Abstract
A recent HIV-1 molecular epidemiology survey in Singapore identified a novel CRF01_AE/B recombinant form, which accounted for 13 (11.9%) of 109 patient samples. Peripheral blood mononuclear cell DNA from three of these 13 patients was used to generate near full-length sequences to characterize the novel CRF01_AE/B recombinant form. The three isolates had a recombinant structure composed of CRF01_AE and subtype B, and shared identical breakpoints. As the three patients were not epidemiologically linked, this recombinant form has been designated CRF51_01B. Identification of the novel recombinant forms indicates ongoing active HIV-1 transmission in Singapore.
The emergence of HIV-1 recombinant forms is a marker of ongoing, active transmission.1 A prerequisite for recombination is coinfection of a single cell, and by implication the human host, with genetic material originating from at least two HIV-1 viruses belonging to different strains. In Thailand and Malaysia, emergence of distinct CRF01_AE/B circulating recombinant forms (CRFs) was documented mainly among injection drug users.1–4
A recent HIV-1 molecular epidemiology study in Singapore was performed to determine if there was any cross-border transmission of the newly identified CRFs.5 Unexpectedly, a novel recombinant form, with subtype B in protease and gp41 and CRF01_AE in gp120, was identified in 13 (11.9%) of 109 individuals tested.
From January 2011 to March 2011, three patients from the prior study at the Singapore Communicable Disease Centre outpatient clinic, identified as infected with the novel recombinant, were contacted to participate in this follow-up study.5 Demographic information (age, gender, and ethnicity) and transmission risk factor data were obtained from clinical chart review. The National Healthcare Group ethics committee approved this study.
After obtaining written, informed consent, peripheral venous blood was drawn for peripheral blood mononuclear cell (PBMC) isolation using Ficoll-Paque Plus (GE Healthcare, Waukesha, WI). PBMC DNA was extracted using the Qiagen DNA Mini Kit (Qiagen, Valencia, CA) and used for near full-length proviral amplification using a hot start nested PCR method. As previously described, approximately 9 kb of the HIV-1 genome was amplified using primers MSF12b/OFMR1 for the first-round amplification and primers F2NST/UNINEF7 for the second-round amplification.6,7 Amplified products were sequenced using the ABI 3130 automated sequencer (Applied Biosystems Inc, Foster City, CA). Sequences were analyzed, edited, and assembled with Sequencher 4.6 (Gene Codes Corporation, Ann Arbor, MI). Phylogenetic analysis was performed using MacGDE and MEGA3.8–10 Reference sequences were downloaded from the Los Alamos HIV Sequence Database.11 Recombination breakpoints were determined using SimPlot v.3.5.1, RIP, and jpHMM.12–14
All three patients were Singapore citizens of Chinese ethnicity (Table 1). The ages of the patients were 23, 36, and 48 years. The transmission risk factor reported was homosexual in two and bisexual in one. Two of the three patients reported sexual exposures only in Singapore. Information on the location of sexual exposure for one of the patients was unavailable. There was no obvious epidemiologic link between the study participants.
Table 1.
Demographic Characteristics for the Three Study Patients
| 11SG.HM021 | 11SG.HM062 | 11SG.HM091 | |
|---|---|---|---|
| Age (years) | 23 | 48 | 36 |
| Gender | Male | Male | Male |
| Ethnicity | Chinese | Chinese | Chinese |
| Transmission risk factor | Homosexual | Bisexual | Homosexual |
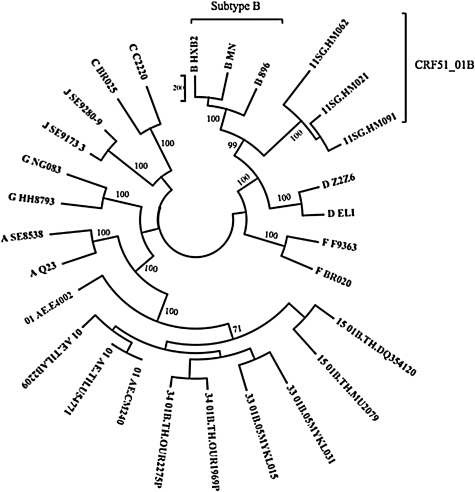
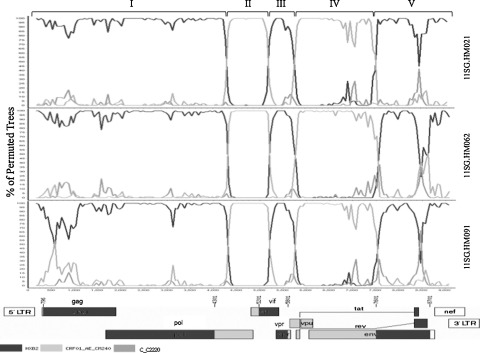
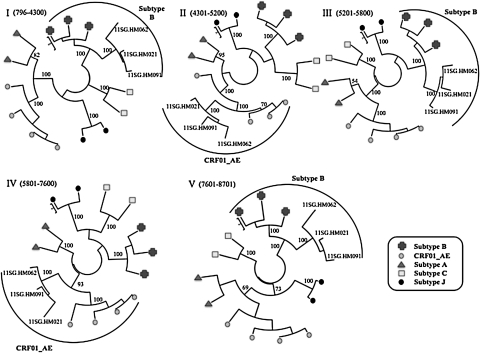
Using phylogenetic analysis, near full-length sequences of the three patients formed a monophyletic branch supported by a bootstrap value of 100%, separate from global subtypes and known CRFs in Southeast Asia (Fig. 1). SimPlot analysis revealed that the breakpoints corresponded to HXB2 nucleotide positions 4300, 5200, 5800, and 7600 (Fig. 2). The breakpoint were identical in all three isolates. Similar results were obtained using RIP and jpHMM. Phylogenetic analysis of the subgenomic fragments confirmed the four breakpoints identified (Fig. 3). The genomic fragments 796 to 4300, 4301 to 5200, 5201 to 5800, 5801 to 7600, and 7601 to 8701 branched with reference subtype B, CRF01_AE, subtype B, CRF01_AE, and subtype B, respectively.
FIG. 1.
Maximum parsimony tree representing the relationship of the three near full-length sequences from Singapore with reference sequences. The scale denotes the number of informative site mutations. Bootstrap analysis was performed with 100 replicates. The taxon name of the study sequences represents the year of sample collection (11) followed by country (SG) and unique study identifier (e.g., HM021). The bootstrap value at the node of the newly sequenced strains is shown.
FIG. 2.
Bootscan analysis of the three Singapore isolates. The subtype reference strains used were Subtype B, HXB2 (black), CRF01_AE, CM240 (light gray), and subtype C, C2220 (gray). A genome alignment of 8701 nts in length, based on HXB2, was used following end trimming. The bootscan window was 350 bp with a step of 50 bp. The recombinant gene mosaic structure is shown at the bottom of the figure with nucleotide positions corresponding to the reference HXB2 sequence.
FIG. 3.
Phylogenetic trees for genome segments of the three Singapore isolates. Using methods described in Fig. 1, phylogenetic analysis was performed based on recombination breakpoints shown in Fig. 2. The genome segments correspond to the following HXB2 positions: (I) nts 796 to 4300; (II) nts 4301 to 5200; (III) nts 5201 to 5800; (IV) nts 5801 to 7600; and (V) nts 7601 to 8701. The bootstrap value at the node of the newly sequenced strains is shown.
In genomic regions I, III, and V, the Singapore strains form a cluster distinct from the reference subtype B strains, supported by strong bootstrap values. A potential reason for this would be that the subtype B progenitor of the CRF51_01B recombinant had diverged significantly from the reference B strains used. However, full-length subtype B sequences from Singapore to test this hypothesis are not available. Another potential reason could be the presence of CRF01_AE sequences within the genomic fragments as non-B segments less than 100 bp would potentially be undetected as a separate recombinant fragment. As the new strains were phylogenetically more similar to “western” B than B-prime reference sequences, "western" B sequences were used for the genomic region trees (data not shown).
This study confirmed the presence of a novel circulating recombinant form, named CRF51_01B, in Singapore.15 The breakpoints identified in 11SG.HM021, 11SG.HM062, and 11SG.HM091 are identical among the three samples but were distinct from the four CRF01_AE/B CRFs described to date in Thailand and Malaysia (Fig. 1). As CRF51_01B has only been described in Singapore and accounted for 11.9% of HIV-1 infections in our previous molecular epidemiology survey, it is likely that CRF51_01B was generated within Singapore and is currently established as a circulating strain. In our prior study, this CRF had a higher prevalence among homosexuals and bisexuals compared to subtype B and CRF01_AE.5 Continual monitoring of the molecular epidemiology of HIV-1 in Singapore will provide vital insight concerning local and transnational transmission.
Acknowledgments
A Singapore National Medical Research Training Fellowship grant provided salary support for O.T. Ng. We acknowledge the physicians, staff, and patients of the outpatient service at the Communicable Disease Centre, Singapore, who made this study possible. Additional support was provided by the Division of Intramural Research, NIAID, NIH. We thank Dr. Mark I.C. Chen, Mr. Ridzwan Abdullah, and Ms. Tan Pei Ling for help with patient recruitment and sample collection. O.T.N. and L.M.E. contributed equally to this work. The sequences analyzed in this study have been deposited in GenBank under accession numbers JN029801, JN029802, and JN029803.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Tovanabutra S. Watanaveeradej V. Viputtikul K, et al. A new circulating recombinant form, CRF15_01B, reinforces the linkage between IDU and heterosexual epidemics in Thailand. AIDS Res Hum Retroviruses. 2003;19:561–567. doi: 10.1089/088922203322230923. [DOI] [PubMed] [Google Scholar]
- 2.Tee KK. Li XJ. Nohtomi K. Ng KP. Kamarulzaman A. Takebe Y. Identification of a novel circulating recombinant form (CRF33_01B) disseminating widely among various risk populations in Kuala Lumpur, Malaysia. J Acquir Immune Defic Syndr. 2006;43:523–529. doi: 10.1097/01.qai.0000242451.74779.a7. [DOI] [PubMed] [Google Scholar]
- 3.Tovanabutra S. Kijak GH. Beyrer C, et al. Identification of CRF34_01B, a second circulating recombinant form unrelated to and more complex than CRF15_01B, among injecting drug users in northern Thailand. AIDS Res Hum Retroviruses. 2007;23:829–833. doi: 10.1089/aid.2006.0300. [DOI] [PubMed] [Google Scholar]
- 4.Li Y. Tee KK. Liao H, et al. Identification of a novel second-generation circulating recombinant form (CRF48_01B) in Malaysia: A descendant of the previously identified CRF33_01B. J Acquir Immune Defic Syndr. 2010;54:129–136. doi: 10.1097/QAI.0b013e3181d82ce5. [DOI] [PubMed] [Google Scholar]
- 5.Ng OT. Munshaw S. Lamers SL, et al. Molecular epidemiology of HIV type 1 in Singapore and identification of novel CRF01_AE/B recombinant forms. AIDS Res Hum Retroviruses. 2011. Feb 14, [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 6.Salminen MO. Carr JK. Burke DS. McCutchan FE. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses. 1995;11:1423–1425. doi: 10.1089/aid.1995.11.1423. [DOI] [PubMed] [Google Scholar]
- 7.Carr JK. Salminen MO. Koch C, et al. Full-length sequence and mosaic structure of a human immunodeficiency virus type 1 isolate from Thailand. J Virol. 1996;70:5935–5943. doi: 10.1128/jvi.70.9.5935-5943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacDGE: Genetic Data Environment for MacOSX. [May 13;2011 ]. http://macgde.bio.cmich.edu/ http://macgde.bio.cmich.edu/
- 9.Kumar S. Tamura K. Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 10.Saitou N. Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 11.HIV Sequence Database. [May 13;2011 ]. http://www.hiv/lanl.gov/content/sequence/HIV/mainpage.html http://www.hiv/lanl.gov/content/sequence/HIV/mainpage.html
- 12.jpHMM. [Jul 11;2011 ]. http://jphmm.gobics.de/ http://jphmm.gobics.de/
- 13.Lole KS. Bollinger RC. Paranjape RS, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.RIP. [Jul 11;2011 ]. http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html http://www.hiv.lanl.gov/content/sequence/RIP/RIP.html
- 15.Robertson DL. Anderson JP. Bradac JA, et al. HIV-1 nomenclature proposal. Science. 2000;288:55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]