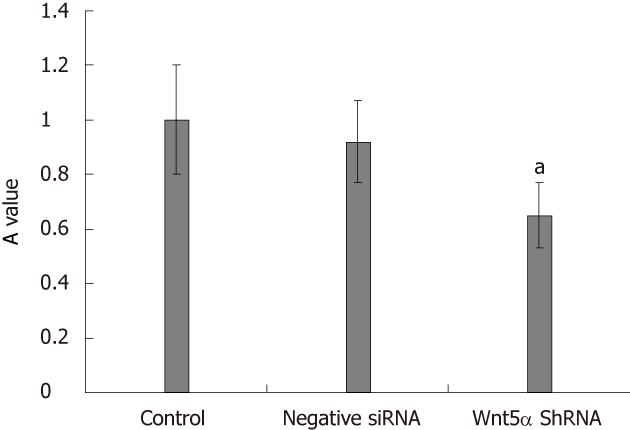
Knock down of Wnt5a decreases the cell proliferation. The role of Wnt5a in regulating LX-2 proliferation was determined by 3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay. Compared with the negative siRNA and control group, Wnt5a ShRNA could inhibit the proliferation of LX-2, aP < 0.05.
Figure 5.
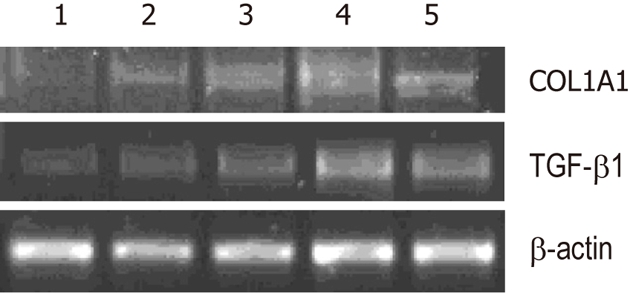
Knockdown of Wnt5a decreases expressions of α-1 type I collagen and transforming growth factor-β1. Experiments were repeated three times and representative results are shown. Compared with the negative siRNA, vector and control groups, the expressions of α-1 type I collagen (COL1A1) and transforming growth factor-β1 (TGF-β1) mRNA were downregulated remarkably in the Wnt5a shRNA group (P < 0.05). 1: Wnt5a ShRNA multiplicity of infection (MOI) 20; 2: Wnt5a ShRNA MOI 10; 3: Negative siRNA; 4:Vector; 5: Control.
Upregulation of Wnt5a in a CCl4-induced fibrosis model
Next, we investigated whether Wnt5a was induced during liver fibrosis in vivo. Compared with the normal group, 8 wk after CCl4 treatment, collagen deposition was increased, pseudolobules formed, and numerous inflammatory cells infiltrated the portal area. Western blotting revealed that Wnt5a was upregulated as significantly as αSMA after 8 wk of CCl4 treatment (P < 0.01) (Figure 6).
Figure 6.
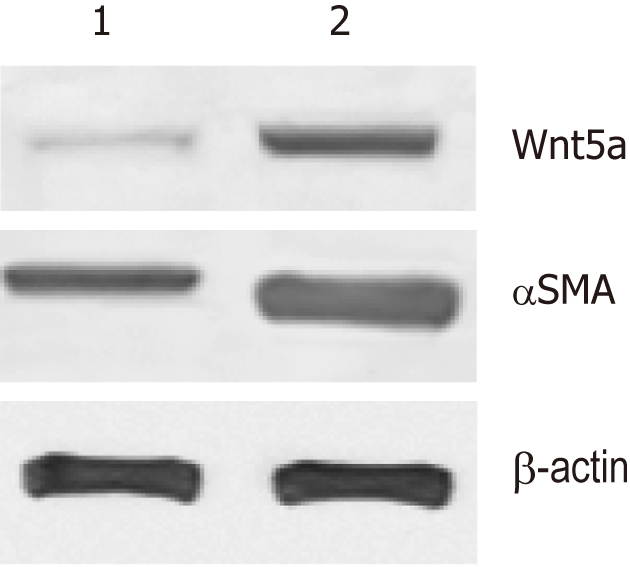
Expression of Wnt5a in fibrotic livers determined with Western blotting. The bands were semi-quantitatively evaluated by densitometric analysis. Wnt5a protein was upregulated as significantly as αSMA in the CCl4-induced liver fibrosis model when compared with the normal rats (P < 0.01). 1: Normal group; 2: Model group.
DISCUSSION
To discover novel genes involved in activation of HSCs, we compared gene expression profile between quiescent and culture activated HSCs. We showed that 2566 genes displayed at least a 2-fold increase or decrease in expression. Of these, 1396 genes were upregulated, while 1170 genes were downregulated in culture-activated HSCs. GO classification showed the most enriched GO terms included response to wounding, wound healing and regulation of cell growth. Analysis of KEGG pathways revealed that Wnt signaling pathway participates in HSCs activation. We were particularly interested in Wnt proteins in view of their potential role in HSCs activation.
The Wnt signaling pathway is a network of proteins best known for their roles in embryogenesis and cancer, but also involved in normal physiological processes in adult animals[12]. There are at least three different Wnt pathways: the canonical pathway, the planar cell polarity (PCP) pathway and the Wnt/Ca2+ pathway. In the canonical Wnt pathway, Wnt ligands bind to the receptor of frizzled family, which transduce signal to β-catenin causing β-catenin to enter the nucleus and activate the target genes. Wnt-Ca2+ signaling is mediated through G proteins and phospholipases and leads to transient increases in cytoplasmic free calcium that subsequently activate the kinase protein kinase C and CamKII and the phosphatase calcineurin[13-15].
With KEGG pathway analyses, we found that the canonical pathway, the PCP pathway and the Wnt/Ca2+ pathway participated in the activation of HSCs. In recent years, researches have shown that activation of Wnt signaling pathway was closely related with the development and progress of fibroproliferative diseases, such as pulmonary fibrosis[16-19], renal fibrosis[20-22], post-infarct cardiac remodeling[23] and hypertrophic scars[24,25], which makes it easy to think that Wnt pathway may have a definite effect on liver fibrosis. It is reported that Wnt/β-catenin signaling contributes to “antiadipogenic” activation of HSCs and may be used as a new target for liver fibrogenesis[26].
The study about the role of Wnt5a in HSCs activation is still in its initial stage. Jiang et al[27] reported that expressions of Wnt5a and its receptor Frizzled 2 were highly upregulated in culture-activated HSCs in vitro. In addition, the expressions of Wnt5a and Frizzled2 in HSCs isolated from the mouse liver fibrosis model induced by bile duct ligation were higher as compared with HSCs isolated from normal mice. It suggests that Wnt5a signaling pathway may be involved in differentiation of quiescent HSCs into myofibroblasts. Shackel et al[28] reported the highly expression of Wnt5a in primary biliary cirrhosis. We found that Wnt5a was upregulated in activated HSCs in comparison with quiescent HSCs as well as its downstream factors such as Frizzled2 and CamKII. Changes in gene expression profiles observed in the microarrays were validated by qRT-PCR analysis. The direct evidence of Wnt5a on the activation of HSCs has not been reported. Because of this, we treated human HSCs line LX-2 with Wnt5a ShRNA, and found that knockdown of Wnt5a gene expression in LX-2 led to significantly impaired proliferation, downregulated expressions of type I collagen and TGF-β1. Wnt5a was upregulated in fibrotic liver of a CCl4 -induced fibrosis rat model.
In summary, our data suggest that Wnt5a may play a role in HSCs activation and liver fibrogenesis, thus revealing the mechanism of HSCs activation from a new perspective, which may provide a novel means and therapeutic target for the prevention and treatment of liver fibrosis.
COMMENTS
Background
Hepatic stellate cells (HSCs) activation is one of the key steps in the development of liver fibrosis. However, the molecular mechanisms underlying the activation of HSCs are not fully understood. It is reported that the expression levels of Wnt5a and its downstream signaling molecules were significantly increased in activated HSCs and experimental liver fibrosis model, indicating that Wnt5a signaling pathways may be involved in liver fibrosis. But the direct evidence of Wnt5a on the activation of HSCs has not been reported. In this study, the authors aimed to identify differentially expressed genes in quiescent and activated HSCs and explore the role of Wnt5a in the activation of HSCs.
Research frontiers
Chip technology is rapid and concise in detecting mRNA expression. Previous studies have reported the differentially expressed gene profiles between quiescent and activated HSCs with gene chip. But, intense bioinformatics analyses and functional assays are deficient. No study has provided the direct evidence of Wnt5a in the mechanism of HSCs activation.
Innovations and breakthroughs
Bioinformatic analysis of the differentially expressed gene profiles between quiescent and activated HSCs reveals the role of Wnt5a signaling pathway in HSCs activation. This is the first study addressing the role of Wnt5a in the mechanism of HSCs activation.
Applications
This study suggests that Wnt5a participates in the HSCs activation and liver fibrogenesis, which may provide a novel means and therapeutic target for the prevention and treatment of liver fibrosis.
Terminology
Wnt signaling molecules are involved in diverse developmental and pathological processes. Based on the different intracellular events activated after Wnt-Frizzled binding, Wnt signaling is divided into two subclasses: the canonical pathway and the non-canonical pathway. Wnt5a is a non-canonical member of the Wnt family. In the Wnt5a signaling pathway, Wnt5a and Frizzled binding triggers the intra-cytoplasmic release of calcium. Increased intracellular calcium concentration activates calcineurin and calcium calmodulin mediated kinase II. Wnt5a signaling takes part in wound healing, regulation of cell proliferation, vasculature development and the formation of cell polarity.
Peer review
The research is the in vitro experiments showing that Wnt5a ShRNA decreases proliferation, downregulates expression of collagen type I and transforming growth factor beta, that probes into a direct Wnt5a participation in the mechanism of HSC activation.
Footnotes
Supported by Research Grant for Health Science and Technology of Pudong Health Bureau of Shanghai, No. PKJ2009-Y16
Peer reviewer: Maria Concepción Gutiérrez-Ruiz, PhD, Department of Health Sciences, Universidad Autónoma Metropolitana-Iztapalapa, DCBS, Av San Rafael Atlixco 186, Colonia Vicentina, México, DF 09340, Mexico
S- Editor Cheng JX L- Editor Ma JY E- Editor Xiong L
References
- 1.Atzori L, Poli G, Perra A. Hepatic stellate cell: a star cell in the liver. Int J Biochem Cell Biol. 2009;41:1639–1642. doi: 10.1016/j.biocel.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Li JT, Liao ZX, Ping J, Xu D, Wang H. Molecular mechanism of hepatic stellate cell activation and antifibrotic therapeutic strategies. J Gastroenterol. 2008;43:419–428. doi: 10.1007/s00535-008-2180-y. [DOI] [PubMed] [Google Scholar]
- 3.De Minicis S, Seki E, Uchinami H, Kluwe J, Zhang Y, Brenner DA, Schwabe RF. Gene expression profiles during hepatic stellate cell activation in culture and in vivo. Gastroenterology. 2007;132:1937–1946. doi: 10.1053/j.gastro.2007.02.033. [DOI] [PubMed] [Google Scholar]
- 4.Woo SW, Hwang KI, Chung MW, Jin SK, Bang S, Lee SH, Lee SH, Chung HJ, Sohn DH. Gene expression profiles during the activation of rat hepatic stellate cells evaluated by cDNA microarray. Arch Pharm Res. 2007;30:1410–1418. doi: 10.1007/BF02977365. [DOI] [PubMed] [Google Scholar]
- 5.Sancho-Bru P, Bataller R, Gasull X, Colmenero J, Khurdayan V, Gual A, Nicolás JM, Arroyo V, Ginès P. Genomic and functional characterization of stellate cells isolated from human cirrhotic livers. J Hepatol. 2005;43:272–282. doi: 10.1016/j.jhep.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 6.Fathke C, Wilson L, Shah K, Kim B, Hocking A, Moon R, Isik F. Wnt signaling induces epithelial differentiation during cutaneous wound healing. BMC Cell Biol. 2006;7:4. doi: 10.1186/1471-2121-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao B, Song H, Bishop K, Elliot G, Garrett L, English MA, Andre P, Robinson J, Sood R, Minami Y, et al. Wnt signaling gradients establish planar cell polarity by inducing Vangl2 phosphorylation through Ror2. Dev Cell. 2011;20:163–176. doi: 10.1016/j.devcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Masckauchán TN, Agalliu D, Vorontchikhina M, Ahn A, Parmalee NL, Li CM, Khoo A, Tycko B, Brown AM, Kitajewski J. Wnt5a signaling induces proliferation and survival of endothelial cells in vitro and expression of MMP-1 and Tie-2. Mol Biol Cell. 2006;17:5163–5172. doi: 10.1091/mbc.E06-04-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grumolato L, Liu G, Mong P, Mudbhary R, Biswas R, Arroyave R, Vijayakumar S, Economides AN, Aaronson SA. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 2010;24:2517–2530. doi: 10.1101/gad.1957710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiskirchen R, Gressner AM. Isolation and culture of hepatic stellate cells. Methods Mol Med. 2005;117:99–113. doi: 10.1385/1-59259-940-0:099. [DOI] [PubMed] [Google Scholar]
- 11.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 12.Lie DC, Colamarino SA, Song HJ, Désiré L, Mira H, Consiglio A, Lein ES, Jessberger S, Lansford H, Dearie AR, et al. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 13.Bejsovec A. Wnt pathway activation: new relations and locations. Cell. 2005;120:11–14. doi: 10.1016/j.cell.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 14.MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 16.Vuga LJ, Ben-Yehudah A, Kovkarova-Naumovski E, Oriss T, Gibson KF, Feghali-Bostwick C, Kaminski N. WNT5A is a regulator of fibroblast proliferation and resistance to apoptosis. Am J Respir Cell Mol Biol. 2009;41:583–589. doi: 10.1165/rcmb.2008-0201OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Königshoff M, Balsara N, Pfaff EM, Kramer M, Chrobak I, Seeger W, Eickelberg O. Functional Wnt signaling is increased in idiopathic pulmonary fibrosis. PLoS One. 2008;3:e2142. doi: 10.1371/journal.pone.0002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim TH, Kim SH, Seo JY, Chung H, Kwak HJ, Lee SK, Yoon HJ, Shin DH, Park SS, Sohn JW. Blockade of the Wnt/β-catenin pathway attenuates bleomycin-induced pulmonary fibrosis. Tohoku J Exp Med. 2011;223:45–54. doi: 10.1620/tjem.223.45. [DOI] [PubMed] [Google Scholar]
- 19.Henderson WR, Chi EY, Ye X, Nguyen C, Tien YT, Zhou B, Borok Z, Knight DA, Kahn M. Inhibition of Wnt/beta-catenin/CREB binding protein (CBP) signaling reverses pulmonary fibrosis. Proc Natl Acad Sci USA. 2010;107:14309–14314. doi: 10.1073/pnas.1001520107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Surendran K, McCaul SP, Simon TC. A role for Wnt-4 in renal fibrosis. Am J Physiol Renal Physiol. 2002;282:F431–F441. doi: 10.1152/ajprenal.0009.2001. [DOI] [PubMed] [Google Scholar]
- 21.He W, Kang YS, Dai C, Liu Y. Blockade of Wnt/β-catenin signaling by paricalcitol ameliorates proteinuria and kidney injury. J Am Soc Nephrol. 2011;22:90–103. doi: 10.1681/ASN.2009121236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hwang I, Seo EY, Ha H. Wnt/beta-catenin signaling: a novel target for therapeutic intervention of fibrotic kidney disease. Arch Pharm Res. 2009;32:1653–1662. doi: 10.1007/s12272-009-2200-3. [DOI] [PubMed] [Google Scholar]
- 23.Colston JT, de la Rosa SD, Koehler M, Gonzales K, Mestril R, Freeman GL, Bailey SR, Chandrasekar B. Wnt-induced secreted protein-1 is a prohypertrophic and profibrotic growth factor. Am J Physiol Heart Circ Physiol. 2007;293:H1839–H1846. doi: 10.1152/ajpheart.00428.2007. [DOI] [PubMed] [Google Scholar]
- 24.Bayle J, Fitch J, Jacobsen K, Kumar R, Lafyatis R, Lemaire R. Increased expression of Wnt2 and SFRP4 in Tsk mouse skin: role of Wnt signaling in altered dermal fibrillin deposition and systemic sclerosis. J Invest Dermatol. 2008;128:871–881. doi: 10.1038/sj.jid.5701101. [DOI] [PubMed] [Google Scholar]
- 25.Sato M. Upregulation of the Wnt/beta-catenin pathway induced by transforming growth factor-beta in hypertrophic scars and keloids. Acta Derm Venereol. 2006;86:300–307. doi: 10.2340/00015555-0101. [DOI] [PubMed] [Google Scholar]
- 26.Cheng JH, She H, Han YP, Wang J, Xiong S, Asahina K, Tsukamoto H. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am J Physiol Gastrointest Liver Physiol. 2008;294:G39–G49. doi: 10.1152/ajpgi.00263.2007. [DOI] [PubMed] [Google Scholar]
- 27.Jiang F, Parsons CJ, Stefanovic B. Gene expression profile of quiescent and activated rat hepatic stellate cells implicates Wnt signaling pathway in activation. J Hepatol. 2006;45:401–409. doi: 10.1016/j.jhep.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Shackel NA, McGuinness PH, Abbott CA, Gorrell MD, McCaughan GW. Identification of novel molecules and pathogenic pathways in primary biliary cirrhosis: cDNA array analysis of intrahepatic differential gene expression. Gut. 2001;49:565–576. doi: 10.1136/gut.49.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]