Abstract
Throughout the world, infants and children with HIV-1 infection are increasingly surviving into adolescence and adulthood. As HIV Nef is an important determinant of the pathogenic potential of the virus, we examined nef alleles in a cohort of extreme long-term survivors of HIV infection (average age of 16.6 years) to determine if Nef defects might have contributed to patient survival. HIV nef gene sequences were amplified for phylogenetic analysis from 15 adolescents and adults infected by mother-to-child transmission (n=10) or by blood transfusion (n=5). Functional analysis was performed by inserting patient-derived nef sequences into an HIV-derived vector that permits simultaneous evaluation of the impact of the Nef protein on MHC-I and CD4 cell surface expression. We found evidence of extensive nef gene diversity, including changes in known functional domains involved in the downregulation of cell surface MHC-I and CD4. Only 3 of 15 individuals (20%) had nef alleles with a loss of the ability to downregulate either CD4 or MHC-I. Survival into adulthood with HIV infection acquired in infancy is not uniformly linked to loss of function in nef. The Nef protein remains a potential target for immunization or pharmacologic intervention.
Introduction
In both resource-rich and developing areas of the world, untreated pediatric HIV-1 infection is characterized by early onset of disease and poor survival. Before antiretroviral medications were available, most infected children in the United States developed symptomatic disease within 1 year and 50% progressed to AIDS by 5 years of age.1,2 In early reports, the median and mean ages of survival for perinatally infected children in the United States and Europe were estimated to be 8.0 and 9.4 years, respectively.2,3 Higher early mortality rates have been described for infants in countries hardest hit by the AIDS pandemic; using individual data from seven randomized mother-to-child transmission (MTCT) intervention trials, Newell et al. estimated overall mortality rates in African children born to HIV-infected mothers to be as high as 35% at 1 year and more than 50% at 2 years of age.4
A variety of viral, host, and treatment factors determine the tempo of disease progression in children and adults. Clearly, antiretroviral therapy and improved strategies for the prevention and treatment of opportunistic infections have dramatically reduced morbidity and mortality in HIV-1-infected children, and survival into adulthood is now the norm in the United States.5,6 However, in some cases, mutations in the HIV-1 nef gene have been associated with marked delay in the natural progression of disease in untreated individuals. The nef gene encodes the 27-kDa cytoplasmic Nef protein with several effects that enhance the pathogenic potential of HIV. Among these, Nef reduces the surface expression of MHC-I A and B molecules of infected cells, allowing escape from detection by cytotoxic T cells.7,8 Nef also binds to both CD4 and components of the intracellular trafficking machinery, inducing CD4 internalization from plasma membranes and lysosomal degradation. This may impair CD4 T cell function, limit repeated HIV infection of individual cells, and facilitate the egress of virions. Furthermore, Nef appears to reduce expression of CD28 and CXCR4 and to augment the infectivity of viral particles, enhancing viral entry and replication.9
Specific structural features and motifs have been identified in Nef that are responsible its diverse biochemical activities. N-terminal myristoylation promotes Nef protein interactions with cellular membranes and is essential to its activity. A polyproline (PXXPXR) motif and a pair of downstream cationic residues (RR) interact with protein kinases involved in cellular activation, a function that may be required for viral replication in resting cells.10,11 A three amino acid (FPD) motif is responsible for dimerization, an event required for downregulation of both MHC-I and CD4. Beyond dimerization, other conserved motifs have been identified that must be maintained for downregulation of CD4 and MHC-I. A series of acidic residues (EEEE) binds to PACS-1, a molecule involved in endosome to Golgi trafficking, potentially leading to the retention of MHC -I molecules in the trans-Golgi apparatus.9 The pathway for CD4 downregulation involves Nef interactions with the CD4 through a tryptophan–leucine (WL) motif, simultaneous binding at two nearby leucine residues to the AP-1 and AP-2 proteins of the adaptor complex in clathrin-coated pits, and with the V-ATPase proton pump that ultimately results in CD4 endocytosis.7
How these biochemical activities of Nef actually affect pathogenesis is not fully clear, but Nef is clearly a determinant of lentivirus virulence. This was first shown in the SIV-rhesus macaque model; full gene deletions prevented the maintenance of high viral loads12 but nef mutants engineered with a premature stop signal quickly reverted back to wild type, suggesting a strong selection pressure for functional forms of Nef protein. In a later experiment, macaques were infected with SIV containing deletions that selectively ablated MHC-I downregulation; the virus rapidly evolved to develop new motifs for this function, demonstrating its importance in vivo.13
The impact of nef point mutations on the outcome of disease in children has been examined in a small number of reports.14,15 In one, nef mutations were identified in eight long-term survivors of perinatal infection. Virus from two of these patients had large deletions; one of these children was asymptomatic with undetectable plasma HIV RNA with no antiretroviral therapy at 10 years of age, and the other had only moderately symptomatic disease at 12 years of age. These reports involved patients likely infected with subtype B HIV-1, but one recent report16 examined nef sequences from 24 children from South Africa with slow progression of disease (median age 6.7 years), extending these observations to pediatric HIV-1 infection with subtype C virus. In this study, missense mutations and small deletions in the amino terminus of the nef gene were identified that would likely interfere with the biochemical activities of Nef, but functional studies were not performed.16 In a recent genetic analysis involving six children ranging in age from 7 to 10 years old, two South African children infected with subtype C virus were also found to have mutations in nef predicted to alter function.17
We hypothesized that some HIV-infected children survived from infancy to adolescence and adulthood because they were initially infected with nef-defective HIV variants, slowing the progression of disease until the availability of potent antiretroviral therapy in the late 1990s. A related possibility was considered, that the relative immaturity of cellular immune function in early childhood would render dispensable Nef functions required for immune evasion, allowing HIV to acquire mutations that impair its function. To examine these possibilities, we examined viral nef gene sequences and their function in a cohort of adolescents and adults with HIV infection acquired by perinatal mother-to-child transmission and transfusion in early childhood.
Materials and Methods
Study subjects
We studied a cohort of 15 adolescents who were enrolled at Children's Hospital Los Angeles (CHLA) between 2001 and 2003, including 13 described in a prior study of T cell parameters.18 Eight of these 13 were subsequently enrolled in a study of the impact of HIV replication on thymopoiesis19 and followed into early adulthood (19.2 to 21.4 years, median 21.1 years). These studies were approved by Institutional Review Boards of CHLA and the University of California, Los Angeles (UCLA) and informed consent was obtained from study participants or their parent or guardians. Information on CD4 lymphocyte counts and HIV-1 viral load and other demographic variables (age, gender, mode of infection, treatment histories) were collected by chart review (Table 1). Four of the individuals studied were infected by transfusions given during neonatal care; patient 1-02 was only known to have received a transfusion in the first 2 years of life. Patients 1-05 and 1-06 are identical twins who acquired HIV-1 through the same blood transfusion; their clinical course has been extensively described.20 The remaining youths were documented to have been born to HIV-infected women and were all diagnosed before 4 years of age by conventional methods; none of these had other known risk factors for HIV infection. The majority of the adolescents and adults studied were receiving therapy consisting of regimens containing three or four antiretroviral agents at the time of blood sampling.
Table 1.
Clinical Characteristics of the Study Population
| Characteristic | Total N=15 |
|---|---|
| Age at entry | 16.3 (2.2) |
| Gender (F/M) | 9F/6M |
| CDC classification | |
| N | 1 |
| A | 0 |
| B | 5 |
| C | 9 |
| Log plasma viral load (copies/ml) | 3.9 (1.0) |
| Race/ethnicity | |
| Black | 6 |
| Hispanic/Latino | 6 |
| White | 3 |
| CD4+ (cells/μl) | 349 (207) |
| CD4% | 17.9 (16.0) |
| CD8+ (cells/μl) | 778 (807) |
| CD8% | 49.3 (54) |
Values shown are the mean with standard deviation in parentheses.
Sequence analysis of patient nef alleles
HIV nef alleles were amplified by PCR from peripheral blood mononuclear cell (PBMC) DNA at limiting dilution, determined by serial 2-fold dilutions to the point at which less than 50% of reactions generated a product.
In all, 124 nef sequences, each 509 nucleotides long (amino acids 31–199, HXB2 numbering), were aligned with NL4-3 and Consensus B nef from the LANL HIV database using CLUSTAL W. The alignment was manually edited in the proper reading frame using BioEdit. The edited alignment was used to reconstruct the phylogeny using both neighbor-joining and maximum likelihood methods as implemented in PHYLIP 3.6. The neighbor-joining tree was statistically evaluated using 1000 bootstrap replicates.
Production of pseudotyped HIV vectors to express patient-derived nef alleles
The ability of patient-derived nef alleles to downregulate MHC-I and CD4 was analyzed by expressing the Nef proteins from an HIV vector, as recently described.21 In brief, patient nef alleles were amplified in bulk from cellular DNA using primers containing XbaI and BspEI tails, digested with these enzymes, and subsequently cloned into the plasmid AA1305#18, replacing the original nef allele. This plasmid contains an HIV genome with deletions that abrogate expression of the vpr, vpu, and envelope coding domains; the cDNA for the murine CD24 (mCD24)/heat-stable antigen (HSA) has been inserted in the place of the vpr reading frame. To generate infectious virus containing patient nef alleles, single-round infectious pseudotyped virus was produced by cotransfecting 293T cells with the whole-genome plasmid AA1305#18 and a plasmid encoding the envelope glycoprotein from VSV (VSV-G) using FuGENEHD (Roche). Supernatants from these cells were collected on days 2 and 3 posttransfection. Virus production was quantified by p24 antigen ELISA (PerkinElmer, Waltham, MA). Transfections typically produced greater than 200 ng/ml of p24. Nef gene sequences from recombinant viruses used for functional testing clustered with the sequences isolated directly from plasma from the same individual.
Functional analysis of nef alleles
The ability of Nef proteins to downregulate HLA A*02 on infected cells was assessed as previously reported.21 Briefly, the T1 lymphocyte line was infected at 37°C for 4 h with ∼0.5 ml of pseudotyped virus stocks containing 200–500 ng p24 antigen. Parallel infections were performed with viruses carrying either wild-type HIVNL4-3 nef (positive control for both CD4 and MHC-I downregulation), M20A nef (positive control for CD4 downregulation and negative control for MHC-I downregulation), LL>AA nef (negative control for CD4 downregulation and positive control for MHC-I downregulation), and ΔNef (negative control for both CD4 and MHC-I downregulation). On day 4 postinfection 2×105 cells were stained with antimurine CD24/HSA-FITC antihuman CD4-APC (BD Pharmingen) and antihuman HLA A*02-PE, washed twice, and fixed with 1% paraformaldehyde. At least 5×104 live cells were counted using a FACSCalibur flow cytometer, and data were analyzed using FlowJo software (Tree Star, Ashland, OR). Maximum levels of HLA A*02 and CD4 were determined using the ΔNef mutant. The relative efficiency of HLA A*02 or CD4 downregulation function of patient nef alleles was calculated based on MFI (median fluorescence intensity), as follows: Downregulation=(MFIΔ Nef−MFIpatient)/(MFIΔ Nef - MFINL4-3). By definition, the efficiencies of ΔNef and wild type NL4-3 nef are 0 and 100%, respectively.
Results
Patients
Fifteen HIV-infected adolescents were enrolled in this study, with a median age of 16.5 years at study (range: 13.3 to 20.2). Using PCR methods described by others,22 we determined that none of these youths was a carrier of the Δ32-base-pair deletion within the coding region of CCR5 (data not shown). All but one had developed moderately symptomatic HIV infection or AIDS by the time of enrollment (Table 1). At the time of collection of specimens, 13 were receiving antiretroviral therapy. Patient 1-01 had never received antiretroviral therapy, and was the only nonprogressor enrolled. Patient 1-03 had been treated unsuccessfully with several antiretroviral regimens in the past and had recently had treatment suspended due to a hospitalization for pancreatitis. Two individuals had undetectable HIV plasma viral loads while the other 13 had viral loads ranging from 509 to 1,046,000 copies per ml.
Sequence analysis of patient nef alleles
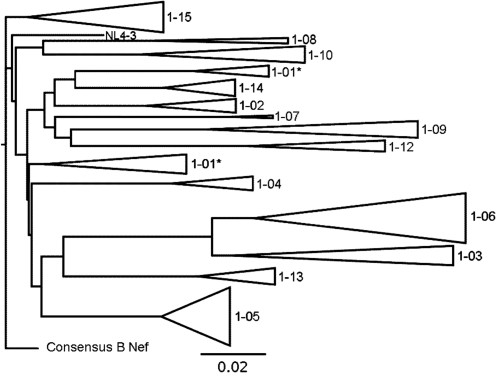
Phylogenetic trees were constructing using 124 nef sequences from the 15 subjects in order to determine their relationship to each other, as well as to NL4-3 and the Clade B consensus nef. An average of eight sequences was obtained from each subject (median=7), aligned with NL4-3 Nef and Clade B consensus nef, then used to construct both a neighbor-joining (NJ) (see Fig. 1) and a maximum likelihood (ML) tree. The NJ tree was evaluated with 1000 bootstrap replicates. Sequences from individual patients clustered together with high (>95%) support for all subjects except subject 1-01. None of the patient sequences clustered with the laboratory strain HIVNL4-3. Subject sequences were approximately 3–8% divergent from Clade B consensus nef, as is expected for typical Clade B isolates. There was approximately 1–3% diversity within the cluster of sequences from each subject, as expected for typical HIV quasispecies. Similar results were obtained with maximum likelihood trees (data not shown). Overall, the results of the phylogenetic analysis indicate that the sequences isolated from the plasma of the study subjects and cloned for functional testing are unique Clade B quasispecies, without evidence of laboratory contamination.
FIG. 1.
Analysis of nef gene alleles from HIV-1-infected youths. A phylogenetic tree is shown representing 124 sequences from 15 subjects. An average of eight sequences were obtained from each subject, aligned together with NL4-3 and Clade B consensus nef, and then used to construct a neighbor-joining tree. The tree was evaluated with 1000 bootstrap replicates. Sequences from individual patients clustered together with high (>95%) support for all subjects except subject 1-01. Sequences isolated from recombinant viruses used for functional testing are marked with an asterisk; all others are sequences isolated directly from plasma.
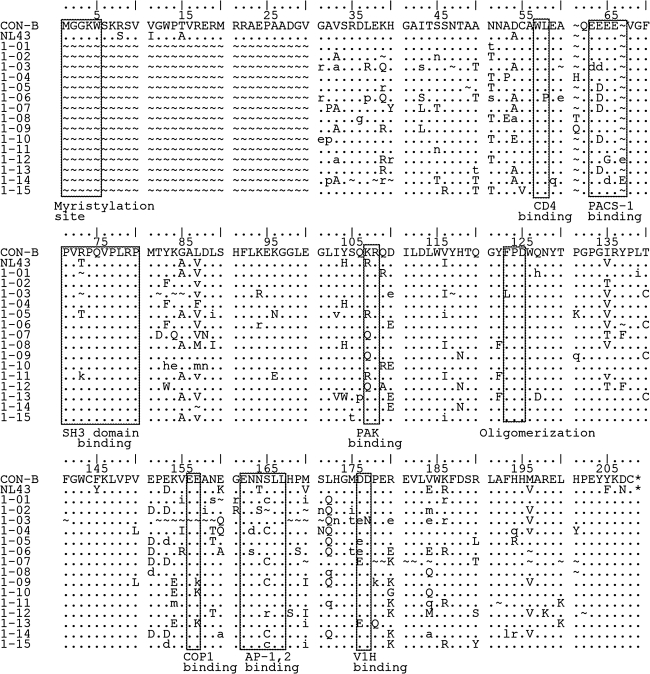
There was substantial variability in the predicted amino acid sequences of the nef alleles in our study participants (Fig. 2). Patient 1-03 contained a deletion of nef predicted to remove the COP1 binding domain mediating retention in endosomes, as well as mutations in the V-ATPase binding domain required for CD4 endocytosis. The same patient had a substitution (F to L) in the FPD dimerization motif required for downregulation of both MHC-I and CD4. As described previously, Patients 1-05 and 1-06 were identical twins who acquired HIV in the first month of life via a blood transfusion from a common donor.20 Viral sequences from patient 1-06 had a WL to WP substitution, potentially disrupting CD4 internalization. This substitution was not present in virus from the twin designated 1-05, suggesting functional differences in the two nef alleles. Nef sequences from patient 1-09 predicted a two amino acid deletion in the polyproline region involved in MHC-I downregulation.
FIG. 2.
Comparison of amino acid sequence alignment of consensus Nef sequences from patients to wild-type consensus B nef. The boxes highlight residues implicated in Nef interactions with cellular proteins.
Functional studies of patient-derived nef alleles
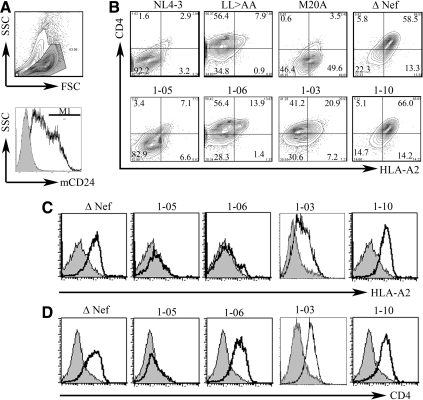
Reporter viruses representing the viral quasispecies were generated to examine the ability of patient-derived nef alleles to downregulate MHC-I and CD4 expression, using flow cytometry to measure these molecules on the surface of cells expressing a reporter virus that encoded the murine CD24 antigen (Fig. 3).21 Controls included wild-type HIVNL4-3 and three mutants of this laboratory strain: a nef null-mutant “Δnef” and two variants with nef genes containing mutations previously shown to selectively interfere with either MHC-I (M20A) or CD4 downregulation (LL>AA).21
FIG. 3.
Downregulation of cell surface MHC-I and CD4 molecules by patient-derived Nef proteins. T1 cells were infected by pseudotyped HIV vectors expressing nef sequences derived from the study subjects and downregulation of cell surface HLA A*02 and CD4 molecules was analyzed using flow cytometry. (A) In the upper panel, viable T1 cells were selected by examining forward and side scatter properties. In the lower panel, infected cells (M1 region) were detected by expression of mCD24 marker. The shaded area represents mock-infected T1 cells in a representative histogram. (B) Representative plots demonstrate HLA- A*02 (horizontal axis) and CD4 (vertical axis) downregulation in T1 cells (gated) infected by vectors expressing Nef from wild-type HIV (positive control) and four study subjects: the twins, 1-05 and 1-06, 1-03, and 1-10. Negative controls include LL>AA mutant (loss of CD4 downregulation), M20A mutant (loss of MHC-I downregulation), and a ΔNef mutant. Numbers represent percentages of cells in each quadrant of total infected T1 cells. (C, D) Histograms demonstrate functional loss of both HLA A*02 (C) and CD4 (D) downregulation by Nef from 1-03 and 1–10, functional loss of CD4 downregulation (D) in Nef from 1-06, and retained functions of Nef from 1-05. These results are representative of at least three independent experiments for each study subject.
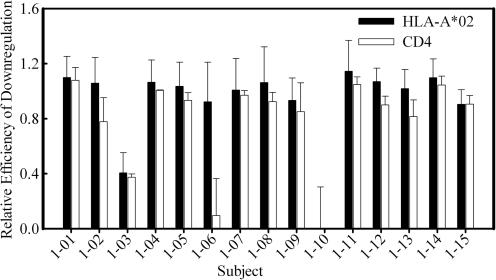
Although we noted an abundance of polymorphisms and deletions in patient nef alleles, only three patients were found to harbor HIV nef sequences that altered or ablated Nef function (Fig. 3). The reporter virus containing 1–10 nef alleles had no ability to downregulate either MHC-1 or CD4, even though 1–10 nef sequences contained no mutations in known functional motifs. In contrast, nef alleles from two adolescents (1-03 and 1-06) had mutations that interfered with CD4 downregulation. In the case of 1-03, MHC-I downregulation also appeared to be partially lost, a finding consistent with the presence of deletions in the COP1-binding domain. Nef from patient 1-06 did not reduce CD4 cell surface expression, likely because of disruption of the WL dipeptide motif noted above (Figs. 3 and 4). The ability to downregulate both MHC-I and CD4 was retained in the rest of study subjects (Fig. 4). To our surprise, nef from patient 1-09 retained wild-type function even though it contained several mutations in functional motifs.
FIG. 4.
Summary of functional analyses of patient Nef mutants. The bar graph demonstrates relative efficiencies of HLA-A*02 (black bars) and CD4 (open bars) downregulation by Nef from all 15 subjects in the study. Reference Nef strains are ΔNef and wild-type NL4-3, and relative efficiencies of ΔNef and NL4-3 are set at 0 and 1, respectively. Bars represent means±standard deviations of three to five independent measurements.
Discussion
In this cohort of extremely long-term survivors of perinatal HIV-1 infection, 124 distinct HIV nef sequences from 15 patient samples were analyzed. Deletions and truncations were uncommon, comprising less than 20% of alleles in a given patient sample. Since the majority of these nef gene sequences encoded a full-length Nef protein, amino acid sequences were analyzed for smaller mutations within functional motifs for MHC-I downregulation, CD4 downregulation, and cellular activation. Comparison of the consensus of the sequences from each patient to that of the consensus B HIV-1 Nef protein revealed that a subset of the cohort was infected with HIV with mutations in key functional residues of the viral protein. However, sequence analysis did not accurately predict the functional properties of the nef alleles. Proviral DNA from only 3 of the 15 patient specimens contained mutations that impaired Nef-mediated downregulation of CD4 or MHC-I molecules, and the nef genes from one of those (1–10) had no mutations in key functional residues.
In this study, we found that the Nef sequences found in subject 1-06 lost the ability to downregulate CD4 at some point. The twin subjects 1-05 and 1-06 were initially infected by a common source, infusion of a contaminated blood unit, and experienced a nearly identical clinical course, including the development of AIDS-defining illnesses and severe CD4 lymphocytopenia.20 At the time of study, both had low CD4 T cell concentrations (340 and 297 cells/μl, respectively) and had recently begun new antiretroviral regimens. HIV sequence analysis performed during our previous study of these twins20 revealed that in 1995 a mutation was already present in the HIV Nef sequences from 1-06 that explains the loss of CD4 downregulation, suggesting that this function of Nef may be dispensable once severe CD4 depletion has occurred. Similarly, HIV Nef in subject 1-03 had lost CD4 downregulation and partially MHC-I downregulation. This individual was 20 years old at enrollment, with severe CD4 depletion (<10 cells/μl) and ongoing plasma viral load despite multidrug antiretroviral therapy. By contrast, HIV Nef in subject 1-10 (17 years of age at entry with CD4+ T cells of 691 cells/μl) had completely lost both CD4 and MHC class I downregulation despite the absence of clearly deleterious mutations in the nef gene. This phenotype has been described on occasion in adults,23 and indicates that loss of Nef function may occur simply by the accumulation of multiple mutations.
Our study is subject to several limitations. Clearly, the small sample size makes it difficult to extrapolate these data to all HIV-infected adolescents. However, the medical histories of these participants in this study are typical of those born prior to the introduction of antiretroviral therapy in 1987: most had survived AIDS-defining illnesses in the past and had already failed one or more antiretroviral regimens. It is also important to note that our functional analysis focused on two of the best described functions of Nef, but did not examine the potential impact of nef sequence polymorphisms on the in vitro replication of HIV. Thus, our focus on the loss of Nef-mediated downregulation of MHC-I or CD4 may underestimate the true impact of Nef polymorphisms on long-term survival of perinatally infected individuals.
Nonetheless, the study of the retention of key HIV Nef residues in long-term survivors, especially those surviving perinatal infection, reinforces the intrinsic importance of this viral protein. In addition, retention of specific amino acid residues or motifs needed for Nef function suggests the potential importance of Nef as a vaccine target, as the selective pressure induced by a vaccine could impair virus replication, slowing disease progression following infection. Furthermore, retention of Nef activity in most of these adolescents and young adults suggests that targeting Nef with small molecule inhibitors could be a potentially useful therapeutic strategy.
Acknowledgments
Financial support for this project was provided by grants from NIAID to P.K. (AI051996), O.Y. (AI051970), and M.L.(AI083083). Work by P.K. was also supported by an Elizabeth Glaser Pediatric AIDS Foundation Scientist Award. Flow cytometry and HIV p24 assays were performed using core laboratory facilities supported by the UCLA Center for AIDS Research (CFAR)(NIAID AI028697) and by the UCLA AIDS Institute and the UCLA Council of Bioscience Resources. A subset of sequence analysis data was presented in preliminary form at a meeting of the Society for Pediatric Research/American Pediatric Society, Seattle, WA, May 2003.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Bamji M. Thea DM. Weedon J, et al. Prospective study of human immunodeficiency virus 1-related disease among 512 infants born to infected women in New York City. The New York City Perinatal HIV Transmission Collaborative Study Group. Pediatr Infect Dis J. 1996;15:891–898. doi: 10.1097/00006454-199610000-00012. [DOI] [PubMed] [Google Scholar]
- 2.Barnhart HX. Caldwell MB. Thomas P, et al. Natural history of human immunodeficiency virus disease in perinatally infected children: An analysis from the Pediatric Spectrum of Disease Project. Pediatrics. 1996;97:710–716. [PubMed] [Google Scholar]
- 3.Galli L. de Martino M. Tovo PA. Gabiano C. Zappa M. Predictive value of the HIV paediatric classification system for the long-term course of perinatally infected children. Int J Epidemiol. 2000;29:573–578. doi: 10.1093/intjepid/29.3.573. [DOI] [PubMed] [Google Scholar]
- 4.Newell ML. Coovadia H. Cortina-Borja M. Rollins N. Gaillard P. Dabis F. Mortality of infected and uninfected infants born to HIV-infected mothers in Africa: A pooled analysis. Lancet. 2004;364:1236–1243. doi: 10.1016/S0140-6736(04)17140-7. [DOI] [PubMed] [Google Scholar]
- 5.Brady MT. Oleske JM. Williams PL, et al. Declines in mortality rates and changes in causes of death in HIV-1-infected children during the HAART era. J Acquir Immune Defic Syndr. 2010;53:86–94. doi: 10.1097/QAI.0b013e3181b9869f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gortmaker SL. Hughes M. Cervia J, et al. Effect of combination therapy including protease inhibitors on mortality among children and adolescents infected with HIV-1. N Engl J Med. 2001;345:1522–1528. doi: 10.1056/NEJMoa011157. [DOI] [PubMed] [Google Scholar]
- 7.Geyer M. Fackler OT. Peterlin BM. Structure–function relationships in HIV-1 Nef. EMBO Rep. 2001;2:580–585. doi: 10.1093/embo-reports/kve141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laguette N. Bregnard C. Benichou S. Basmaciogullari S. Human immunodeficiency virus (HIV) type-1, HIV-2 and simian immunodeficiency virus Nef proteins. Mol Aspects Med. 2010;31:418–433. doi: 10.1016/j.mam.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Arhel NJ. Kirchhoff F. Implications of Nef: host cell interactions in viral persistence and progression to AIDS. Curr Top Microbiol Immunol. 2009;339:147–175. doi: 10.1007/978-3-642-02175-6_8. [DOI] [PubMed] [Google Scholar]
- 10.Greenberg M. DeTulleo L. Rapoport I. Skowronski J. Kirchhausen T. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr Biol. 1998;8:1239–1242. doi: 10.1016/s0960-9822(07)00518-0. [DOI] [PubMed] [Google Scholar]
- 11.Mangasarian A. Piguet V. Wang JK. Chen YL. Trono D. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J Virol. 1999;73:1964–1973. doi: 10.1128/jvi.73.3.1964-1973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kestler HW., 3rd Ringler DJ. Mori K, et al. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 13.Swigut T. Alexander L. Morgan J, et al. Impact of Nef-mediated downregulation of major histocompatibility complex class I on immune response to simian immunodeficiency virus. J Virol. 2004;78:13335–13344. doi: 10.1128/JVI.78.23.13335-13344.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geffin R. Wolf D. Muller R, et al. Functional and structural defects in HIV type 1 nef genes derived from pediatric long-term survivors. AIDS Res Hum Retroviruses. 2000;16:1855–1868. doi: 10.1089/08892220050195810. [DOI] [PubMed] [Google Scholar]
- 15.Rousseau C. Abrams E. Lee M. Urbano R. King MC. Long terminal repeat and nef gene variants of human immunodeficiency virus type 1 in perinatally infected long-term survivors and rapid progressors. AIDS Res Hum Retroviruses. 1997;13:1611–1623. doi: 10.1089/aid.1997.13.1611. [DOI] [PubMed] [Google Scholar]
- 16.Walker PR. Ketunuti M. Choge IA, et al. Polymorphisms in Nef associated with different clinical outcomes in HIV type 1 subtype C-infected children. AIDS Res Hum Retroviruses. 2007;23:204–215. doi: 10.1089/aid.2006.0080. [DOI] [PubMed] [Google Scholar]
- 17.Tzitzivacos DB. Tiemessen CT. Stevens WS. Papathanasopoulos MA. Viral genetic determinants of nonprogressive HIV type 1 subtype C infection in antiretroviral drug-naive children. AIDS Res Hum Retroviruses. 2009;25:1141–1148. doi: 10.1089/aid.2009.0080. [DOI] [PubMed] [Google Scholar]
- 18.Pham T. Belzer M. Church JA, et al. Assessment of thymic activity in human immunodeficiency virus-negative and -positive adolescents by real-time PCR quantitation of T-cell receptor rearrangement excision circles. Clin Diagn Lab Immunol. 2003;10:323–328. doi: 10.1128/CDLI.10.2.323-328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JC. Boechat MI. Belzer M, et al. Thymic volume, T-cell populations, and parameters of thymopoiesis in adolescent and adult survivors of HIV infection acquired in infancy. AIDS. 2006;20:667–674. doi: 10.1097/01.aids.0000216366.46195.81. [DOI] [PubMed] [Google Scholar]
- 20.Yang OO. Church J. Kitchen CM, et al. Genetic and stochastic influences on the interaction of human immunodeficiency virus type 1 and cytotoxic T lymphocytes in identical twins. J Virol. 2005;79:15368–15375. doi: 10.1128/JVI.79.24.15368-15375.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ali A. Realegeno S. Yang OO. Lewis MJ. Simultaneous assessment of CD4, MHC-I downregulation by Nef primary isolates in the context of infection. J Virol Methods. 2009;161:297–304. doi: 10.1016/j.jviromet.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buseyne F. Janvier G. Teglas JP, et al. Impact of heterozygosity for the chemokine receptor CCR5 32-bp-deleted allele on plasma virus load and CD4 T lymphocytes in perinatally human immunodeficiency virus-infected children at 8 years of age. J Infect Dis. 1998;178:1019–1023. doi: 10.1086/515660. [DOI] [PubMed] [Google Scholar]
- 23.Lewis MJ. Balamurugan A. Ohno A. Kilpatrick S. Ng HL. Yang OO. Functional adaptation of Nef to the immune milieu of HIV-1 infection in vivo. J Immunol. 2008;180:4075–4081. doi: 10.4049/jimmunol.180.6.4075. [DOI] [PubMed] [Google Scholar]