Abstract
AIM: To investigate the function of the KISS-1 gene in gastric carcinoma cells and to explore its potential mechanism.
METHODS: A KISS-1 eukaryotic expression vector was constructed and transfected into BGC-823 cells. Resistant clones were obtained through G418 selection. reverse transcription-polymerase chain reaction and western blotting were used to detect KISS-1 and matrix metalloproteinase-9 (MMP-9) expression in transfected cells. The growth of transfected cells was investigated by 3-(4, 5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) proliferation assays, and the cells’ invasive potential was analyzed by basement membrane (Matrigel) invasion assays. The anti-tumor effects of KISS-1 were tested in vivo using allografts in nude mice.
RESULTS: The expression level of KISS-1 mRNA and protein in BGC-823/KISS-1 transfected cells were significantly higher than in BGC-823/pcDNA3.1 transfected cells (P < 0.05) or the parental BGC-823 cell line (P < 0.05). The expression level of MMP-9 mRNA and protein in BGC-823/KISS-1 were significantly less than in BGC-823/pcDNA3.1 (P < 0.05) or BGC-823 cells (P < 0.05). MTT growth assays show the proliferation of BGC-823/KISS-1 cells at 48 h (0.642 ± 0.130) and 72 h (0.530 ± 0.164) were significantly reduced compared to BGC-823/pcDNA3.1 (0.750 ± 0.163, 0.645 ± 0.140) (P < 0.05) and BGC-823 cells (0.782 ± 0.137, 0.685 ± 0.111) (P < 0.05). Invasion assays indicate the invasive potential of BGC-823/KISS-1 cells (16.50 ± 14.88) is significantly reduced compared to BGC-823/pcDNA3.1 (20.22 ± 14.87) (P < 0.05) and BGC-823 cells after 24 h (22.12 ± 16.12) (P < 0.05). In vivo studies demonstrate the rate of pcDNA3.1-KISS-1 tumor growth is significantly slower than pcDNA3.1 and control cell tumor growth in nude mice. Furthermore, tumor volume of pcDNA3.1-KISS-1 tumors (939.38 ± 82.08 mm3) was significantly less than pcDNA3.1 (1250.46 ± 44.36 mm3) and control tumors (1284.36 ± 55.26 mm3) (P < 0.05). Moreover, the tumor mass of pcDNA3.1-KISS-1 tumors (0.494 ± 0.84 g) was significantly less than pcDNA3.1 (0.668 ± 0.55 g) and control tumors (0.682 ± 0.38 g) (P < 0.05).
CONCLUSION: KISS-1 may inhibit the proliferation and invasion of gastric carcinoma cells in vitro and in vivo through the downregulation of MMP-9.
Keywords: KISS-1, Matrix metalloproteinase-9, BGC-823 cells, Proliferation, Metastasis, Nude mice
INTRODUCTION
KISS-1 has been identified as a human melanoma metastasis suppressor gene using subtractive hybridization between the metastatic human melanoma cell line C8161 and non-metastatic variants generated after microcell-mediated transfer of chromosome 6 into C8161[1]. The KISS-1 gene maps to chromosome 1 bands q32-q41 and encodes a largely hydrophobic 145-amino-acid protein[2]. The 54-amino-acid, C-terminally amidated fragment of the KISS-1 protein (amino acids 68-121) is termed metastin, while the full-length protein is called kisspeptin[2,3]. Expression of KISS-1 is detectable in normal heart, brain, liver and lung. In human tumors, KISS-1 expression is weak or undetectable. The KISS-1 gene product functions as tumor metastasis suppressor and is reported to act after binding with hOT7T175, an orphan G-protein-coupled receptor. Previous studies demonstrated KISS-1 could suppress metastasis of human malignant melanoma and human breast carcinoma cells without affecting tumorigenicity[4,5]. Although the loss of KISS-1 expression has been associated with tumor progression and poor prognosis in various cancers, the mechanism underlying this activity is still unknown. Investigating the role of KISS-1 in other cancers and identifying its potential mechanism in suppressing tumor metastasis will require additional experiments.
MATERIALS AND METHODS
Construction of recombinant expression vectors
Two primers, K1(5’-CGAAGCTTATGAACTCACTGGTTTCT-3’, carrying a HinDI site, underlined) and K2(5’-CTGGATCCTCACTGCCCCGCACCTG-3’, carrying an BamHI site, underlined), were designed for the KISS-1 gene’s CDS (sequence coding for amino acids in protein) domain. This sequence was amplified by quantitative reverse transcription-polymerase chain reaction (RT-PCR) with primer K1 and K2 from 50 g of human gastric tissue derived from 5 cm of the edge of gastric carcinoma lesions. Quantitative RT-PCR conditions were as follows: denaturing at 94 °C for 1 min, annealing at 48 °C for 30 s, and extension at 72 °C for 1 min. The PCR was run for 30 cycles. Final extension was performed at 72 °C for 10 min. The fragment carrying both the HinDI and BamHI site was acquired by quantitative RT-PCR. This PCR produced a product of 454 bp. The product was subjected to a double digestion with HinDI and BamHI enzymes, and the digested DNA product was ligated into a 5.4-kb fragment of pcDNA3.1 (Invitrogen, United States) that was digested with the same enzymes. The ligated product was transformed into JM109 cells. Restriction endonuclease analysis, quantitative RT-PCR and plasmid sequencing were performed to validate the recombinant plasmid reading frame.
Cell culture and transfection
The BGC-823 cells (a gift from Zheng Zhou University) were cultured in RPMI 1640 medium supplemented with 10% bovine calf serum, 100 U/mL penicillin and 100 U/mL streptomycin. Cells were cultured at 37 °C in a humidified incubator with 5% CO2. NIH3T3 cells cultured in the same conditions were used as controls. Twelve hours before transfection, cells were seeded onto 24-well plates at a density of 1-2 × 105 cells per well. Cells were transfected when plate confluence was approximately 90%-95%. The cells were transfected with 1.25 μL/well of PcDNA3.1-KISS-1 vector using 1 μL Lipofectamine (Invitrogen). The culture medium was replaced with a selection medium containing G418 (at concentrations ranging from 100 μg/mL to 1000 μg/mL, Alexis Biochemicals) forty-eight hours later. When stably transfected cells were obtained, the cells were continuously maintained in 200 μg/mL of G418. BGC-823 cells were transfected with the empty pcDNA3.1 vector as a control.
Reverse transcription-polymerase chain reaction
Total RNA was isolated from cells using Trizol purification (Gibco). After denaturing RNA at 94 °C for 5 min, 500 ng of RNA was transcribed into cDNA. Next, cDNA was amplified using the following primers: KISS-1 sense (5’-ATGAACTCACTGGTTTCTTGGCAG-3’), KISS-1 antisense (5’-TCACTGCCCCGCACCTG-3’); MMP-9 sense (5’-AGGAGCGGCTCTCCAAGAAG-3’) and MMP-9 antisense (5’-GGGCACTGCAGGATGTCATAG-3’). Duplex amplification was performed using a thermocycler for 30 cycles according to the following program: 1 min at 94 °C, 30 s at 48 °C (KISS-1), 60 °C (MMP-9) and 1 min at 72 °C. PCR fragments were separated by electrophoresis on a 1.5% agarose gel, with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (315 bp) as an internal standard. PCR products were quantitated using SYNGENE gel analysis software, and GAPDH was used to normalize the data.
Western blotting
For Western blotting protein analysis, 40 μg of total protein was separated by 10%-15% gel gradient SDS-PAGE under reducing conditions. Proteins were subsequently transferred to a polyvinylidene fluoride (PVDF) membrane. The PVDF membrane was incubated with blocking buffer (PBS containing 5% non fat milk) for 2 h at room temperature. Primary antibodies were either rabbit anti-KISS-1 (Boshide, China) or goat anti-MMP-9 (Santa Cruz, United States). Membranes were incubated overnight at 4 °C with gentle shaking. The membrane was washed twice with PBS for 5 min and incubated with secondary antibodies (Zhongshan, China) for 2 h at room temperature. After washing, KISS-1 and MMP-9 were detected using a chemiluminescence reaction. The results were analyzed with TotalLab2.0 software. Protein levels were normalized to β-actin protein.
Cell proliferation assay
Cells were seeded at 5 × 103 cells per well in a 96-well plate and cultured in the presence of 10% fetal bovine serum for 24, 48 or 72 h. The cells were pulsed with 3-(4, 5-dimethylthiazolyl-2)-2, 5-diphenyltetrazolium bromide (MTT, Sigma, United States) 20 μL/well [5 mg/mL in phosphate-buffered saline (PBS)] to measure cell growth. The purple-blue MTT formsazan precipitate was dissolved in 200 μL of DMSO and swirled for 30 min. Absorbance (UA) was measured at 570 nm with a spectrophotometer. Experiments were repeated five times.
Proliferation inhibition rates (%) = (1-UAE/UAC) ×100% (UAE: Average UA value of experimental group; UAC: Average UA value of control group)
Invasion assays
For invasion assays, transwell polycarbonate filters (8-μm pore size, Millipore, United States) were coated with 100 μL of matrigel (Sigma, United States) at a dilution of 1:20 in serum-free medium and were then air-dried for 24 h. 5 × 105 cells in 400 μL of complete medium were seeded into the upper chamber. Next, 200 μL of medium was added to the lower chamber, and the plate was incubated at 37 °C in a 5% CO2 incubator for 24 h. Cells on the lower surface of the filter were stained with hexamethylpararosaniline and counted. The experiment was performed in triplicate for three different cell lines.
The invasion index is defined as the number of cells that migrated through the 8 μm pores of the filter in the experimental group divided by the number of cells that migrated through the filter in the control group × 100%.
The cell invasion inhibition rate is equal to the number of cells that migrated through the filter in the control group minus the number of cells that migrated through the filter in the experimental group divided by the number of cells that migrated through the filter in the control group × 100%.
Tumor development in nude mice
Fourteen 4-6 wk old athymic female nude BALB/c mice (weighing 18-22 g) were purchased from the Experimental Animal Center of Shanghai, Academia Sinica. These mice were bred in specific pathogen-free conditions and were randomly divided into three groups of 8 mice per group. Three different cell lines (pcDNA3.1-KISS-1, pcDNA3.1, BGC-823) were each suspended at a concentration of 5 × 107 cells per 0.1 mL serum-free RMPI 1640. Cells were injected subcutaneously into the right forelimb. Tumor diameters were measured every 3 d. Tumor volume was calculated according to the following formula: volume (V) = 0.5236AB2 (A and B represent the long and short tumor diameter respectively). The in vivo growth curve of cancer cells was drawn. The animals were sacrificed 45 d after inoculation, and tumors were weighed. Inhibition rate (%) = [tumor volume (mass) of control group-tumor volume (mass) of experiment group/ tumor volume (mass) of control group] × 100%.
Statistical analysis
Data were analyzed using the SPSS13.0 software. Analysis of variance was conducted followed by independent-sample t-tests. A P value less than 0.05 was considered to be statistically significant.
RESULTS
Construction and identification of recombinant plasmids
Full-length human KISS-1 CDS domain cDNA was inserted into a pcDNA3.1 vector at the HinDI/BamHIsite to form the recombinant plasmid pcDNA3.1-KISS-1. Recombinant pcDNA3.1-KISS-1 vectors containing the KISS-1 gene were selected through α-complementation. The expression vector was further analyzed by digestion with the restriction endonucleases HinDI/BamHI. The resulting fragment sizes were consistent with the expected fragments predicted by the expression vector map.
Transfection of the KISS-1 gene increased expression of KISS-1 and downregulated matrix metalloproteinase-9 expression in BGC-823 cells
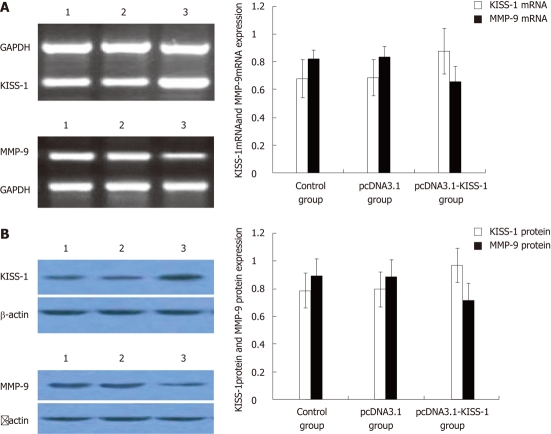
Strong expression of KISS-1 was observed in pcDNA3.1-KISS-1 transfected BGC-823 cells by RT-PCR and Western blotting. BGC-823 cells transfected with pcDNA3.1-KISS-1 also showed a significant reduction in MMP-9 expression (P < 0.05). The expression of KISS-1 in pcDNA3.1-transfected BGC-823 cells and control BGC-823 cells remained low (P < 0.05) (Figure 1).
Figure 1.
The expressions of KISS-1 and matrix metalloproteinase-9 mRNA and protein in BGC-823 cells. A: The expression level of KISS-1 mRNA was distinctly higher and the expression level of matrix metalloproteinase-9 (MMP-9) mRNA was distinctly lower in pcDNA3.1-KISS-1 group than that in pcDNA3.1 group (P < 0.05) and control group (P < 0.05); B: The expression level of KISS-1 protein was distinctly higher and the expression level of MMP-9 protein was distinctly lower in pcDNA3.1-KISS-1 group than that in pcDNA3.1 group (P < 0.05) and control group (P < 0.05). Lane 1: Control group; Lane 2: pcDNA3.1 group; Lane 3: pcDNA3.1-KISS-1group; MMP-9: Matrix metalloproteinase-9; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
Transfection of KISS-1 gene suppressed cell growth
At 48 and 72 h after transfection, the absorbance values (570 nm) of pcDNA3.1-KISS-1-transfected BGC-823 cells were significantly different from those of the pcDNA3.1-transfected (P < 0.05) and control BGC-823 cells (P < 0.05). KISS-1 transfection decreased cell proliferation by 17.90% compared to pcDNA3.1-transfected cells and by 22.63% compared to control BGC-823 cells. These results indicate KISS-1 protein expression in BGC-823 cells strongly inhibits cell proliferation (Table 1).
Table 1.
The influence on the proliferation ability of BGC-823 cells transfected with pcDNA3.1-KISS-1
| Group |
24 h |
48 h |
72 h |
|||
| UA value | Inhibition rate (%) | UA value | Inhibition rate (%) | UA value | Inhibition rate (%) | |
| Control group | 0.744 ± 0.126 | 0 | 0.782 ± 0.137 | 0 | 0.685 ± 0.111 | 0 |
| pcDNA3.1 group | 0.713 ± 0.222 | 4.17 | 0.750 ± 0.163 | 4.09 | 0.645 ± 0.140 | 5.84 |
| pcDNA3.1-KISS-1 group | 0.691 ± 0.131 | 7.12 | 0.642 ± 0.130a | 17.90 | 0.530 ± 0.164a | 22.63 |
P < 0.05 vs control group and pcDNA3.1 group.
Effects of KISS-1 transfection on cell invasion
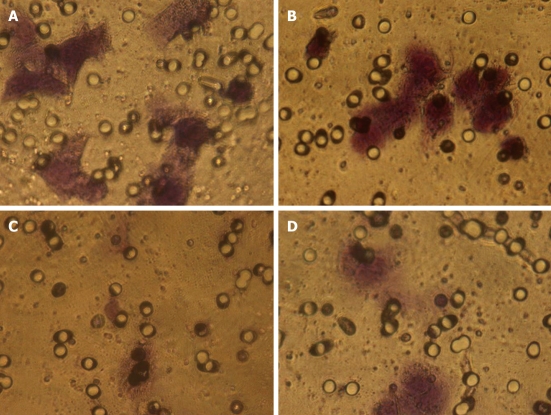
In order to invade surrounding tissue, epithelial cancer cells must degrade the underlying basement membrane. The numbers of cells that digested Matrigel and migrated through the 8-μm pores in the filter were counted after 24 h (Figure 2). The total number of pcDNA3.1-KISS-1 transfected cells that migrated through the transwell polycarbonate filter was significantly less than the number of migrating cells in the pcDNA3.1 transfected group and control group (P < 0.05). These data suggest that transfection with the KISS-1 expression vector results in a significant decrease in invasive capacity (Table 2).
Figure 2.
Suppression effects on invasion potency of BGC-823 by KISS-1 transfection. A: Control group; B: Group transfected with pcDNA3.1; C, D: Group transfected with pcDNA3.1-KISS-1.
Table 2.
The influence on the invasion potency of BGC-823 cells transfected by KISS-1
| Group |
24 h |
||
| Cells numbers | Invasion index (%) | Invasion ability inhibition rates (%) | |
| Control group | 22.12 ± 16.12 | 100.00 | 0.0 |
| pcDNA3.1 group | 20.22 ± 14.87 | 91.33 ± 12.14 | 8.75 ± 11.18 |
| pcDNA3.1-KISS-1 group | 16.50 ± 14.88ac | 74.66 ± 17.14ac | 25. 38 ± 16.25a |
P < 0.05 vs transfected with pcDNA3.1 group;
P < 0.05 vs control group.
Tumor growth
Tumor volume was assessed 7 d after injection of tumor cells and every 3 d thereafter until 45 d. Time curves show the percentage increase in tumor size (Figure 3). The tumor volume and mass in the pcDNA3.1-KISS-1 transfected group were both significantly less than the tumor volume and mass in the pcDNA3.1-transfected group and the control group (P < 0.05). The tumor volume and mass inhibition rates for the pcDNA3.1-KISS-1 transfected group were 26.86% and 27.57%, respectively. No lung or liver metastatic nodules were observed. As a result of KISS-1 transfection, tumor growth was reduced (Table 3).
Figure 3.
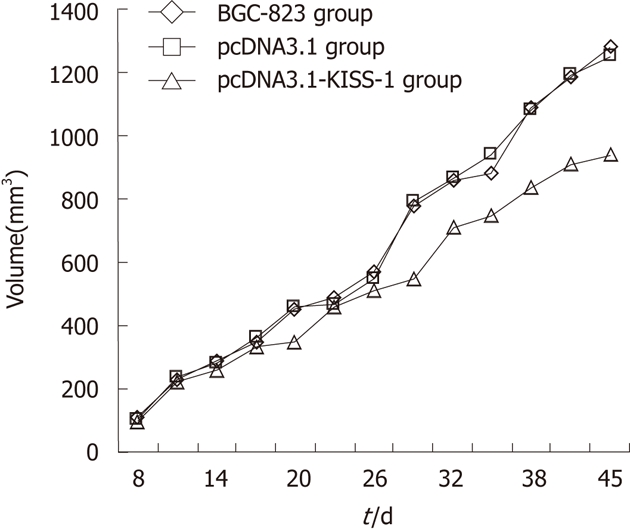
Inhibition effects on growth of BGC-823 by KISS-1 transfection in vivo. The tumor volume in pcDNA3.1-KISS-1 transfected group was significantly lower compared with pcDNA3.1 transfected group and control group.
Table 3.
The comparison of average tumor volume and tumor weight and inhibition rate in groups
| Group (n = 8) | Tumor volume (mm3) | Inhibitory rate (%) | Tumor weight (g) | Inhibitory rate (%) |
| BGC-823 group | 1284.36 ± 55.26 | - | 0.682 ± 0.38 | - |
| pcDNA3.1 group | 1250.46 ± 44.36 | 2.64 | 0.668 ± 0.55 | 2.05 |
| pcDNA3.1-KISS-1 group | 939.38 ± 82.08a | 26.86 | 0.494 ± 0.84a | 27.57 |
P < 0.05 vs BGC-823 group and pcDNA3.1 group.
DISCUSSION
The vast majority of gastric cancer deaths result from complications caused by tumor cell metastasis rather than as a consequence of the original tumor growth. Metastasis is a multistep process involving complex interactions between tumor cells and host cells. To become metastatic, primary tumor cells must migrate from the tumor, dissociate from the tumor mass and travel to nearby or distant secondary sites. Single cells, homotypic clusters of cells, or heterotypic emboli subsequently arrest at a distant site with the use of both organ specific and nonspecific mechanisms. Next, these cells invade the surrounding tissue and respond to growth signals at the secondary site[6-9]. Interference at any one of these steps can block the metastatic cascade and prevent the formation of metastatic tumor growths. Consequently, there is increasing interest in researching the metastatic process to identify possible ways to inhibit metastatic tumor progression.
Metastasis suppressor genes, which inhibit the spread of cancers to secondary sites, have become the target of mounting clinical and basic cancer research. Recently, the KISS-1 gene was reported to be a novel metastasis suppressor gene in human melanoma and breast carcinoma cells[4,5]. KISS-1 was found to reduce tumor cell invasive and migratory properties without affecting their tumorigenicity[5]. Kisspeptin, the product of the KISS-1 gene, belongs to the RF-amide family of peptides that possess an Arg-Phe-amide sequence motif at the C terminus. This family of peptides is involved in the control of reproduction and tumor metastasis in mammals. In 2001, kisspeptin was first shown to be the endogenous ligand for the orphan G protein-coupled receptor GPR54 (or KISS1R). Kisspeptin is also called metastin in consideration of its suppressive effects on tumor growth and tumor metastasis[2]. Blocked or reduced expression of KISS-1 has been found in a variety of tumor metastasis, including breast cancer, bladder cancer and pancreatic cancer[4,10,11]. These studies suggest that KISS-1 is a human metastasis suppressor gene, and loss of KISS-1 and its receptor may correlate with human tumor progression and metastasis. The mechanism of KISS-1 suppression is still unknown. Recent publications suggest possible mechanisms for KISS-1 metastasis suppression. Metastin (this peptide is encoded from the KISS-1 gene) induces Ca2+ in receptor-transfected CHO cells[2], as well as phosphorylation of ERK1/2 and weak phosphorylation of p38/MAPK but not of SAPK/JNK3. Metastin inhibits motility, chemotaxis, and invasion in vitro[2,12], possibly by repressing the transcription of MMP-9 via the induction of cytosolic IκBα[13]. Metastin also induces excessive formation of focal adhesions and stress fibers in hOT7T175-transfected B16/BL6 and induces the phosphorylation of FAK and paxillin[13], possibly through Rho[14]. Our previous work has shown that loss of KISS-1 gene expression in tumors has a significant correlation with lymph node metastasis from gastric carcinoma. The purpose of this study was to determine whether KISS-1 might function as a metastasis suppressor and to investigate a possible mechanism for this action in gastric carcinoma.
We constructed a eukaryotic vector containing the full-length human KISS-1 cDNA and transfected it into BGC-823 cells. Selection with G418 enabled stably transfected cells expressing high levels of KISS-1 protein to be obtained. In this study, we demonstrate a significant reduction in the growth of BGC-823 cells transfected with an exogenous KISS-1 gene. Additionally, the colony-forming ability of KISS-1-transfected cells was reduced in vitro, and tumor growth in nude mice was inhibited. Using a reconstituted basement membrane assay (Matrigel), we showed a reduction in the number of invading KISS-1 transfected BGC-823 cells. This finding indicates that the propensity for local/regional invasion and distant metastasis of gastric cancer may be dependent on its ability to invade the basement membrane. These results are similar to those reported by Lee et al[4] using a breast carcinoma cell line.
Cancer mortality is largely due to distant metastases and subsequent organ failure. Metastasis involves the degradation of the basement membrane and stromal extracellular matrix (ECM) and subsequent migration into the adjacent blood vessels. This phenomenon results in tumor growth in distant organs[8,15]. Degradation of the basement membrane and ECM involve the secretion of several proteases, such as one or more members of the MMP family[16-18]. MMP-9 can cleave ECM proteins to promote cell invasion and mobility and has been described as a negative prognostic marker of metastasis and disease-free interval[19-21]. We transfected KISS-1 cDNA into BGC-823 cells and subsequently monitored the expression of MMP-9 by Western blotting and RT-PCR. Our results show KISS-1 gene transfection decreases the expression of MMP-9 and suggests that KISS-1 could downregulate expression of MMP-9.
In summary, KISS-1 is able to be exogenously expressed in eukaryotic cells. Its transient expression significantly suppresses the growth and invasion of tumor cells. Importantly, our data suggests that the effect of KISS-1 on tumor growth and invasion might occur by decreasing expression of MMP-9. In the future, additional experiments will be required to determine a role for KISS-1 in other cancers and to explore the exact mechanism of KISS-1 function.
COMMENTS
Background
KISS-1 has been identified as a putative metastasis-suppressor gene in human melanomas and breast-cancer cell lines. However, the mechanism of how KISS-1 works and the effects of a synthesized truncated KISS-1 protein on the proliferation and invasive ability remain unknown. Our previous studies have shown that the loss of KISS-1 expression was correlated with lymph node metastasis in gastric carcinoma tissue.
Research frontiers
This study investigated the function of KISS-1 in gastric carcinoma cells and explored its possible mechanism.
Innovations and breakthroughs
This study explored KISS-1 inhibits the proliferation and invasion of gastric carcinoma cells in vitro and in vivo and concluded the mechanism is mediated through the downregulation of matrix metalloproteinase-9.
Applications
By understanding the relationship between KISS-1 and tumor-cell proliferation and invasion in gastric carcinoma, this study may be useful in the clinical management of patients with gastric carcinoma.
Peer review
This topic is an import topic for research and the manuscript is good.
Footnotes
Supported by The Natural Science Foundation, No. 2011A 310005; and the Science and Technique Foundation of Henan Province, No. 112102310206
Peer reviewers: Balraj Mittal, PhD, Professor, Department of Genetics, Sanjay Gandhi Medical Institute, Lucknow 226014, India; Fariborz Mansour-Ghanaei, MD, Professor of Medicine, Division of Gastroenterology and Hepatology, Director, Gastrointestinal and Liver Diseases Research Center, Guilan University of Medical Sciences, Sardar Jangle Ave., Rasht 41448-95655, Iran
S- Editor Shi ZF L- Editor A E- Editor Xiong L
References
- 1.Lee JH, Miele ME, Hicks DJ, Phillips KK, Trent JM, Weissman BE, Welch DR. KiSS-1, a novel human malignant melanoma metastasis-suppressor gene. J Natl Cancer Inst. 1996;88:1731–1737. doi: 10.1093/jnci/88.23.1731. [DOI] [PubMed] [Google Scholar]
- 2.Lee YR, Tsunekawa K, Moon MJ, Um HN, Hwang JI, Osugi T, Otaki N, Sunakawa Y, Kim K, Vaudry H, et al. Molecular evolution of multiple forms of kisspeptins and GPR54 receptors in vertebrates. Endocrinology. 2009;150:2837–2846. doi: 10.1210/en.2008-1679. [DOI] [PubMed] [Google Scholar]
- 3.Pinilla L, Castellano JM, Romero M, Tena-Sempere M, Gaytán F, Aguilar E. Delayed puberty in spontaneously hypertensive rats involves a primary ovarian failure independent of the hypothalamic KiSS-1/GPR54/GnRH system. Endocrinology. 2009;150:2889–2897. doi: 10.1210/en.2008-1381. [DOI] [PubMed] [Google Scholar]
- 4.Lee JH, Welch DR. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res. 1997;57:2384–2387. [PubMed] [Google Scholar]
- 5.Lee JH, Welch DR. Identification of highly expressed genes in metastasis-suppressed chromosome 6/human malignant melanoma hybrid cells using subtractive hybridization and differential display. Int J Cancer. 1997;71:1035–1044. doi: 10.1002/(sici)1097-0215(19970611)71:6<1035::aid-ijc20>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 6.Rojiani MV, Alidina J, Esposito N, Rojiani AM. Expression of MMP-2 correlates with increased angiogenesis in CNS metastasis of lung carcinoma. Int J Clin Exp Pathol. 2010;3:775–781. [PMC free article] [PubMed] [Google Scholar]
- 7.Shengbing Z, Feng LJ, Bin W, Lingyun G, Aimin H. Expression of KiSS-1 gene and its role in invasion and metastasis of human hepatocellular carcinoma. Anat Rec (Hoboken) 2009;292:1128–1134. doi: 10.1002/ar.20950. [DOI] [PubMed] [Google Scholar]
- 8.Fidler IJ, Ellis LM. The implications of angiogenesis for the biology and therapy of cancer metastasis. Cell. 1994;79:185–188. doi: 10.1016/0092-8674(94)90187-2. [DOI] [PubMed] [Google Scholar]
- 9.Nicolson GL. Cancer progression and growth: relationship of paracrine and autocrine growth mechanisms to organ preference of metastasis. Exp Cell Res. 1993;204:171–180. doi: 10.1006/excr.1993.1022. [DOI] [PubMed] [Google Scholar]
- 10.Masui T, Doi R, Mori T, Toyoda E, Koizumi M, Kami K, Ito D, Peiper SC, Broach JR, Oishi S, et al. Metastin and its variant forms suppress migration of pancreatic cancer cells. Biochem Biophys Res Commun. 2004;315:85–92. doi: 10.1016/j.bbrc.2004.01.021. [DOI] [PubMed] [Google Scholar]
- 11.Sánchez-Carbayo M, Cordon-Cardo C. Applications of array technology: identification of molecular targets in bladder cancer. Br J Cancer. 2003;89:2172–2177. doi: 10.1038/sj.bjc.6601406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hori A, Honda S, Asada M, Ohtaki T, Oda K, Watanabe T, Shintani Y, Yamada T, Suenaga M, Kitada C, et al. Metastin suppresses the motility and growth of CHO cells transfected with its receptor. Biochem Biophys Res Commun. 2001;286:958–963. doi: 10.1006/bbrc.2001.5470. [DOI] [PubMed] [Google Scholar]
- 13.Yan C, Wang H, Boyd DD. KiSS-1 represses 92-kDa type IV collagenase expression by down-regulating NF-kappa B binding to the promoter as a consequence of Ikappa Balpha -induced block of p65/p50 nuclear translocation. J Biol Chem. 2001;276:1164–1172. doi: 10.1074/jbc.M008681200. [DOI] [PubMed] [Google Scholar]
- 14.Mandache E, Gherghiceanu M, Serafinceanu C, Penescu M, Mircescu G. Myofibroblast involvement in tubular basement membrane remodeling in type II diabetic nephropathy. Rom J Morphol Embryol. 2011;52:75–79. [PubMed] [Google Scholar]
- 15.Itoh Y, Nagase H. Matrix metalloproteinases in cancer. Essays Biochem. 2002;38:21–36. doi: 10.1042/bse0380021. [DOI] [PubMed] [Google Scholar]
- 16.Hornebeck W, Maquart FX. Proteolyzed matrix as a template for the regulation of tumor progression. Biomed Pharmacother. 2003;57:223–230. doi: 10.1016/s0753-3322(03)00049-0. [DOI] [PubMed] [Google Scholar]
- 17.Worthley DL, Giraud AS, Wang TC. The extracellular matrix in digestive cancer. Cancer Microenviron. 2010;3:177–185. doi: 10.1007/s12307-010-0053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J. Matrix metalloproteinases and tissue inhibitor of metalloproteinases are essential for the inflammatory response in cancer cells. J Signal Transduct. 2010;2010:985132. doi: 10.1155/2010/985132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leppä S, Saarto T, Vehmanen L, Blomqvist C, Elomaa I. A high serum matrix metalloproteinase-2 level is associated with an adverse prognosis in node-positive breast carcinoma. Clin Cancer Res. 2004;10:1057–1063. doi: 10.1158/1078-0432.ccr-03-0047. [DOI] [PubMed] [Google Scholar]
- 20.Ahmed MM, Mohammed SH. Matrix metalloproteinases 2 and 9 in situ mRNA expression in colorectal tumors from Iraqi patients. Indian J Pathol Microbiol. 2011;54:7–1. doi: 10.4103/0377-4929.77316. [DOI] [PubMed] [Google Scholar]
- 21.Uchima Y, Sawada T, Nishihara T, Umekawa T, Ohira M, Ishikawa T, Nishino H, Hirakawa K. Identification of a trypsinogen activity stimulating factor produced by pancreatic cancer cells: its role in tumor invasion and metastasis. Int J Mol Med. 2003;12:871–878. [PubMed] [Google Scholar]