Summary
Cyclin-dependent-kinases comprise the conserved machinery that drives progress through the cell cycle, but how they do this in mammalian cells is still unclear. To identify the mechanisms by which cyclin-cdks control the cell cycle, we performed a time-resolved analysis of the in vivo interactors of cyclins E1, A2 and B1 by quantitative mass spectrometry. This global analysis of context-dependent protein interactions reveals the temporal dynamics of cyclin function in which networks of cyclin-cdk interactions vary according to the type of cyclin and cell cycle stage. Our results explain the temporal specificity of the cell cycle machinery, thereby providing a biochemical mechanism for the genetic requirement for multiple cyclins in vivo, and reveal how the actions of specific cyclins are coordinated to control the cell cycle. Furthermore, we identify key substrates (Wee1 and c15orf42/Sld3) that reveal how cyclin A is able to promote both DNA replication and mitosis.
Introduction
How the cell orchestrates its precise duplication and division into two new cells is an important, unanswered question. In eukaryotes, cyclins activate the cyclin-dependent kinases (cdks) to control progress through DNA replication and mitosis, but how they achieve this is unclear because insufficient human targets are known to explain all of the cyclin/cdk-dependent events. Furthermore, cyclin-cdks must trigger these events in the correct order: DNA replication and mitosis must occur sequentially. To understand how the cell cycle is controlled requires the identification of the proteins that are regulated by cyclin-cdks, and the timing of these interactions.
In fission yeast, one cyclin-cdk, cdc13p-cdc2p, can trigger both DNA replication and mitosis: low kinase levels trigger DNA replication whereas subsequent higher levels trigger mitosis (Fisher and Nurse, 1996). This led Stern and Nurse to propose, a ‘threshold-model’ whereby cdc13p-cdc2p phosphorylates its DNA replication substrates at lower kinase levels than its mitotic substrates (Stern and Nurse, 1996), and Coudreuse and Nurse recently confirmed this model in fission yeast (Coudreuse and Nurse, 2010). This model could work in a number of ways: substrates could contain high affinity binding sites for the cyclin-cdks, e.g. the Cy motif that binds to the ‘hydrophobic patch’ of some cyclins (Schulman et al, 1998). Alternatively, substrates (e.g. Wee1) could have multiple sites, of which the first to be phosphorylated have no effect, and higher levels of kinase activity are required to phosphorylate the lower affinity sites that do have a biological effect (Kim and Ferrell, 2007). A third alternative is that substrates could bind to antagonistic phosphatases and thereby require much higher levels of kinase to remain phosphorylated.
In contrast, budding yeast utilize multiple G1-phase Cln cyclins and mitotic B-type Clb cyclins with a single essential cdk, Cdc28 (Bloom and Cross, 2007). Yet there is redundancy in this machinery: overexpressing Clb5 rescues viability of cells lacking all Clns, but yeast lacking Clb5 are also viable (Epstein and Cross, 1992). In the absence of Swe1 (the homolog of Wee1), early expression of Clb2 alone can direct phosphorylation of all essential Clb targets (Hu and Aparicio, 2005). Despite this overlap in abilities, in vitro Clb5/Cdk1 phosphorylates 24% of a set of 150 cdk substrates more efficiently than Clb2/Cdk1 (Loog and Morgan, 2005). These data support a second model; that multiple cyclins evolved with different binding affinities and thus differences in phosphorylation targets.
In animal cells, Cdk1 is the only essential cdk, whereas there are two essential cyclins, A and B (Kalaszczynska et al., 2009; Kozar et al., 2004; Geng et al., 2003; Murphy et al., 1997; Brandeis et al., 1998; Santamaría et al., 2007). Animal cells have evolved a division of labour between multiple cyclins: cyclin A is required for DNA replication and cyclin B for mitosis. (Moreover, in at least one type of somatic cell, though not in embryonic and adult stem cells, cyclin E can compensate for cyclin A (Kalaszczynska et al., 2009)). By contrast, neither D- nor E-type cyclins are strictly essential for cell division (Kalaszczynska et al., 2009; Kozar et al., 2004; Geng et al., 2003; Murphy et al., 1997; Brandeis et al., 1998). E-type cyclins are only essential in endoreplicating cells and important in cells re-entering the cell cycle from quiescent phase (Geng et al., 2003). Thus, the second model of cyclin function may explain the genetic data in mammals better than a threshold model.
To test the second model, a number of studies have been carried out in budding yeast extracts or using purified animal cyclin-cdks in vitro to compare directly different cyclin/cdk complexes, but found that different cyclin complexes show similar phosphorylation efficiencies for the majority of substrates (Loog and Morgan, 2005; Errico et al., 2010). In vitro studies may not fully recapitulate the in vivo affinities of cyclin-cdks for different proteins, e.g.: some targets may only be available at specific times in the cell cycle, because either they are only present in specific phases, or the cyclin-cdk only has access to them at a precise time or place (Moore, Kirk and Hunt, 2003). Sub-cellular localization, however, is unlikely to be the sole explanation because all cyclin-cdks continuously shuttle between the nucleus and the cytoplasm (Jackman et al., 2002; Yang et al., 1998; Toyoshima et al., 1998; Hagting et al., 1998). Moreover, in late G1 and early S phases both cyclin E and cyclin A are predominantly nuclear but cyclin E cannot replace cyclin A. Similarly, in G2 phase there is a significant population of cyclin A in the cytoplasm with cyclin B1, yet mouse cells genetically null for cyclin B1 arrest in G2 phase (B. Strauss, M. Zernicka-Goetz, and JP, unpublished results).
To understand how the human cell cycle is coordinated by cyclin-cdks we sought a means to identify interacting proteins that preserved both potential intrinsic cyclin specificity in vivo and the temporal dynamics of the cell cycle. Since the core cell cycle machinery of mammalian cells requires at least one A- or E-type cyclin and one B-type cyclin (Kalaszczynska et al., 2009; Kozar et al., 2004; Geng et al., 2003; Murphy et al., 1997; Brandeis et al., 1998) (only specialized tissues require D-type cyclins) we focused on cyclins E1, A2 and B1. Here, we have performed a time-resolved proteomic analysis of the core cell cycle components, which has provided insights into the regulatory networks of animal cyclin-cdks and revealed unexpected aspects of the coordination between the different cyclin-cdk modules.
Results and Discussion
Cyclins bind overlapping but distinct sets of protein complexes in vivo
To determine how the cyclin-cdks control the cell cycle in vivo, we purified specific cyclin/cdk complexes at defined stages of the cell cycle and analysed them by quantitative mass spectrometry using SILAC (Stable Isotope Labeling by Amino Acids in Cell Culture (Ong et al., 2002)). This approach has substantial advantages for analyzing the mechanics of the cell cycle because in addition to identifying cyclin/cdk regulators, many substrates bind relatively tightly to the ‘hydrophobic patch’ of the cyclin and are recovered by immunoprecipitation (Archambault et al., 2004; Schulman, Lindstrom and Harlow, 1998). Moreover, some cyclins have cdk-independent roles in the cell cycle (Geng et al., 2007), which requires identification of both substrates and other interacting proteins.
We used stable cell lines expressing human cyclin E1, cyclin A2, or cyclin B1, all tagged with Venus-YFP to analyze their localization, and a 3xFLAG tag for immunoprecipitation to eliminate problems inherent in using endogenous antibodies (notably differences in immunoprecipitation efficiency and spurious interactions introduced by cross-reaction). Tagged plant cell cycle proteins have also been used recently to identify new cyclin/cdk complexes (Van Leene et al., 2010). In our study all tagged human cyclins exhibited their characteristic sub-cellular localizations (Jackman et al., 2002; Draviam et al., 2001), were correctly degraded (Supplemental Movies 1-3) and efficiently bound their kinase partners (Supplemental Table 1). Furthermore, tagged cyclin A bound a similar set of proteins to endogenous cyclin (Supplemental Table 2a) of which a number were confirmed in a non-transformed cell line (Supplemental Table 2b).
Each cell line was labelled with light, intermediate, or heavy-mass isotope versions of lysine and arginine, and used for control or cyclin immunoprecipitations from different cell cycle phases (Figure 1A). Cyclins were immunoprecipitated from the phases in which they were present: cyclin E from G1 and S; cyclin A from S and G2; cyclin B from G2 and mitosis. Cells were released from a double thymidine block for one hour, six hours, twelve hours, or seventeen hours for S, G2, M, and G1 samples, respectively (Supplemental Figure 1). The Eg5 motor protein inhibitor, dimethylenastron, was added to the mitotic samples to accumulate cells in mitosis. Immunoprecipitates were pooled, digested with trypsin and analyzed by mass spectrometry to distinguish proteins arising from different cell cycle phases (Figure 1A). Quantitative ratios were generated using MaxQuant software from at least three biological replicates and used to identify interactors enriched in cyclin immunoprecipitates relative to controls (Cox and Mann, 2008) (Figure 1B-D, Supplemental Tables 1 and 5.)
Figure 1.
Identification of the protein interaction network of human cyclins by immunoprecipitation and quantitative mass spectrometry. A) Schematic of the experimental strategy. HeLa cells expressing tagged cyclins were metabolically labeled with three different combinations of isotopic lysine and arginine, synchronized and immunoprecipitated with control or anti-FLAG antibodies. Immunoprecipitates were pooled and analyzed simultaneously by mass spectrometry. Peptides arising from each cell population were quantified by measuring the relative intensity of light (K0/R0), intermediate (K4/R6) and heavy (K8/R10) peaks with MaxQuant. Example schema is for cyclin A: intermediate-labeled S phase cells, and heavy-labeled cells G2 phase. For cyclin E, intermediate-labeled cells were G1 phase and heavy-labeled cells were S phase. For cyclin B, intermediate-labeled cells were G2 phase and heavy-labeled cells were M phase. B) Cyclin E complexes were isolated from G1 and S phase cells and analysed by mass spectrometry. Identified proteins are shown. Solid colored lines indicate the cut-off score for the ratio of FLAG to control IP for either G1 phase or S phase (M/L: medium/light cell populations; H/L: heavy/light populations). Each point indicates one protein identified and its color reflects the average S/G1 phase ratio (the heavy/medium ratio) of the replicate experiments. Grey points indicate the identified protein was nonspecific (not enriched above the control IP.) Purple points indicate the protein was identified in both phases, yellow, red, blue, or green shading indicates the interaction was enriched in G1, S, G2, or M phase, respectively. Note that ratios could not be calculated for all interactors as very specific interactors appear solely as heavy or medium-weight peaks with no light peak from the control sample. C) Cyclin A complexes were isolated from S and G2 phase cells. D) Cyclin B complexes were isolated from G2 and M phase cells. For complete protein identification and quantification see Supplemental Tables 1 & 5.
A total of 295 proteins were reproducibly enriched over controls in at least two of three replicates: 73 with cyclin E1, 227 with cyclin A2, and 192 with cyclin B1 (Figures 2-3, Supplemental Table 1b.) Only thirty four (11%) were identified with all three cyclins, including Cdk1, Cdk2 and known cdk regulators (e.g.SCFSkp2 components Skp1and Skp2, and Plk1). Therefore, very few, if any proteins, bound non-specifically or via the common tag, and, since each cyclin bound both Cdk1 and Cdk2, this showed that cyclins provided specificity to the interaction networks.
Figure 2.
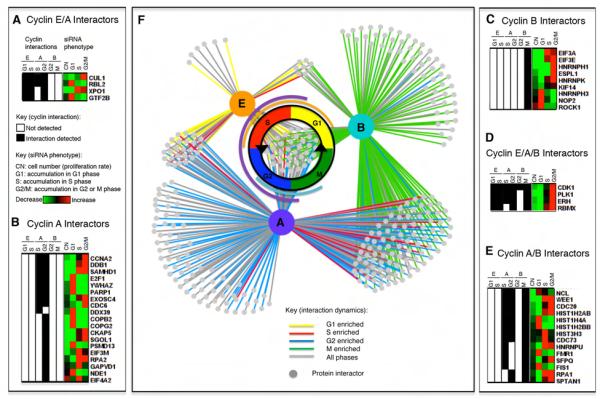
Different interactors bind distinct cyclins and contribute to cell cycle progression. Cyclin interaction data was compared to phenotypic data from siRNA depletion studies. On the left, black shading indicates that the protein in that row was identified with that cyclin (E, A, or B) in that cell cycle phase (G1, S, G2, or M). On the right, increases or decreases in cell number (CN) or in the proportion of cells abnormally accumulating in different cell cycle phases (G1, S, or G2/M) after siRNA depletion is indicated by red (increase) to green (decrease) shading, according to data from Kittler et al. (see Supplemental Table 1). A) Interactors that bind both cyclins E and A. B) Cyclin A-specific interactors. C) Cyclin B-specific interactors. D) Interactors that bind all three cyclins. E) Interactors that bind cyclins A and B. F) The cyclin interactome shows the specificity of cyclin interactions. Many interactions are temporally dynamic and enriched in one cell cycle phase, indicated by yellow (G1), red (S), blue (G2), or green (M) connecting lines.
Figure 3.
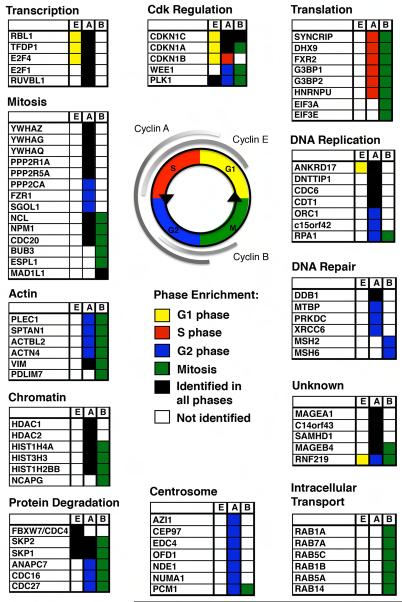
Cyclin/cdk complexes bind proteins of varied biological function and these proteins are enriched in specific cell cycle phase contexts and with specific cyclins. No shading indicates that the protein was not identified with that cyclin or was not enriched above the control. Black shading indicates identification in all phases analyzed with no enrichment in any one phase. Phase-enriched hits (ratio <0.5 or >2) are colored according to the schema shown (yellow G1, red S, blue G2, green M). A subset of representative interactors is shown; for complete data refer to Supplemental Tables 1 and 5.
To analyze the sensitivity of our method, we examined published cyclin interactors in six databases: MINT, IntAct, DIP, BioGRID, HPRD, and MIPS/MPact (Wu et al., 2009). We selected those interactions supported by more than one publication to serve as a “gold standard” dataset (Supplemental Figure 2). Our experiments detected 76% (31/41) of those interactors indicating a high level of sensitivity, but the majority of our cyclin-protein interactions had not been previously described. Our proteomic strategy also identified known substrates at a similar rate to in vitro kinase assay strategies (Supplemental Figure 3) (Blethrow et al., 2008; Chi et al., 2008; Errico et al., 2010).
Importantly, our experiments preserved in vivo specificity because those proteins previously shown as specific to a particular cyclin were identified only with that cyclin, e.g. Cdc6 only bound cyclin A (Petersen et al., 1999), separase only bound cyclin B (Holland and Taylor, 2006), Fbxw7/Cdc4 (responsible for cyclin E ubiquitylation) only bound cyclin E (Strohmaier et al., 2001; Koepp et al., 2001) and anaphase promoting complex/cyclosome (APC/C) components were identified with cyclins A and B, the targets and regulators of the APC/C (Kraft et al., 2003; King et al., 1995; Sudakin et al., 1995).
To dissect the function of the cyclin interactors, we looked up the phenotype described for cells depleted of individual interactors by siRNA treatment (Kittler et al., 2007) (Figure 2, Supplemental Table 1b.) Proteins with a cell division defect (reduced proliferation or accumulation in one cell cycle phase) were highly enriched (98/295) compared to all the siRNAs tested (2155/17837; Z-test=10.8, p< 10−4), confirming the functional relevance of the novel interactors we identified (Kittler et al., 2007). Yet only 15 of those 98 proteins with observed cell cycle defects were previously described cyclin interactors or substrates. Therefore, these newly identified cyclin binding partners play functional roles in human cell cycle progression. Moreover, the siRNA screen was not saturated, therefore, the other cyclin interactors we identified could also be important for the cell cycle.
We used two recent analyses of global protein phosphorylation across the cell cycle (Dephoure 2008; Olsen et al., 2010) to analyze the phosphorylation of cdk sites (S/T*P) within our cyclin interactor network (Supplemental Table 1b.) 155 of the 295 cyclin interactors (53%) contain cdk sites that can be phosphorylated in vivo. Cyclin/cdk substrates often contain RxL motifs that mediate binding to the ‘hydrophobic patch’ of the cyclin (Schulman et al., 1998), and the crystal structure of a Cdc6 peptide bound to Cyclin A-Cdk2 indicates that the minimum distance between the target serine and the RxL motif is 15 residues (Cheng et al., 2006). Therefore, we looked for RxL motifs 15 to 30 or 15 to 60 amino acids distal to S/TP sites (Supplemental Table 1b). The RxL motif was present 15 to 30 residues upstream in 80 interactors (including APC/C subunits Apc1 and Apc7, separase, Ki67, p21 and Cdc6), and 15 to 60 residues upstream in 140 interactors (140/295; 47%) (Supplemental Table 1b).
A biochemical basis for Cyclin E redundancy
Strikingly few proteins bound specifically to cyclin E compared to cyclins A (or B), indicating that cyclin E may have a more restricted substrate specificity in vivo, even though it phosphorylated a similar range of substrates in vitro (Errico et al., 2010). Several interactors were enriched in G1 (p21/Cdkn1a, p57/Cdkn1c, E2F4, Rbl2 and Ubr4) or S phases (Ccnh) (Figure 1B). The G1 enrichment of E2F and Rb-like proteins with cyclin E was reported previously (Lees et al., 1992), confirming the ability of our approach to identify cell cycle changes in protein interactions. Cyclin E bound primarily to E2F4 and to p130 of the Rb-family, components of the dREAM complex that maintains the quiescent state (Litovchick et al., 2007), which may be relevant to cyclin E’s role in promoting re-entry to the cell cycle.
Although cyclin E could promote entry to S phase (Lew et al., 1991; Koff et al., 1991), genetic studies showed that cyclin E was dispensable for cell division (Geng et al., 2003). Our analyses reconciled these observations and provided a biochemical explanation for the redundancy of cyclin E, because the majority of proteins identified in complex with cyclin E also bound to cyclin A (58/72; 81%) (Figure 3.) These included proteins implicated in the control of transcription in late G1 and S phase such as Tfdp1, E2F4, Rbl1, Rbl2/p130, Rbbp4 and NPAT. By contrast, just nine proteins were identified as common interactors of only cyclins E and B (Supplemental Table 1b.) Cyclin E is primarily required for endoreplication, therefore it will be interesting to analyse the proteins it binds in these specialized cell cycles.
Cyclin A controls DNA replication via human Sld3
The genetic requirement for cyclin A (Murphy et al., 1997) and its dual role in promoting S phase and mitosis have been enigmatic because other cyclins are present in the phases where cyclin A is required—cyclin E in late G1 and S phase, and cyclin B in G2 phase. Our data shed light on this because some cyclin A targets also interacted with either cyclin E (25%; 58/228) or cyclin B (56%; 127/228), but a significant subset of proteins were specific for cyclin A (34%; 77/228), demonstrating an intrinsic difference in the biological properties of the cyclins. These cyclin A-specific proteins included DNA replication factors (Cdt1, Cdc6 and Orc1), chromatin remodelling enzymes (Hdac1, Hdac2 and Phf8), and a number of uncharacterized proteins (Figure 3, Supplemental Figure 4). In G2 phase, cyclin A bound to proteins required for DNA repair by the non-homologous end-joining pathway (NHEJ) including Xrcc5/Ku80, Xrcc6/Ku70, Prkdc/DNA-PK and Ddb1, indicating that cyclin A/cdk might regulate the balance between the NHEJ and homologous end-joining pathways that varies during the cell cycle (Huertas, 2010).
Comparing the set of cyclin A interactors with an analogue-sensitive Cdk2 kinase screen (Chi et al., 2008) corroborated several novel substrates (e.g. Thrap3, Ankrd17, Nde1), but many known cyclin A substrates (Cdc6, Orc1, E2F1, p107/Rbl1, p21/Cdkn1a, Fzr1/Cdh1) were identified only in our analysis (Supplemental Figure 3.) Therefore, our approach is ideally suited to identify both regulators and substrates of cyclin/cdks.
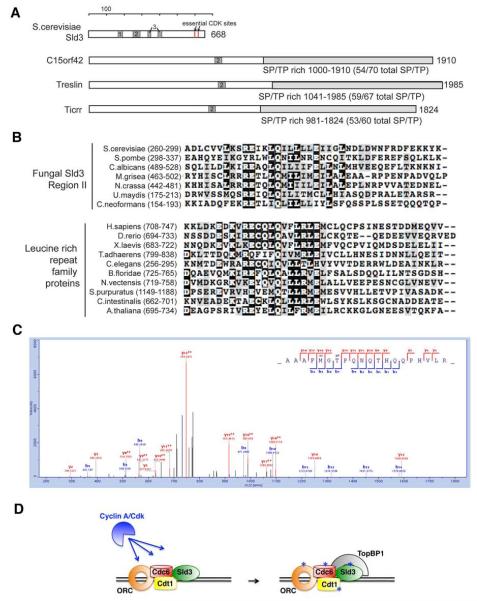
Since we identified most of the known cdk targets required for DNA replication, it was possible that new DNA replication factors also specifically co-immunoprecipitated with cyclin A. Uncharacterized proteins that met those criteria included c15orf42, c14orf43, Znf185, and MageA1. The budding yeast DNA replication factor, Sld3, is an essential cdk target (Tanaka et al., 2007; Zegerman and Diffley, 2007) but a homologue had not been identified in animal cells (Supplemental Table 4). We identified homology between our c15orf42 and Sld3 by six iterations of psi-BLAST with three regions of Saccharomyces cerevisiae Sld3 conserved among fungi (Figure 4). This homology has recently been noted by others (Sanchez-Pulido et al., 2010). The essential cdk sites in yeast Sld3 mapped to the C-terminus, and the C-terminus of c15orf42 and its homologs also had a large number of potential cdk sites (Figure 4), a number of which (S441, S838, S923, T1260) were phosphorylated when bound to cyclin A/cdk (Figure 4, Supplementary Table 3.) Moreover, a number of these sites were sensitive to the addition of a cdk inhibitor in vivo (Table 1, Supplemental Table 6.) Two further lines of evidence led us to conclude that c15orf42 is human Sld3. First, depleting c15orf42 from HeLa cells reduced DNA replication and consequently increased the proportion of S phase cells (Supplementary Figure 5.) Second, yeast Sld3 binds TopBP1 after phosphorylation by cyclin/cdk, and the fish and frog homologues of c15orf42, ticrr and treslin, and c15orf42 itself, were found to bind TopBP1 (Sansam et al., 2010; Kumagai et al., 2010). Moreover, depleting ticrr and treslin also reduced DNA replication (Sansam et al., 2010; Kumagai et al., 2010). In contrast, the frog Gemc1 protein had been proposed as the vertebrate Sld3 homolog but we did not identify a Gemc1 homolog in any of our experiments (Balestrini et al., 2010). The Sld3 DNA replication partner in yeast, Sld2, was identified as a Clb5 -specific target in vitro (Loog and Morgan, 2005), thus cyclin specificity in targeting DNA replication factors may be a conserved aspect of cyclin function.
Figure 4.
Cyclin A-specific interactor c15orf42 is the human homologue of DNA replication factor Sld3. A) Comparison of cdk sites in S. cerevisiae Sld3, human c15orf42, Xenopus treslin, and zebrafish ticrr. B) c15orf42 has homology to yeast Sld3. Black shading indicates identical amino acid residues. C) Cyclin A-associated c15orf42 is phosphorylated at cdk sites in vivo. A representative phosphopeptide fragmentation spectra of c15orf42 shows phosphorylation at T1260. See also Supplemental Tables 3 and 6. D) A model of human DNA replication including c15orf42/Sld3. All colored proteins were identified in purifications of cyclin A protein complexes.
Table 1.
The phosphorylation of cyclin A interactors is sensitive to cdk inhibition.
| Protein Names |
Positio n |
Phosphopeptide Sequence | Average ratio (+ inhibitor/− inhibitor) |
|---|---|---|---|
| c14orf43 | S923 | RE(S)PSEERLEPK | 0.43 |
| c15orf42 | S441 S599 S1623 |
HVLQTAVAD(S)PR LNVKAQKLHPDG(S)PDVAGEK SLSKPEPTYV(S)PPCPR |
0.63 0.24 0.33 |
| CDC20 | S41 T157 |
KAKEAAGPAP(S)PMR VLYSQKA(T)PGSSR |
0.53 0.37 |
| CDC6 | S45 T67 S74 S419 |
LEPTNVQTVTC(S)PR LGDDNLCN(T)PHLPPC(S)PPKQGKK LGDDNLCN(T)PHLPPC(S)PPKQGKK SQTILKPLSECK(S)PSEPLIPK |
0.60 0.39 0.39 0.33 |
| DNTTIP1 | S161 | QAEEECAHRG(S)PLPK | 0.41 |
| OFD1 | S789 | HSLSIPPVS(S)PPEQK | 0.65 |
| PHF8 | T1007 | ST(T)PMAPGVFLTQR | 0.57 |
| RBL1 | S640 S749 S762 |
DMQPL(S)PISVHER VK(S)PVSLTAHSLIGASPK SPVSLTAHSLIGA(S)PK |
0.46 0.28 0.36 |
| THRAP3 | S253 | SPALK(S)PLQSVVVR | 0.66 |
| TRERF1 | S491 | AQPG(S)PESSGQPK | 0.51 |
Thirty minutes prior to cell lysis, 300nM Cdk I/II inhibitor III was added to the cells used for cyclin A immunoprecipitation experiments. A selection of inhibitor sensitive phosphopeptides are shown. See also Table S6 for the full dataset.
Roles for Cyclin A in G2
Unlike its role in DNA replication, the role of cyclin A in G2 phase is far less clear. Our analysis both indicated potential G2 phase roles and gave insights into the temporal specificity by which cyclin A promotes S phase and mitosis. We found a number of cyclin A interactors exclusively, or more than two-fold more abundantly, in G2 phase (69/228; 30%). As well as several uncharacterized proteins (Rnf219, Znf185) they included several centrosomal proteins (Nde1, Ckap5, Ofd1, Cep97, plus Cep131 & Pcm1 detected in at least one experiment, Figure 3, Supplemental Tables 1a and 5). Ofd1 was phosphorylated at a potential cdk site, S789, which was sensitive to cdk inhibition (Table 1, Supplemental Table 6). These proteins could be important for the role of cyclin A-cdk in centrosome duplication. Alternatively, they may be important to bind cyclin A to centrosomes in G2 phase (Supplementary Movie 2 and (den Elzen and Pines, 2001)).
Notably, cyclin A co-immunoprecipated components of the protein phosphatase 2A complex (Ppp2r1a, Ppp2ca/b, Ppp2r5a) whereas other cyclins did not (Figure 3, Supplementary Table 1a). More peptides of each PP2A component were identified with cyclin A in G2 phase than S phase, and the Saccharomyces cerevisiae ortholog of Ppp2r5a, Rts1, is phosphorylated by cdk in vitro (Supplemental Table 4). Since PP2A limits entry to mitosis in frog extracts (Mochida et al., 2009), these results indicate that cyclin A/cdk may contribute to mitotic entry by controlling PP2A.
Cyclin A may promote mitotic entry by inhibitory phosphorylation of Wee1
Given the major role for protein kinases at mitosis, we looked for those bound by cyclin A in G2 phase, and identified Plk1, Ndr1 and Wee1. Wee1 inhibits cyclin B1/Cdk1 by phosphorylating Cdk1 to prevent mitosis. Yet cyclin B/Cdk1 is the main regulator of Wee1—cyclin B/Cdk1 phosphorylates Wee1 to inactivate and/or promote its degradation (Watanabe et al., 2005; Kim and Ferrell, 2007). How an initial small pool of cyclin B/Cdk1 is activated is unclear, but recent evidence supports the hypothesis that cyclin A may contribute because it binds and phosphorylates Wee1 (Li et al., 2010).
Our experiments showed that from S to G2 phase cyclin A interacted with an increasing proportion of Wee1, mirroring the increase in Wee1 levels observed in G2 (Olsen et al., 2010). Supporting the hypothesis that cyclin A/cdk-dependent phosphorylation of Wee1 promoted mitotic entry, cyclin A-associated Wee1 in G2 phase was phosphorylated at the conserved ‘ultrasensitive’, cdk-dependent phosphosites (T187 and T190, Supplemental Table 3) that inhibit Wee1 activity (Kim and Ferrell, 2007).
We confirmed that these sites were phosphorylated directly by cyclin A/cdk complexes by in vitro phosphorylation assays using immunoprecipitated cyclin A/cdk and recombinant Wee1 (Supplemental Figure 6), in the presence and absence of a cdk inhibitor, and analysis by mass spectrometry (Table 2, Supplemental Table 7a.). Four phosphosites, including T187 and T190, were sensitive to cdk inhibition (Table 2, Supplemental Table 7a). To show that these sites were phosphorylated in vivo in a cdk-dependent manner, we immunoprecipitated G2 phase Wee1 in the presence and absence of cdk inhibitors. We identified a number of proline-directed phosphorylation sites on Wee1 by mass spectrometry, five of which were sensitive to cdk inhibition (Table 2 and Supplemental Table 7b). These data, together with those of Li and colleagues, support a model in which cyclin A contributes to mitotic entry through inhibitory phosphorylation of Wee1 in late G2 to tip the balance of the cyclin B/Cdk1—Wee1 feedback loop in favour of active cyclin B/Cdk1 and inactive Wee1.
Table 2.
In vitro and in vivo phosphorylation of Wee1 by cyclin A/cdk in G2 phase.
| Wee1 phosphorylation sites | Wee1 immuno- precipitation |
Wee1 in vitro phosphorylation |
|
|---|---|---|---|
| Site | Phosphopeptie Sequence |
Average ratio (+ inhibitor/− inhibitor) |
Average ratio (+ inhibitor/− inhibitor) |
| S150 | CGGPGDA(S)PRGCGAR | 0.57 | |
| S165 | R(S)PRPDHPGTPPHKTFR | 0.68 | |
| S165/T173 | R(S)PRPDHPG(T)PPHKTFR | 0.28 | 0.38 |
| T173 | SPRPDHPG(T)PPHKTFR | 0.66 | 0.54 |
| T187/T190 | LFD(T)PH(T)PK | 0.47 | 0.59 |
| T187/T190 | LFD(T)PH(T)PKSLLSK | 0.34 | 0.46 |
| T190 | LFDTPH(T)PKSLLSKAR | 0.11 | |
Proline directed phosphorylation sites in Wee1 are reduced in abundance by Cdk I/II inhibitor III treatment in G2 phase Wee1 immunoprecipitates. Wee1 was also phosphorylated in vitro using immunoprecipitated cyclin-A/cdk with and without the addition of the cdk inhibitor roscovitine. See also Figure S6 and Table S7.
Cyclin B coordinates changes in the cell architecture at mitosis
The activation of cyclin B/Cdk1 is the direct trigger for mitotic entry (Gavet and Pines, 2010) and this kinase is likely to effect many of the changes in the cell at mitosis. Recently, we correlated changes in cell architecture with different levels of cyclin B/cdk1 activity (Gavet and Pines, 2010): low levels trigger cell rounding and APC/C activation, whereas higher levels are required for nucleolar and nuclear envelope disassembly. Our analysis of proteins binding to cyclin B1/cdk in mitosis now revealed likely substrates for these processes (Figure 3.)
We corroborated several substrates proposed from in vitro kinase assays (Hnrnpk, Cttn, Mki67, Ubap2l) (Errico et al., 2010; Blethrow et al., 2008) and identified as many known substrates of cyclin B/cdk as did the in vitro assays (Supplemental Figure 3). Our data provide candidates for cyclin B targets in each aspect of mitotic entry. For example, for cell rounding, which involves actin cytoskeletal remodelling, changes in osmotic pressure, and changes in intracellular trafficking (Boucrot and Kirchhausen, 2007; Kunda and Baum, 2009;Stewart et al., 2011) we identified a number of mitosis-enriched cyclin B-specific interactors related to the control of the actin cytoskeleton (24/191; 13%), (including actinins Actn1 and Actn4, Coro1c, Flna, Sptan1, Sptbn1, Pdlim7 and Plec1, whose Cdk-dependent phosphorylation diminished its ability to cross-link intermediate filaments during mitosis (Foisner et al., 1996)) and many Rab proteins involved in trafficking (Figure 3.)
Immediately upon activation, a substantial proportion of cyclin B/Cdk1 moves into the nucleus, followed by visible signs of chromosome condensation (Gavet and Pines, 2010). Condensins, the likely mediators of chromosome condensation, were identified as cyclin/cdk substrates (Kimura et al., 1998; Abe et al., 2011) and we identified the condensin subunit Ncapg specifically interacting with cyclin B1 in mitosis. Nucleolar disassembly also followed the arrival of cyclin B/Cdk1 activity in the nucleus and we identified three nucleolar proteins, Ncl, Npm1, and Nop2 with cyclin B, all of which were phosphorylated in vivo at consensus cdk sites (Peter et al., 1990; Dephoure et al., 2008). Lamin A/C is also a canonical cyclin B1/Cdk1 substrate (Ward and Kirschner, 1990; Heald and McKeon, 1990; Peter et al., 1990), but although we identified it in cyclin B immunoprecipitates, the enrichment over background was not high enough to meet our stringent threshold for genuine interactors. Some bona fide interactors, particularly transiently binding proteins, can be excluded when the cut-off ratio is set to a stringent level to eliminate non-specific proteins (Boulon et al., 2010). Thus, other proteins identified in our analyses that fall below this threshold may be genuine interactors or substrates (Supplemental Table 5.) Nevertheless, phosphorylated lamin peptides were identified at a two-fold higher level in mitotic cyclin B immunoprecipitates than in control immunoprecipitates or G2 phase cyclin B immunoprecipitates (Supplemental Table 3), and were phosphorylated at serine 22, the consensus cdk site required for lamin disassembly (Heald and McKeon, 1990). That the lamins did not bind with high affinity to cyclin B1/Cdk1 agreed with our finding that a higher level of cyclin B/Cdk1 activity was required for nuclear lamina disassembly than for most other changes in the mitotic cell. Thus, the ‘threshold model’ could apply to the regulation of events by a specific cyclin-cdk within a single phase of the cell cycle.
Finally, after nuclear envelope breakdown we, and others (Bentley et al., 2007), observed a population of cyclin B1 at unattached kinetochores, and in our proteomic analyses we detected cyclin B bound to components of the spindle assembly checkpoint (SAC), including Mad1, Mad2, BubR1 and Bub3 (Supplemental Table 1a). This raised the possibility that cyclin B/cdk might have a role in the SAC and indeed we find that cyclin B/cdk is required to maintain the SAC (Jackman, M., Pardo, M., Choudhary, J.S., Pines, J., manuscript in preparation.)
The genetic requirement for cyclin B1 is supported by our observation that almost one third (55/191) of the proteins identified with cyclin B did not interact with other cyclin/cdk complexes; 56% (31/55) of these were phosphorylated at consensus cdk sites in vivo (Olsen et al., 2010; Dephoure et al., 2008) (Supplemental Table 1b.) During G2 phase a large pool of inactive cyclin B/Cdk1 builds up, and it was striking that most proteins did not bind to cyclin B1/Cdk1 until it was activated (148/191; 77%) (Figure 2.) To our knowledge, there are no previous indications that cyclin B1/Cdk1 activation promotes binding to its substrates.
Coordination between cyclins A and B is revealed by a ‘handover’ of interactors at mitosis
Our results provide direct evidence for the model that cyclins contribute biochemical specificity to cyclin/cdk complexes through binding a unique subset of the proteome in vivo. Unexpectedly we found that many interactors were shared between more than one cyclin/cdk complex in a context-dependent manner: the same substrate was bound by different cyclins but at different times in the cell cycle, even when the cyclins were present in the same cell cycle phase (Figure 2). Thus, these results identify an additional aspect of cyclin function—coordination between cyclins in targeting a common set of proteins at different times in the cell cycle—that can explain how multiple cyclin/cdk complexes phosphorylate common proteins in vitro. This was most clearly illustrated by the interaction networks for cyclins A and B.
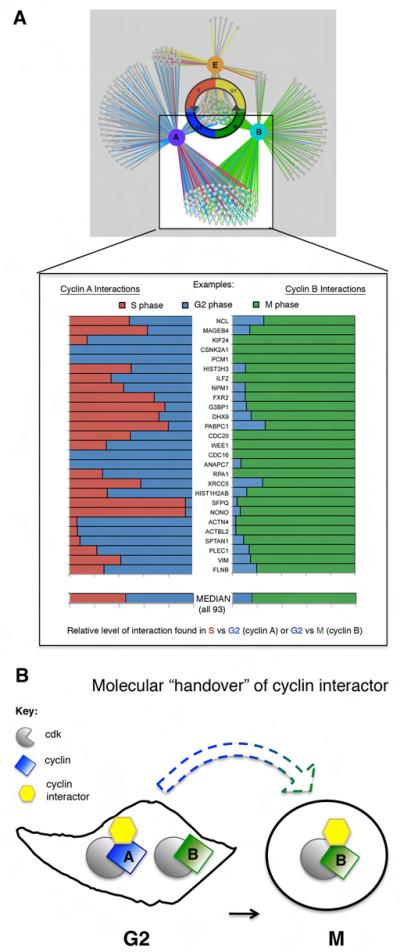
Since cyclin A drives cells through interphase, and cyclin B regulates mitosis, we were surprised to identify the largest overlap in interactors between cyclins A and B (66%; 127/191 of cyclin B interactors, Supplementary Table 1b.) The shared interactors were nuclear and cytoplasmic, consistent with both cyclin/cdk complexes shuttling between the nucleus and cytoplasm (Jackman et al., 2002; Yang et al., 1998). Of proteins that interact with both cyclins A and B, 48/93 (52%) are annotated as cytoplasmic whilst the rest are nuclear or both nuclear and cytoplasmic (Supplemental Table 5). These shared interactions did not take place simultaneously in G2 phase, when both cyclins A and B were at near maximal levels. Instead the majority of shared proteins interacted first with cyclin A in interphase and only later with cyclin B after entry into mitosis (88%; 112/127; Figure 5.) This pattern of sequential interaction parallels both the activation of cyclin A- and B-cdks and the degradation of the cyclins: cyclin A starts to disappear immediately upon mitotic entry whilst cyclin B remains until chromosomes are attached to the spindle at metaphase (Supplementary Movies 2 and 3) (Geley et al., 2001; den Elzen and Pines, 2001; Clute and Pines, 1999).
Figure 5.
Cyclins A and B share many protein partners but the interactions are temporally coordinated. A) The protein interaction networks of cyclins A and B overlap. B) Interactors specific to cyclins A and B bind to cyclin A in interphase (red, S, and blue, G2) and cyclin B primarily in mitosis (green). Each horizontal bar represents one protein and a representative subset of total cyclin A and B shared interactors is shown (complete data in Supplemental Table 1; median calculated from complete set of quantified proteins.) The width of the bar in each color represents the relative amount of protein immunoprecipitated from each phase (calculated from the H/M ratio). The median contributions from S (red) and G2 (blue) for the total quantified cyclin A interactors are nearly 1 to 1 (median G2/S ratio= 1.2) whereas the same interactors are mitotic enriched with cyclin B (median M/G2 ratio=5.4).
We analyzed the relationship between cyclin interaction patterns to gain further insights into how the temporal order of events is regulated by a progressive increase in cyclin B/Cdk1 activity (Gavet and Pines, 2010). A high threshold of cyclin B activity is required for late events in prophase, including chromosome condensation and both nucleolar and nuclear envelope breakdown, and most proteins corresponding to these late events were only identified bound to cyclin B and exclusively in mitosis, e.g.: the condensin Ncapg, the lamin interactor Lap2 and the nucleolar proteins Nop2 and Ki67. By contrast, early prophase events, such as cell rounding and APC/C activation, require a lower threshold of cyclin B/cdk activity, and proteins potentially involved in these processes, e.g.: spectrins (Fowler and Adam, 1992), plectin (Foisner et al., 1996), vimentin (Yamaguchi et al., 2005), filamins (Cukier et al., 2007), and APC/C subunits, were identified with both cyclin B in mitosis and cyclin A in G2 phase. One interpretation of these data is that early events in mitosis can occur at lower thresholds of cyclin B/cdk activity because they have been primed in G2 by cyclin A/cdk.
In conclusion, we have shown that in vivo, key human cyclins, cyclins E1, A2 and B1 act via dual mechanisms—first, regulating a unique set of targets, and second, coordinating to regulate a shared subset of cell cycle proteins (Figure 5, Supplemental Figure 7). Thus, the cell cycle is controlled by both biochemical and temporal specificity in cyclin interactions and cell cycle phase coordination between the cyclins. Until now the global role of dynamic and context-dependent protein interactions has been difficult to analyze in biological systems. The ability to compare protein interaction networks of other cell cycle regulators or post-translational modifications in multiple cell cycle phases using quantitative proteomics will lead to a more comprehensive understanding of cell cycle progression and the dynamics of protein regulation in general.
Experimental Procedures
Cell lines
Tetracycline-inducible stable cell lines from the HeLa Flp-In system (Invitrogen) expressed human cyclins E1, A2, or B1 tagged with Venus-YFP and 3xFLAG. Cells were cultured in DMEM lacking lysine and arginine (Dundee Cell Products) supplemented with 1kDa dialyzed FBS (PAA) and light (Sigma) or isotopically enriched (Cambridge Isotope Laboratories) lysine and arginine.
Cell synchronization
Cells were synchronized by double thymidine block followed by release into normal medium for 16.5 hours (G1), 1 hour (S), or 6 hours (G2), or release into medium containing the Eg5 inhibitor dimethylenastron to block cells in prometaphase (M) for 12 hours. Expression of the tagged cyclin was induced by addition of tetracycline 24 hours prior to harvesting cells. Synchronization was confirmed by propidium iodide staining and flow cytometry.
Immunoprecipitation and mass spectrometry
Protein extracts were prepared by lysing cells using a nitrogen cavitation chamber (Parr) followed by ultracentrifugation. Cyclin complexes were immunoprecipitated using anti-FLAG antibody M2 (Sigma) covalently coupled to Protein G Dynabeads (Invitrogen) and eluted with FLAG peptide (Sigma). Immunoprecipitates were resolved on protein gels, trypsinized and analyzed by LC-MS/MS using LTQ-FT Ultra/LTQ Orbitrap Velos mass spectrometers (Thermo). Peptides were identified and quantified using MaxQuant software (Cox and Mann, 2008). See Extended Experimental Procedures for more details.
Supplementary Material
Highlights.
Quantitative proteomic strategy reveals dynamics of cell cycle protein interactions
Cyclins confer biochemical specificity to cyclin/cdk interaction networks
Cyclin A phosphorylates Sld3/c15orf42 and Wee1 to promote S phase and mitosis
Cyclins A and B coordinate to bind a set of proteins sequentially in G2 phase and mitosis
Acknowledgements
We thank the Pines and Choudhary labs for helpful discussions throughout the project, and Charles Bradshaw for identifying yeast orthologs. We are grateful to Jörg Mansfeld for help with the in vitro kinase assays and Lars Koop for the anti-cyclin A phospho serine 154 antibody. This work was supported by a programme grant from Cancer Research UK to JP and core grant support to the Gurdon Institute and the Sanger Institute from The Wellcome Trust. FWP was supported by a Marshall Scholarship, a Cambridge University Overseas Research Studentship and a U.S. National Science Foundation Graduate Research Fellowship. AL was supported by a Herschel-Smith studentship.
References
- Abe S, Nagasaka K, Hirayama Y, Kozuka-Hata H, Oyama M, Aoyagi Y, Obuse C, Hirota T. The initial phase of chromosome condensation requires Cdk1-mediated phosphorylation of the CAP-D3 subunit of condensin II. Genes Dev. 2011;25:863–74. doi: 10.1101/gad.2016411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault V, Chang EJ, Drapkin BJ, Cross FR, Chait BT, Rout MP. Targeted proteomic study of the cyclin-Cdk module. Mol Cell. 2004;14:699–711. doi: 10.1016/j.molcel.2004.05.025. [DOI] [PubMed] [Google Scholar]
- Balestrini A, Cosentino C, Errico A, Garner E, Costanzo V. GEMC1 is a TopBP1-interacting protein required for chromosomal DNA replication. Nat Cell Biol. 2010;12:484–91. doi: 10.1038/ncb2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley AM, Normand G, Hoyt J, King RW. Distinct sequence elements of cyclin B1 promote localization to chromatin, centrosomes, and kinetochores during mitosis. Mol Biol Cell. 2007;18:4847–58. doi: 10.1091/mbc.E06-06-0539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. Proc Natl Acad Sci U S A. 2008;105:1442–7. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J, Cross FR. Multiple levels of cyclin specificity in cell-cycle control. Nat Rev Mol Cell Biol. 2007;8:149–60. doi: 10.1038/nrm2105. [DOI] [PubMed] [Google Scholar]
- Boucrot E, Kirchhausen T. Endosomal recycling controls plasma membrane area during mitosis. Proc Natl Acad Sci U S A. 2007;104:7939–44. doi: 10.1073/pnas.0702511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S, Ahmad Y, Trinkle-Mulcahy L, Verheggen C, Cobley A, Gregor P, Bertrand E, Whitehorn M, Lamond AI. Establishment of a protein frequency library and its application in the reliable identification of specific protein interaction partners. Mol Cell Proteomics. 2010;9:861–79. doi: 10.1074/mcp.M900517-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M, Rosewell I, Carrington M, Crompton T, Jacobs MA, Kirk J, Gannon J, Hunt T. Cyclin B2-null mice develop normally and are fertile whereas cyclin B1-null mice die in utero. Proc Natl Acad Sci U S A. 1998;95:4344–9. doi: 10.1073/pnas.95.8.4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng KY, Noble ME, Skamnaki V, Brown NR, Lowe ED, Kontogiannis L, Shen K, Cole PA, Siligardi G, Johnson LN. The role of the phospho-CDK2/cyclin A recruitment site in substrate recognition. J Biol Chem. 2006;281:23167–79. doi: 10.1074/jbc.M600480200. [DOI] [PubMed] [Google Scholar]
- Chi Y, Welcker M, Hizli AA, Posakony JJ, Aebersold R, Clurman BE. Identification of CDK2 substrates in human cell lysates. Genome Biol. 2008;9:R149. doi: 10.1186/gb-2008-9-10-r149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clute P, Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat Cell Biol. 1999;1:82–7. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- Coudreuse D, Nurse P. Driving the cell cycle with a minimal CDK control network. Nature. 2010;468:1074–9. doi: 10.1038/nature09543. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cukier IH, Li Y, Lee JM. Cyclin B1/Cdk1 binds and phosphorylates Filamin A and regulates its ability to cross-link actin. FEBS Lett. 2007;581:1661–72. doi: 10.1016/j.febslet.2007.03.041. [DOI] [PubMed] [Google Scholar]
- den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol. 2001;153:121–36. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–7. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam VM, Orrechia S, Lowe M, Pardi R, Pines J. The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J Cell Biol. 2001;152:945–58. doi: 10.1083/jcb.152.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein CB, Cross FR. CLB5: a novel B cyclin from budding yeast with a role in S phase. Genes & development. 1992;6:1695. doi: 10.1101/gad.6.9.1695. [DOI] [PubMed] [Google Scholar]
- Errico A, Deshmukh K, Tanaka Y, Pozniakovsky A, Hunt T. Identification of substrates for cyclin dependent kinases. Adv Enzyme Regul. 2010;50:375–99. doi: 10.1016/j.advenzreg.2009.12.001. [DOI] [PubMed] [Google Scholar]
- Fisher DL, Nurse P. A single fission yeast mitotic cyclin B p34cdc2 kinase promotes both S-phase and mitosis in the absence of G1 cyclins. EMBO J. 1996;15:850–60. [PMC free article] [PubMed] [Google Scholar]
- Foisner R, Malecz N, Dressel N, Stadler C, Wiche G. M-phase-specific phosphorylation and structural rearrangement of the cytoplasmic cross-linking protein plectin involve p34cdc2 kinase. Mol Biol Cell. 1996;7:273–88. doi: 10.1091/mbc.7.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler VM, Adam EJ. Spectrin redistributes to the cytosol and is phosphorylated during mitosis in cultured cells. J Cell Biol. 1992;119:1559–72. doi: 10.1083/jcb.119.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavet O, Pines J. Progressive activation of CyclinB1-Cdk1 coordinates entry to mitosis. Dev Cell. 2010;18:533–43. doi: 10.1016/j.devcel.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol. 2001;153:137–48. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Lee YM, Welcker M, Swanger J, Zagozdzon A, Winer JD, Roberts JM, Kaldis P, Clurman BE, Sicinski P. Kinase-independent function of cyclin E. Mol Cell. 2007;25:127–39. doi: 10.1016/j.molcel.2006.11.029. [DOI] [PubMed] [Google Scholar]
- Geng Y, Whoriskey W, Park MY, Bronson RT, Medema RH, Li T, Weinberg RA, Sicinski P. Rescue of cyclin D1 deficiency by knockin cyclin E. Cell. 1999;97:767–77. doi: 10.1016/s0092-8674(00)80788-6. [DOI] [PubMed] [Google Scholar]
- Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. Cyclin E ablation in the mouse. Cell. 2003;114:431–43. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- Hagting A, Karlsson C, Clute P, Jackman M, Pines J. MPF localization is controlled by nuclear export. EMBO J. 1998;17:4127–38. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwell RM, Mull BB, Porter DC, Keyomarsi K. Activation of cyclin-dependent kinase 2 by full length and low molecular weight forms of cyclin E in breast cancer cells. J Biol Chem. 2004;279:12695–705. doi: 10.1074/jbc.M313407200. [DOI] [PubMed] [Google Scholar]
- Heald R, McKeon F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell. 1990;61:579–89. doi: 10.1016/0092-8674(90)90470-y. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Taylor SS. Cyclin-B1-mediated inhibition of excess separase is required for timely chromosome disjunction. J Cell Sci. 2006;119:3325–36. doi: 10.1242/jcs.03083. [DOI] [PubMed] [Google Scholar]
- Hu F, Aparicio OM. Swe1 regulation and transcriptional control restrict the activity of mitotic cyclins toward replication proteins in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2005;102:8910–5. doi: 10.1073/pnas.0406987102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11–6. doi: 10.1038/nsmb.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M, Kubota Y, den Elzen N, Hagting A, Pines J. Cyclin A- and cyclin E-Cdk complexes shuttle between the nucleus and the cytoplasm. Mol Biol Cell. 2002;13:1030–45. doi: 10.1091/mbc.01-07-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M, Lindon C, Nigg EA, Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat Cell Biol. 2003;5:143–8. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- Kalaszczynska I, Geng Y, Iino T, Mizuno S, Choi Y, Kondratiuk I, Silver DP, Wolgemuth DJ, Akashi K, Sicinski P. Cyclin a is redundant in fibroblasts but essential in hematopoietic and embryonic stem cells. Cell. 2009;138:352–65. doi: 10.1016/j.cell.2009.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Ferrell JE. Substrate competition as a source of ultrasensitivity in the inactivation of Wee1. Cell. 2007;128:1133–45. doi: 10.1016/j.cell.2007.01.039. [DOI] [PubMed] [Google Scholar]
- Kimura K, Hirano M, Kobayashi R, Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–90. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- King RW, Peters JM, Tugendreich S, Rolfe M, Hieter P, Kirschner MW. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–88. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Kittler R, Pelletier L, Heninger AK, Slabicki M, Theis M, Miroslaw L, Poser I, Lawo S, Grabner H, Kozak K, et al. Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat Cell Biol. 2007;9:1401–12. doi: 10.1038/ncb1659. [DOI] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Koff A, Cross F, Fisher A, Schumacher J, Leguellec K, Philippe M, Roberts JM. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell. 1991;66:1217–28. doi: 10.1016/0092-8674(91)90044-y. [DOI] [PubMed] [Google Scholar]
- Kozar K, Ciemerych MA, Rebel VI, Shigematsu H, Zagozdzon A, Sicinska E, Geng Y, Yu Q, Bhattacharya S, Bronson RT, et al. Mouse development and cell proliferation in the absence of D-cyclins. Cell. 2004;118:477–91. doi: 10.1016/j.cell.2004.07.025. [DOI] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG. Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell. 2010;140:349–59. doi: 10.1016/j.cell.2009.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P, Baum B. The actin cytoskeleton in spindle assembly and positioning. Trends Cell Biol. 2009;19:174–9. doi: 10.1016/j.tcb.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Lees E, Faha B, Dulic V, Reed SI, Harlow E. Cyclin E/cdk2 and cyclin A/cdk2 kinases associate with p107 and E2F in a temporally distinct manner. Genes & Development. 1992;6:1874. doi: 10.1101/gad.6.10.1874. [DOI] [PubMed] [Google Scholar]
- Lew DJ, Dulić V, Reed SI. Isolation of three novel human cyclins by rescue of G1 cyclin (Cln) function in yeast. Cell. 1991;66:1197. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- Li C, Andrake M, Dunbrack R, Enders GH. A bifunctional regulatory element in human somatic Wee1 mediates cyclin A/Cdk2 binding and Crm1-dependent nuclear export. Mol Cell Biol. 2010;30:116–30. doi: 10.1128/MCB.01876-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CJ, Vassilev A, DePamphilis ML. Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex’s largest subunit (Orc1) from binding to chromatin during mitosis. Mol Cell Biol. 2004;24:5875–86. doi: 10.1128/MCB.24.13.5875-5886.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell. 2007;26:539–51. doi: 10.1016/j.molcel.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Loog M, Morgan DO. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–8. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Mochida S, Ikeo S, Gannon J, Hunt T. Regulated activity of PP2A-B55 delta is crucial for controlling entry into and exit from mitosis in Xenopus egg extracts. EMBO J. 2009;28:2777–85. doi: 10.1038/emboj.2009.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JD, Kirk JA, Hunt T. Unmasking the S-phase-promoting potential of cyclin B1. Science. 2003;300:987–90. doi: 10.1126/science.1081418. [DOI] [PubMed] [Google Scholar]
- Murphy M, Stinnakre MG, Senamaud-Beaufort C, Winston NJ, Sweeney C, Kubelka M, Carrington M, Bréchot C, Sobczak-Thépot J. Delayed early embryonic lethality following disruption of the murine cyclin A2 gene. Nat Genet. 1997;15:83–6. doi: 10.1038/ng0197-83. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Peter M, Nakagawa J, Dorée M, Labbé JC, Nigg EA. In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell. 1990;61:591–602. doi: 10.1016/0092-8674(90)90471-p. [DOI] [PubMed] [Google Scholar]
- Petersen BO, Lukas J, Sørensen CS, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. The EMBO Journal. 1999;18:396. doi: 10.1093/emboj/18.2.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Pulido L, Diffley JF, Ponting CP. Homology explains the functional similarities of Treslin/Ticrr and Sld3. Curr Biol. 2010;20:R509–10. doi: 10.1016/j.cub.2010.05.021. [DOI] [PubMed] [Google Scholar]
- Sansam CL, Cruz NM, Danielian PS, Amsterdam A, Lau ML, Hopkins N, Lees JA. A vertebrate gene, ticrr, is an essential checkpoint and replication regulator. Genes Dev. 2010;24:183–94. doi: 10.1101/gad.1860310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M, Barbacid M. Cdk1 is sufficient to drive the mammalian cell cycle. Nature. 2007;448:811–5. doi: 10.1038/nature06046. [DOI] [PubMed] [Google Scholar]
- Schulman BA, Lindstrom DL, Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc Natl Acad Sci U S A. 1998;95:10453–8. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern B, Nurse P. A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 1996;12:345–50. [PubMed] [Google Scholar]
- Stewart MP, Helenius J, Toyoda Y, Ramanathan SP, Muller DJ, Hyman AA. Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature. 2011;469:226–30. doi: 10.1038/nature09642. [DOI] [PubMed] [Google Scholar]
- Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- Sudakin V, Ganoth D, Dahan A, Heller H, Hershko J, Luca FC, Ruderman JV, Hershko A. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Molecular biology of the cell. 1995;6:185. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–32. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. Nuclear export of cyclin B1 and its possible role in the DNA damage-induced G2 checkpoint. EMBO J. 1998;17:2728–35. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, et al. Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana. Mol Syst Biol. 2010;6:397. doi: 10.1038/msb.2010.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward GE, Kirschner MW. Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell. 1990;61:561–77. doi: 10.1016/0092-8674(90)90469-u. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Arai H, Iwasaki J, Shiina M, Ogata K, Hunter T, Osada H. Cyclin-dependent kinase (CDK) phosphorylation destabilizes somatic Wee1 via multiple pathways. Proc Natl Acad Sci U S A. 2005;102:11663–8. doi: 10.1073/pnas.0500410102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Vallenius T, Ovaska K, Westermarck J, Mäkelä TP, Hautaniemi S. Integrated network analysis platform for protein-protein interactions. Nat Methods. 2009;6:75–7. doi: 10.1038/nmeth.1282. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Goto H, Yokoyama T, Silljé H, Hanisch A, Uldschmid A, Takai Y, Oguri T, Nigg EA, Inagaki M. Phosphorylation by Cdk1 induces Plk1-mediated vimentin phosphorylation during mitosis. J Cell Biol. 2005;171:431–6. doi: 10.1083/jcb.200504091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bardes ES, Moore JD, Brennan J, Powers MA, Kornbluth S. Control of cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev. 1998;12:2131–43. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–5. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.