Abstract
Background
Congenital long-QT syndrome (LQTS) is potentially lethal secondary to malignant ventricular arrhythmias and is caused predominantly by mutations in genes that encode cardiac ion channels. Nearly 25% of patients remain without a genetic diagnosis, and genes that encode cardiac channel regulatory proteins represent attractive candidates. Voltage-gated sodium channels have a pore-forming α-subunit associated with 1 or more auxiliary β-subunits. Four different β-subunits have been described. All are detectable in cardiac tissue, but none have yet been linked to any heritable arrhythmia syndrome.
Methods and Results
We present a case of a 21-month-old Mexican-mestizo female with intermittent 2:1 atrioventricular block and a corrected QT interval of 712 ms. Comprehensive open reading frame/splice mutational analysis of the 9 established LQTS-susceptibility genes proved negative, and complete mutational analysis of the 4 Navβ-subunits revealed a L179F (C535T) missense mutation in SCN4B that cosegregated properly throughout a 3-generation pedigree and was absent in 800 reference alleles. After this discovery, SCN4B was analyzed in 262 genotype-negative LQTS patients (96% white), but no further mutations were found. L179F was engineered by site-directed mutagenesis and heterologously expressed in HEK293 cells that contained the stably expressed SCN5A-encoded sodium channel α-subunit (hNaV1.5). Compared with the wild-type, L179F-β4 caused an 8-fold (compared with SCN5A alone) and 3-fold (compared with SCN5A + WT-β4) increase in late sodium current consistent with the molecular/electrophysiological phenotype previously shown for LQTS-associated mutations.
Conclusions
We provide the seminal report of SCN4B-encoded Navβ4 as a novel LQT3-susceptibility gene.
Keywords: genetics, ion channels, long-QT syndrome
Long-QT syndrome (LQTS) represents the prototypic cardiac channelopathy that affects 1 in 3000 individuals and is characterized by QT prolongation, abnormal ventricular repolarization, and increased propensity for sudden cardiac death as a result of its trademark dysrhythmia of torsade de pointes. Since the first discovery in 1995 that pathogenic mutations in cardiac channels cause LQTS,1,2 hundreds of mutations distributed among 9 genes comprise 75% of LQTS. Five genes encode critical, pore-forming, ion channel α subunits (KCNQ1, KCNH2, SCN5A, KCNJ2, and CACNA1C) and 4 encode ion-channel regulatory proteins (KCNE1, KCNE2, ANKB, and CAV3). However, 25% of patients with LQTS remain genotype-negative after evaluation of all known LQTS-susceptibility genes.3,4
Because genes that encode cardiac channel interacting or auxiliary proteins can affect channel function, they are attractive candidates5 for the cause of LQTS. One set of auxiliary proteins belongs to the sodium channel β subunit (Navβ) gene family, where these Navβ subunits play a critical role in cell adhesion, signal transduction, channel expression at the plasma membrane, and voltage dependence of channel gating.6–9 Presently, 4 different Navβ subunits have been described (SCN1B, SCN2B, SCN3B, and SCN4B).10–14 All are detectable in cardiac tissue,15 but none have been linked to any heritable arrhythmia syndrome.
Case Report
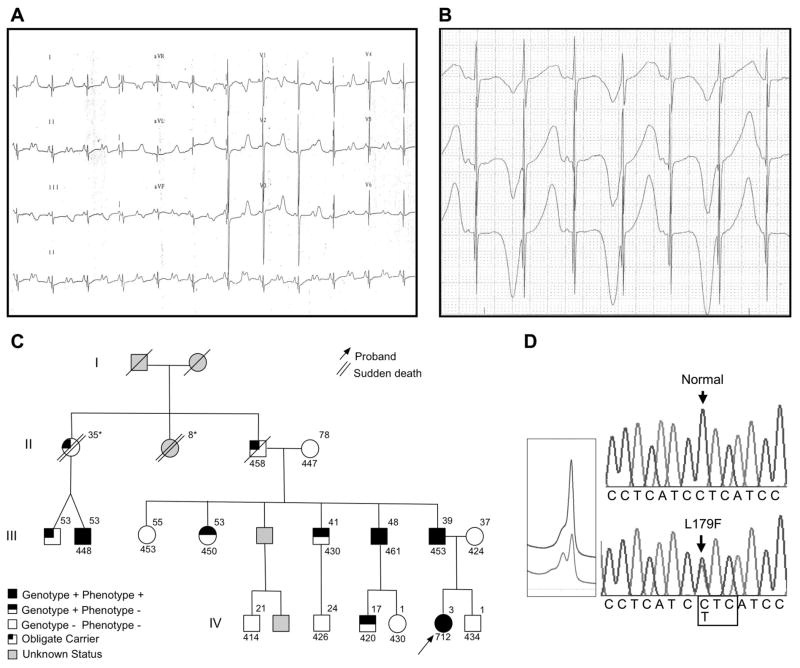
The index case was a 21-month–old Mexican-mestizo girl, referred for evaluation of asymptomatic bradycardia with rates <60 bpm. Her 12-lead ECG revealed profound QT prolongation with a heart rate–corrected QT interval (QTc) of 712 ms and intermittent 2:1 atrioventricular (AV) block (Figure 1A). During 1:1 conduction, macroscopic T-wave alternans were observed (Figure 1B). Her past medical history included fetal bradycardia noted at 24 weeks of gestation and small ventricular septal defect that spontaneously closed by 6 months of age. Despite the severe electrocardiographic phenotype, the patient has remained asymptomatic during her first 5 years of life after placement of an epicardial pacemaker. On further inquiry, a family history of premature, unexpected, and unexplained sudden cardiac deaths that involved 2 paternal great aunts was elucidated (sudden cardiac death at 35 years after delivery of twins and sudden cardiac death at 8 years during exercise) (Figure 1C).
Figure 1.
Clinical characterization and molecular discovery of L179F-SCN4B. A, Electrocardiographic evaluation of 21-month-old index case shows QTc=712 ms and 2:1 AV block. B, Holter evaluation of index case shows T wave alternans during 1:1 conduction. C, Pedigree; affected status was assigned as outlined previously: QTc>440 ms in males and >460 ms in females. QTc values are shown below each individual. Ages at clinical evaluation are shown above each individual. *Age at death. D, DNA sequencing chromatograms demonstrate a leucine (L) to phenylalanine (F) substitution at residue 179. Bottom, Aberrant denaturing high-performance liquid chromatography profile. Top, Wild-type.
On the basis of the ECG findings, the patient’s ECG phenotype was found to be consistent with SCN5A-mediated LQT3, which shows a long isoelectric ST segment with late-onset T wave, and 2:1 AV block, which was shown previously to be associated with defective LQTS-susceptibility genes, in particular the cardiac sodium channel.16–18
Methods
Subjects
The study was performed according to the terms required by the Research Ethics Committee of the National Institute of Cardiology “Ignacio Chávez,” Mexico City, and the Mayo Foundation Institution Review Board; written informed consent was obtained from all participants. Genomic DNA from the index case and all consenting family members was extracted from peripheral blood lymphocytes by use of standard techniques. Control DNA from 200 healthy Mexican-mestizo subjects was obtained from Unidad de Biología Molecular y Medicina Genómica, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City. Mexican-mestizo ethnicity resulted from admixture of racial ancestry that included European (Spanish) and indigenous descent, with smaller contribution from Asian and African groups. Two hundred additional control DNA samples (100 from white donors and 100 from black), obtained from Coriell Cell Repositories (Camden, NJ), were also examined.
In addition, 262 genotype-negative patients (180 females; 96% white; age at diagnosis, 25±16 years; mean QTc, 470±60 ms) with a suspected clinical diagnosis of congenital LQTS, who were referred previously for genetic testing to the Mayo Clinic Windland Smith Rice Sudden Death Genomics Laboratory, were included.
Mutational Analysis
Through polymerase chain reaction, denaturing high-performance liquid chromatography, and direct DNA sequencing, we performed comprehensive open reading frame/splice site mutational analysis of all known LQTS-susceptibility genes (KCNQ1, KCNH2, SCN5A, ANKB, KCNE1, KCNE2, KCNJ2, CACNA1C, and CAV3) using previously published primers, followed by mutational analysis of the 4 Navβ subunits encoded by SCN1B, SCN2B, SCN3B, and SCN4B. The flanking primers used in polymerase chain reaction amplification of the β subunits were designed with Oligo software (Molecular Biology Insights, Inc., Cascade, Colo.); primers, polymerase chain reaction, and denaturing high-performance liquid chromatography conditions are shown in Table 1.
TABLE 1.
Oligonucleotide Primers, Polymerase Chain Reaction, and Denaturing High-Performance Liquid Chromatography Conditions for Mutational Analysis of Navβ Subunits
| Gene-Exon | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | Size, base pairs | MgCl, mmol/L | Thermal Cycling Method† | Gradient 1, %B; Temp °C‡ | Gradient 2, %B; Temp °C |
|---|---|---|---|---|---|---|---|
| SCN1B-1 | CGC CTC TCG CCC CGC TAT TA | CTC CCG CCG CCC CGC CAG TG | 164 | 1* | 2 | 50.5 to 51.5; 67 | 48 to 58; 72 |
| SCN1B-2 | CCT GAC CTG AGC CTG CTG TC | TGC CCT CCC ATG CCG TTC C | 227 | 1* | 2 | 56.6 to 66.6; 65 | |
| SCN1B-3 | CCT TCC CCT CCC TGG CTA C | GGC AGG CAG CAC CCG ACT CA | 287 | 1* | 2 | 56.3 to 66.3; 63 | 52.6 to 62.6; 65 |
| SCN1B-4 | CAG CCT GGG CTA CCC CCT TA | CCC TGG GTG CCC TCC CAC CT | 220 | 1* | 2 | 53.7 to 63.7; 62.5 | |
| SCN1B-5 | CGG TCT GAT GAT GGG GTC AC | TTA CGG CTG GCT CTT CCT TG | 243 | 1* | 2 | 54.8 to 64.8; 63 | |
| SCN2B-1 | CCA TTC CTC CCT TGT AGT TCT | CCC CAT CCT CTT CAC ATT GC | 216 | 2 | 1 | 53.6 to 63.6; 61.5 | |
| SCN2B-2 | CCA ACA CTC CCA GGC ACA G | GAC CAG GGG CTT CAT GCC A | 316 | 2 | 1 | 57.2 to 67.2; 62 | 57.2 to 67.2; 63.5 |
| SCN2B-3 | GGC ATC CTC ACT GTC CTT G | AGG TGG GTG GGA AAG GTC A | 329 | 2 | 1 | 57.5 to 67.5; 63 | 57.5 to 67.5; 64 |
| SCN2B-4 | CAC GGG TAG TGG GGT GAT G | CGA GCA GGC AGG GTC ACT G | 346 | 2 | 1 | 57.9 to 67.9; 63.5 | |
| SCN3B-2 | GCA GTC CTT GAC CGA GGG A | AGA GGC AAG CCA GCC AGA G | 223 | 2 | 1 | 53.9 to 63.9; 59.5 | 53.9 to 63.9; 61.5 |
| SCN3B-3 | CCT CCC TCC TTC TTC TCC AA | CAG GAG CCA GGC TGG GAA C | 316 | 2 | 1 | 57.2 to 67.2; 63.5 | |
| SCN3B-4 | CTG CCC TGT CCC TAA CTG G | TTC CCT GTC CAC AGA GAG C | 358 | 2 | 1 | 57.2 to 67.2; 63.5 | |
| SCN3B-5 | TCC AAT GAC GGC TCT AGG T | GAG CAA GCA TTC TGA AGG TG | 258 | 2 | 1 | 55.3 to 65.3; 59.5 | |
| SCN3B-6 | CTC CTG TGC CCT GCT CCT T | ACA ACC TGC CAT CCA CAT TC | 219 | 2 | 1 | 53.7 to 63.7; 61.5 | |
| SCN4B-1 | GCT GTG CCC AGT ATC CCA T | CCA CCA TCC TCA TTC CGT G | 241 | 2* | 1 | 47.2 to 57.2; 64 | 54.7 to 64.7; 67 |
| SCN4B-2 | CCC GAG GTT GGC ACT GAG G | GGA CCA GAG CGT AGG AGG C | 373 | 2* | 1 | 58.5 to 68.5; 62 | |
| SCN4B-3 | TCT CGG CTA CTT TCT CAC CC | CCT CCC AAG TCC TTC CCA C | 320 | 2 | 2 | 57.3 to 67.3; 61 | |
| SCN4B-4 | GCT CCA GGT TGA CTC TTG CT | GCT GCT GGG AGG ACA GGA G | 326 | 2 | 2 | 57.4 to 67.4; 61.5 | |
| SCN4B-5 | TCC CCC TAC TCT TGC TCC T | GGA CTC TGG TTT CTT GTG CC | 294 | 2 | 2 | 56.5 to 66.5; 63 |
Polymerase chain reaction and other reactions were performed in 20-μl volumes with 50 ng of DNA, 16 pmol of each primer, 200 μM of each dNTP, 50 mmol/l KCl, 10 mmol/l Tris-HCl (pH 8.3), and 1.0 U of Amplitaq Gold (Applied Biosystems, Branchburg, NJ). PCR indicates polymerase chain reaction; DHLPC, denaturing high-performance liquid chromatography.
8% dimethyl sulfoxide was added to the reaction mixture. Polymerase chain reaction amplification was performed with a DNA Engine Tetrad thermal cycler.
Thermal cycling method 1: 94°C for 5 minutes, followed by 5 cycles of 94°C for 20 s, 64°C for 20 s, and 72°C for 30 s; additional 35 cycles of 94°C for 20 s, 62°C for 20 s, 72°C for 30 s, and a final extension of 72°C for 10 minutes. Thermal cycling method 2: 94°C for 15 minutes, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 30 s, and a final extension of 72°C for 10 minutes.
DHPLC was performed with a 5% buffer B/minute gradient. The start and stop % buffer B followed by the temperature at which the gradient was performed is indicated in the table.
Functional Assay
Cloning, mutagenesis, and voltage-clamp techniques were performed as previously described.19 Briefly, the WT-β4 was cloned from human heart cDNA with reverse transcriptase–polymerase chain reaction and 2 primers: 5′-AGAGAACAGGACTATGCCCG-3′ and 5′-TTTCATCATCAGAAAGGGGG-3′. The WT-β4 was sub-cloned into pcDNA3 and confirmed by DNA sequencing analysis. L179F-β4 was engineered by site-directed mutagenesis with the following primers: Navβ4 L179F sense 5′-ATCGGGCTCCTCATCTTC-ATCCTGCTGATCAAG-3′ and Navβ4 L179F antisense 5′-CTTGATCAGCAGGATGAAGATGAGGAGCCCGAT-3′. The WT-β4 or L179F-β4 was subcloned into pIRGFP1 vector, a mammalian expression vector (kindly provided by Dr David Johns from the Johns Hopkins University, Baltimore, Md), which contains IRES and GFP. These WT-β4 and L179F-β4 constructs were expressed heterologously in HEK 293 cells that contain stably expressed SCN5A-encoded sodium channel α subunits (hNaV1.5, hH1c, GeneBank accession #AY148488). Macroscopic sodium current was measured by standard whole-cell-patch-clamp method with use of an Axopatch 200B amplifier and pClamp8.0 software (Axon Instruments, Foster City, Calif). “Steady-state” activation, inactivation, and recovery from inactivation were performed and analyzed as previously described.19,20 The conditioning step for inactivation and recovery was 1 s, and holding potential and recovery potential was −140 mV. Activation and inactivation were fitted to standard Boltzmann function. Recovery time course and the time course of current decay were fitted with a biexponential function. Summary data are shown as the mean±SEM with statistical significance determined by the Student t test.
Biochemical Analysis
Coimmunoprecipitation between SCN5A and β4 subunit was performed with a SCN5A-labeled with HA and β4 subunit labeled with Myc and coexpressed in HEK cells. The preparation was immunoprecipitated with 1 μg of mouse anti-HA monoclonal antibody (Zymed Laboratories, South San Francisco, Calif) and then electrophoretically separated by 12% SDS-PAGE. After immunoblotting was performed with 0.5 μg/mL rabbit anti-Myc polyclonal antibody (Sigma-Aldrich, St Louis, Mo.), immune complexes were developed with the ECL-plus kit (Amersham Biosciences, Piscataway, NJ).
The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as written.
Results
After a negative mutational analysis of the 9 known LQTS-susceptibility genes, we identified a C to T base mutation at position 535, which yielded a novel L179F-SCN4B missense mutation (leucine [L] to phenylalanine [F] at position 179, when the full-length gene product is considerd) by denaturing high-performance liquid chromatography and DNA sequencing (Figure 1D). This mutation was absent in 800 reference alleles, which included 400 ethnicity-matched, Mexico-mestizo alleles. SCN4B mutational analysis of 262 genotype-negative LQTS patients (predominantly white) was negative.
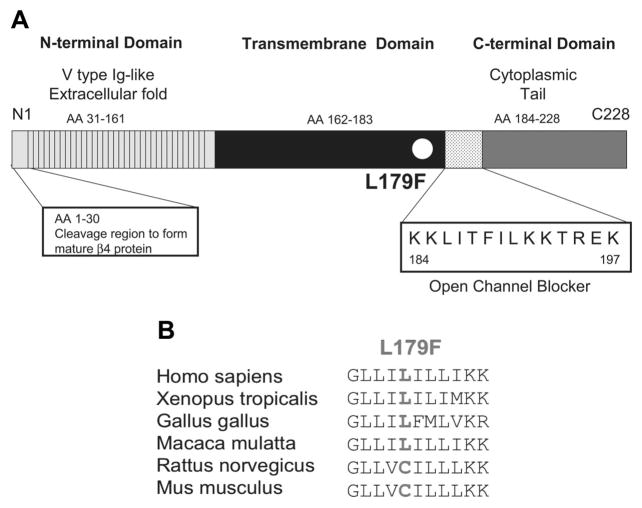
L179 localizes to the transmembrane spanning region (Figure 2A), is relatively conserved across species (Figure 2B), and a substitution with phenylalanine is predicted to alter secondary structure (data not shown). L179F cosegregated properly through a 3-generational pedigree with incomplete penetrance. QTc values and ages are shown in Figure 1C. Although other family members displayed QTc values above normal average, the index case exhibited the only severe ECG phenotype detected on this pedigree, and despite the severe ECG anomalies, the index case has never experienced syncope or torsade de pointes. However, no premortem ECGs were available for either of the sudden-death victims and 1 of the decedents was proven to have been an obligate mutation carrier.
Figure 2.
Location of L179F in the β4 subunit. A, Linear topology of the β4 subunit. Numbering of the amino acid sequence is relative to the full-length gene product. B, L179F conservation across species.
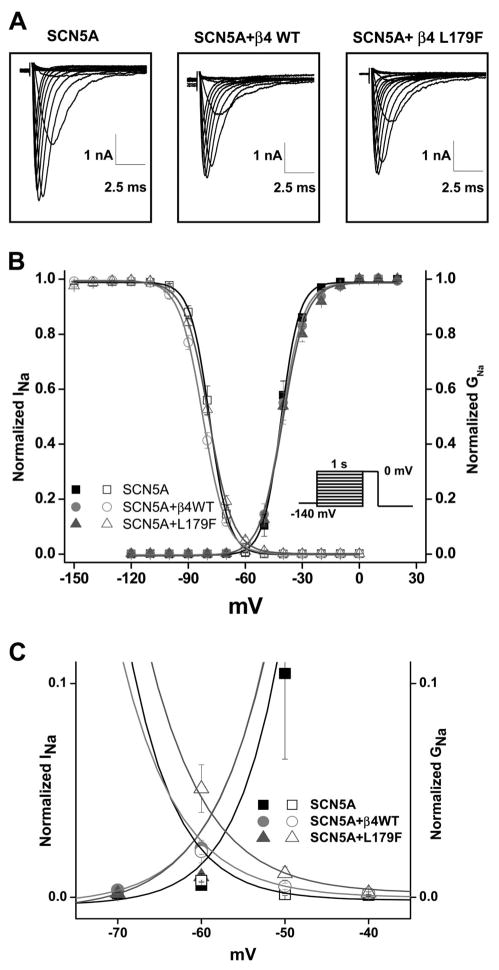
When the wild-type (WT) SCN4B and the L179F mutant SCN4B were transfected transiently into an HEK cell line that stably expressed the most common SCN5A transcript in humans (H558/Q1077del),20 robust sodium current traces were recorded and no significant differences in current amplitude and time course were noted in comparison to SCN5A alone (Figure 3A and Table 2). The steady-state voltage dependence of activation and inactivation of the sodium channel were analyzed with standard protocols and fitted with the use of a Boltzmann function. Voltage dependence of inactivation but not activation was modified by L179F mutant (Figure 3, B and C). This small but significant 3.42-mV positive shift in inactivation (WT-β4, −82.52±0.74 (n=9), to L179F-β4, −79.10±0.59 (n=9); P<0.05) increases the window current and could be arrhythmogenic. More importantly, this voltage shift in inactivation indicates a consistent effect and an interaction of the α subunit with β4 in the heterologous system.
Figure 3.
Functional characterization of L179F in HEK293 cells stably transfected with SCN5A-encoded Nav1.5 α-subunit. A, Representative traces of sodium channel current recorded with the whole-cell configuration of the patch-clamp technique for a clamp step from −140 mV to various potentials. Current amplitude and time course are shown on each panel for the sodium channel alone, sodium channel plus WT-β4, and sodium channel plus β4 mutant. No significant differences in amplitude or current/time course were noted (Summary data in Table 2). B, Steady-state voltage dependence of activation (right plot) and inactivation (left plot) with and without WT-β4 or L179F-β4. Data are means of measurements, and the lines are Boltzmann fits with n numbers and parameters of the fit found in Table 2. C, Detail of the “window” current where the activation curves overlap the inactivation curves. L179F increases window current. INa indicates robust sodium current; GNa, sodium conductance (calculated from peak sodium current [INa] divided by the driving force [V–Na reversal potential]).
TABLE 2.
Summary of Current Density and Gating Parameters
| SCN5A | SCN5A + WT-β4 | SCN5A + L179F-β4 | |
|---|---|---|---|
| INa density | |||
| pA/pF | −425.70±25.58 (n=5) | −378.43±61.81 (n=7) | −341.56±35.30 (n=8) |
| Activation | |||
| V1/2, mV | −43.78±1.28 (n=5) | −43.69±1.61 (n=7) | −43.37±1.13 (n=8) |
| Slope factor | 3.12±0.10 (n=5) | 3.63±0.16* (n=7) | 3.85±0.23* (n=8) |
| Inactivation | |||
| V1/2, mV | −78.77±1.11 (n=10) | −82.28±0.74* (n=9) | −79.10±0.59† (n=9) |
| Slope factor | 4.72±0.09 (n=10) | 6.03±0.23* (n=9) | 6.03±0.35* (n=9) |
| Recovery from inactivation | |||
| τ Fast, ms | 1.52±0.14 (n=12) | 1.48±0.10 (n=18) | 1.80±0.18 (n=22) |
| Amp fast, % | 0.83±0.02 (n=12) | 0.82±0.02 (n=18) | 0.75±0.02*† (n=22) |
| τ Slow, ms | 37.28±3.93 (n=12) | 24.50±4.01* (n=18) | 47.95±7.51† (n=22) |
| Amp slow, % | 0.18±0.01 (n=12) | 0.19±0.01 (n=18) | 0.20±0.01 (n=22) |
| Decay, −20 mV | |||
| τ Fast, ms | 0.68±0.02 (n=5) | 0.77±0.04 (n=6) | 0.80±0.04 (n=8) |
| τ Slow, ms | 2.85±0.26 (n=5) | 3.16±0.31 (n=6) | 3.66±0.27 (n=8) |
All data are represented as mean±SD. INa indicates robust sodium current.
P<0.05 vs SCN5A.
P<0.05 vs WT-β4.
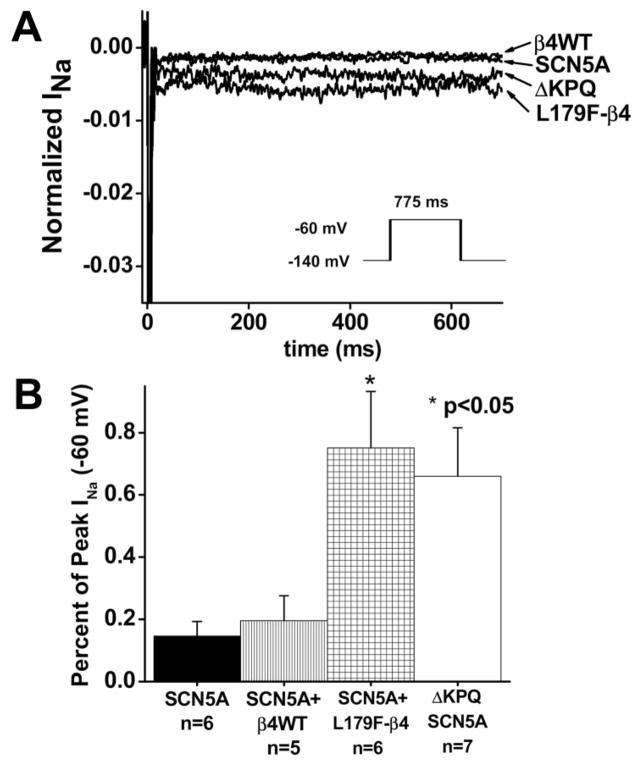
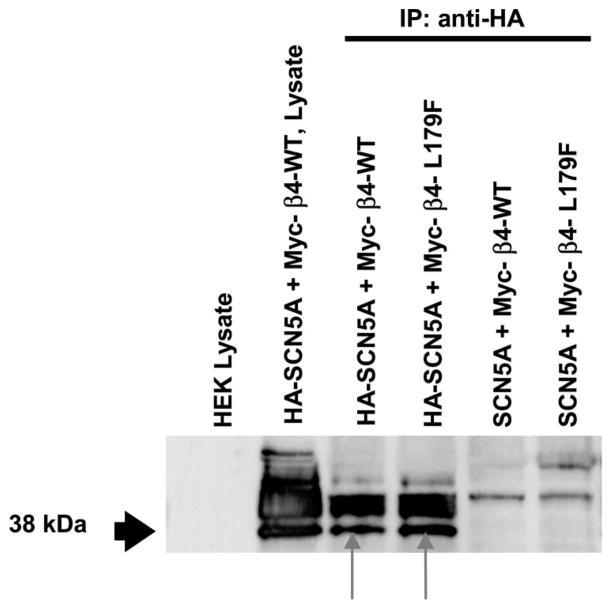
Compared with SCN5A alone, L179F-β4 caused a dramatic 8-fold increase in late sodium current at −60 mV, which is consistent with an effect on terminal repolarization to prolong the QT interval as previously shown for LQT3-associated mutations in SCN5A. In fact, the increase in late sodium current exceeded that of the classic mutation that causes LTQ3 (ΔKPQ) in the pore-forming, SCN5A-encoded α subunit (Figure 4).21 Table 2 presents a summary of current density and the remaining gating parameters. Statistically significant changes in recovery kinetics, particularly a significant increase in the slow Tau of recovery, were noted. Coimmunoprecipitation experiments (Figure 5) showed that both WT-β4 and L179F-β4 were present when SCN5A was specifically immunoprecipitated by a labeled tag.
Figure 4.
Effect of coexpression of L179F-β4 mutant on late sodium current. A, Representive sodium current traces in response to a step to −60 mV for 700 ms from a holding potential of −140 mV (protocol inset) are shown. Leak-subtracted currents were normalized to peak current and are shown on a scale such that peak current is off-scale to emphasize the small late component. B, Summary data showed that L179F-β4 mutant increased late sodium current during the window of terminal repolarization as much as the α subunit sodium channel mutation (ΔKPQ) that causes LQT3.21
Figure 5.
Coimmunoprecipitation of SCN5A and β4 subunit. The lysates were obtained from untransfected HEK cells (column 1) and from cells cotransfected with HA-SCN5A and Myc-β4 (column 2). Columns 3 to 6 are immunoprecipitation protein complexes derived from cotransfection of HA-SCN5A and Myc-β4-WT; HA-SCN5A and Myc-β4-L179F; HA-SCN5A and untagged β4-WT; and HA-SCN5A and untagged β4-L179F, respectively. The 38-kDa line recognizes the β4 subunit, and the bands above 38 kDa show nonspecific binding. Column 1 shows no recognition in control; column 2 shows recognition of the expressed Myc label. In columns 3 and 4, the β4 subunit was recognized in the SCN5A precipitate for both WT and L179F (red arrows). In columns 5 and 6, the β4 subunit was not recognized because the complex of unlabeled SCN5A and Myc-β4 were not precipitated.
Discussion
We provide the first report to detail SCN4B as a novel LQTS-susceptibility gene on the basis of identification of a novel missense mutation (L179F) that cosegregated properly, was absent in 800 reference alleles, and produced a “gain-of-function” Nav1.5 current in a family with no other identifiable LQTS-associated mutations.
L179F cosegregated properly with incomplete penetrance, which is typical for heritable arrhythmia syndromes like LQTS. Genetic testing in LQTS has demonstrated that 40% of carriers of LQTS-related mutations had a QTc within normal limits.4,22 Penetrance, expressivity, and phenotype do not depend solely on the primary LQTS-associated mutation, as these can be influenced by modifier genes.19 Thus far, no modifiers have been identified to account for the extreme QT prolongation and intermittent 2:1 AV block in the index case or the sudden death in the obligate carrier compared with the milder ECG phenotype seen in other genotype-positive living family members. Intermittent functional 2:1 AV block in the setting of LQTS has an incidence of 4% to 5% in pediatric series,23 is usually an isolated disorder, and is associated with a high mortality rate of >50% regardless of treatment.24,25 Symptoms could appear at a very young age; neonatal paroxysmal bradycardia and/or hydrops fetalis are common findings. The phenomenon occurs in the setting of a very long, rate-dependent, effective refractory period.26 QRS complex is usually narrow; however, in spite of this, infrahisian block locations have been documented,26–28 although supra-hisian block is not ruled out and the level of the block could depend on the genotype. Until now 3 genes have been associated with 2:1 AV block: SCN5A, KCNH2, and CACNA1.17,27–38 Our data suggest that perturbations in the SCN4B-encoded β4 subunit constitute another pathogenic mechanism for 2:1 AV block in patients with LQTS.
According to the proband’s family history, the 2 sudden deaths occurred during exercise or after delivery; these triggers have been previously associated with the KCNQ1 and KCNH2 genes, respectively; nevertheless, no mutations in these genes were found in our patient. Although exertional syncope more likely suggests LQT1, and the presence of cardiac events during the postpartum period more likely suggests LQT2, 44% of patients referred for LQTS genetic testing because of exertional syncope and 25% with family history of an event that occurred post-partum did not have LQT1 to LQT6.39 Importantly, the genotype-phenotype relationships for the rare subtypes of LQTS, mainly those that involve ion channel interacting proteins, have not been determined.
Ion channels exist as macromolecular complexes with several auxiliary proteins known as channel interacting proteins that localize to or are involved in the plasma membrane, extracellular matrix, intracellular proteins, cytoskeleton anchoring, and signal transduction.5 In principle, perturbations in any component of this complex may affect the proper function of the pore-forming channel subunit itself. Voltage-gated sodium channels have a pore-forming α subunit and 1 or more auxiliary β subunits.40 Presently, 9 functional α subunits (Nav1.1 to Nav1.9)41 and 4 different Navβ subunits have been identified in humans. Numerous mutations in various Nav α subunits have been associated with inherited diseases such as mutations in the SCN5A-encoded Nav1.5 subunit that precipitates LQT3 and Brugada syndrome. However, among all the β subunits, only β1 has been associated with human disease, namely febrile seizures.42
Navβ subunits are proteins with type I topology characterized by an extracellular N-terminal cleaved region, a transmembrane segment, and a cytoplasmic domain with a C-terminal tail. Navβ subunits contain an extracellular Ig-like fold, often found in cell adhesion molecules43 that target ion channels to the plasma membrane and mediate interactions with signaling molecules. Navβ1 and Navβ3 are similar in sequence and associate noncovalently with α subunits. Navβ2 and Navβ4 are related proteins that are disulfide-linked to α subunits.13,14 Immunocytochemical studies in mouse hearts indicate that the primary cardiac sodium channel in ventricular myocytes is composed of Nav1.5 plus β2 and/or β4 subunits in intercalated disks, whereas other isoforms, such as Nav1.1, Nav1.3, and Nav1.6, are also expressed in heart plus β1 or β3 in the transverse tubules.15 Knockout mice that lack the sodium channel β1-subunit display spontaneous generalized seizures and ataxia,44 whereas mice that lack the β2-subunit have increased susceptibility to seizures.45
The β4 subunit is distinguished by a cytoplasmic tail insert (KKLITFILKKTREK) from amino acid 184 to 197 when the full gene product is considered, or amino acid 154 to 167 when the mature protein after cleavage is considered, as shown in Figure 2A. This cluster of lysine and hydrophobic residues has been implicated recently with a transient and resurgent sodium conduction contributing to the short refractory period seen in the SCN8A-encoded sodium channel of cerebellar Purkinje cells46 and may serve as an endogenous open-channel blocker.13 Enzymatic removal of this endogenous blocker generated a delay in channel recovery. This resurgent current, induced by the β4 cytoplasmic tail, has been demonstrated also in the cardiac isoform in vitro and a putative S6-binding site within the inner cavity of hNav1.5 has been suggested.47 This mechanism is analogous to that seen in patients with SCN5A-mediated LQT3 and raises the possibility that L179F-β4 represents a primary loss-of-function mutation in SCN4B that secondarily precipitates a gain-of-function on Nav1.5. In fact, the accentuation in late sodium current rendered by L179F-β4 is greater than many primary mutations in the SCN5A-encoded α subunit itself. Unlike SCN5A mutations, however, the gain-of-function mechanism in these experiments for L179F-β4 is confined to the window current and would primarily affect and slow terminal repolarization rather than the classic accentuation of late sodium current at more depolarized membrane potentials that would prolong the action potential plateau.
Experiments were performed in heterologous mammalian expression systems, a standard technique for characterization of LQTS mutations. These systems, however, lack the native environment of the cardiac cell and did not include β1, β2, or β3 subunits, or other components of the Na channel macromolecular complex such as caveolin-3. As such, the effects may differ and be more severe in a more complete and native environment. However, direct concordance between the degree of channel dysfunction and the manifest clinical severity is not necessarily present. Nonetheless, the molecular phenotype was consistent with LQT3, and in combination with the clinical data and genetic cosegregation supports the L179F mutation in the sodium channel β4 subunit as disease associated.
In the present report we have provided proof of principle that mutations in SCN4B may contribute to the pathogenic substrate for some cases of LQTS by alteration of the cardiac sodium channel current (Nav1.5). Our study provides the first human disease associated with perturbations in the sodium channel β4 subunit. Notably, mutations in CAV3-encoded caveolin-3 were demonstrated recently as a novel LQTS-susceptibility gene (LQT9).48 In coexpression studies that involved heterologous expression of only the α subunit and mutant caveolin-3, marked accentuation of the late sodium current was observed, similar to that seen here with L179F-β4. Thus, SCN4B (LQT10) joins CAV3 (LQT9) as rare LQTS-susceptibility genes that encode cardiac sodium encode cardiac channel interacting proteins that, when disrupted, yield a LQT3-like molecular/electro-physiological phenotype.
CLINICAL PERSPECTIVE.
Long-QT syndrome (LQTS) is a potentially lethal heritable arrhythmia syndrome that affects an estimated 1 in 3000 persons. Since the sentinel discovery of cardiac channel mutations as its pathogenic basis in 1995, LQTS has been viewed as a “cardiac channelopathy.” To date, 9 LQTS-susceptibility genes have been discovered, and 5 of these genes encode the critical ion channel pore-forming α subunit whereas the other 4 genes encode cardiac channel interacting proteins. Presently, ≈ 20% of LQTS remains genetically elusive and encodes cardiac channel interacting protein–encoding genes represent the latest targets of investigation. Sodium channel βsubunits are crucial regulatory proteins. Four different βsubunits (β1 to β4), encoded by SCN1Bthrough SCN4B, respectively, have been described. All are detectable in cardiac tissue but none have been related to any arrhythmogenic disease. In the present study, we discovered that SCN4Bis a novel albeit rare LQTS-susceptibility gene (LQT10). Consistent with the Towbin Final Common Pathway Hypothesis, a single missense mutation (L179F) in the 228-amino acid that contains β4 subunit conferred a secondary gain-of-function clinical and biophysical phenotype to the sodium channel macromolecular complex. The mutation converted the αsubunit of the otherwise intact NaV1.5 into a channel with accentuated late sodium current that essentially mimicked mutations that cause LQT3.
Acknowledgments
We are grateful to Salvador Ramírez-Jimenez for technical assistance. We are particularly indebted to the index case and family members for their participation in this study.
Sources of Funding
Dr Medeiros received financial support from Consejo Nacional de Ciencia y Tecnología (CONACyT) and Fundación Mexicana para la Salúd A.C. (FUNSALUD). Dr Ye was supported by the Scientist Development Grants from the American Heart Association National Center (0435030N). The Dr. Scholl Foundation, the CJ Foundation for SIDS, the American Heart Association, and the National Institutes of Health (HD42569) support the Mayo Clinic Windland Smith Rice Sudden Death Genomics Laboratory, which is directed by Dr Ackerman.
Footnotes
Disclosures
None.
References
- 1.Curran ME, Splawski I, Timothy KW, Vincent GM, Green ED, Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 2.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–11. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 3.Tester DJ, Will ML, Haglund CM, Ackerman MJ. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2:507–517. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Napolitano C, Priori SG, Schwartz PJ, Bloise R, Ronchetti E, Nastoli J, Bottelli G, Cerrone M, Leonardi S. Genetic testing in the long QT syndrome: development and validation of an efficient approach to genotyping in clinical practice [see comment] JAMA. 2005;294:2975–2980. doi: 10.1001/jama.294.23.2975. [DOI] [PubMed] [Google Scholar]
- 5.Meadows LS, Isom LL. Sodium channels as macromolecular complexes: implications for inherited arrhythmia syndromes. Cardiovasc Res. 2005;67:448–458. doi: 10.1016/j.cardiores.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Malhotra JD, Kazen-Gillespie K, Hortsch M, Isom LL. Sodium channel beta subunits mediate homophilic cell adhesion and recruit ankyrin to points of cell-cell contact. J Biol Chem. 2000;275:11383–11388. doi: 10.1074/jbc.275.15.11383. [DOI] [PubMed] [Google Scholar]
- 7.Meadows L, Malhotra JD, Stetzer A, Isom LL, Ragsdale DS. The intracellular segment of the sodium channel beta 1 subunit is required for its efficient association with the channel alpha subunit. J Neurochem. 2001;76:1871–1878. doi: 10.1046/j.1471-4159.2001.00192.x. [DOI] [PubMed] [Google Scholar]
- 8.Patton DE, Isom LL, Catterall WA, Goldin AL. The adult rat brain beta 1 subunit modifies activation and inactivation gating of multiple sodium channel alpha subunits. J Biol Chem. 1994;269:17649–17655. [PubMed] [Google Scholar]
- 9.Isom LL. Sodium channel beta subunits: anything but auxiliary. Neuroscientist. 2001;7:42–54. doi: 10.1177/107385840100700108. [DOI] [PubMed] [Google Scholar]
- 10.McClatchey AI, Cannon SC, Slaugenhaupt SA, Gusella JF. The cloning and expression of a sodium channel beta 1-subunit cDNA from human brain. Hum Mol Genet. 1993;2:745–749. doi: 10.1093/hmg/2.6.745. [DOI] [PubMed] [Google Scholar]
- 11.Isom LL, De Jongh KS, Patton DE, Reber BF, Offord J, Charbonneau H, Walsh K, Goldin AL, Catterall WA. Primary structure and functional expression of the beta 1 subunit of the rat brain sodium channel. Science. 1992;256:839–842. doi: 10.1126/science.1375395. [DOI] [PubMed] [Google Scholar]
- 12.Isom LL, Ragsdale DS, De Jongh KS, Westenbroek RE, Reber BF, Scheuer T, Catterall WA. Structure and function of the beta 2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell. 1995;83:433–442. doi: 10.1016/0092-8674(95)90121-3. [DOI] [PubMed] [Google Scholar]
- 13.Yu FH, Westenbroek RE, Silos-Santiago I, McCormick KA, Lawson D, Ge P, Ferriera H, Lilly J, DiStefano PS, Catterall WA, Scheuer T, Curtis R. Sodium channel beta4, a new disulfide-linked auxiliary subunit with similarity to beta2. J Neurosci. 2003;23:7577–7585. doi: 10.1523/JNEUROSCI.23-20-07577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan K, Stevens EB, Shah B, Cox PJ, Dixon AK, Lee K, Pinnock RD, Hughes J, Richardson PJ, Mizuguchi K, Jackson AP. Beta 3: an additional auxiliary subunit of the voltage-sensitive sodium channel that modulates channel gating with distinct kinetics. Proc Natl Acad Sci U S A. 2000;97:2308–2313. doi: 10.1073/pnas.030362197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maier SK, Westenbroek RE, McCormick KA, Curtis R, Scheuer T, Catterall WA. Distinct subcellular localization of different sodium channel alpha and beta subunits in single ventricular myocytes from mouse heart. Circulation. 2004;109:1421–1427. doi: 10.1161/01.CIR.0000121421.61896.24. [DOI] [PubMed] [Google Scholar]
- 16.Zhang L, Timothy KW, Vincent GM, Lehmann MH, Fox J, Giuli LC, Shen J, Splawski I, Priori SG, Compton SJ, Yanowitz F, Benhorin J, Moss AJ, Schwartz PJ, Robinson JL, Wang Q, Zareba W, Keating MT, Towbin JA, Napolitano C, Medina A. Spectrum of ST-T-wave patterns and repolarization parameters in congenital long-QT syndrome: ECG findings identify genotypes. Circulation. 2000;102:2849–2855. doi: 10.1161/01.cir.102.23.2849. [DOI] [PubMed] [Google Scholar]
- 17.Wang DW, Viswanathan PC, Balser JR, George AL, Jr, Benson DW. Clinical, genetic, and biophysical characterization of SCN5A mutations associated with atrioventricular conduction block. Circulation. 2002;105:341–346. doi: 10.1161/hc0302.102592. [DOI] [PubMed] [Google Scholar]
- 18.Lupoglazoff JM, Denjoy I, Cheav T, Berthet M, Extramiana F, Cauchemez B, Villain E, Leenhardt A, Guicheney P. Homozygotous mutation of the SCN5A gene responsible for congenital long QT syndrome with 2/1 atrioventricular block. Arch Mal Coeur Vaiss. 2002;95:440–446. [PubMed] [Google Scholar]
- 19.Ye B, Valdivia CR, Ackerman MJ, Makielski JC. A common human SCN5A polymorphism modifies expression of an arrhythmia causing mutation. Physiol Genomics. 2003;12:187–193. doi: 10.1152/physiolgenomics.00117.2002. [DOI] [PubMed] [Google Scholar]
- 20.Makielski JC, Ye B, Valdivia CR, Pagel MD, Pu J, Tester DJ, Ackerman MJ. A ubiquitous splice variant and a common polymorphism affect heterologous expression of recombinant human SCN5A heart sodium channels. Circ Res. 2003;93:821–828. doi: 10.1161/01.RES.0000096652.14509.96. [DOI] [PubMed] [Google Scholar]
- 21.Nagatomo T, Fan Z, Ye B, Tonkovich GS, January CT, Kyle JW, Makielski JC. Temperature dependence of early and late currents in human cardiac wild-type and long QT DeltaKPQ Na+ channels. Am J Physiol. 1998;275:H2016–H2024. doi: 10.1152/ajpheart.1998.275.6.H2016. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz PJ. Clinical applicability of molecular biology: the case of the long QT syndrome. Curr Control Trials Cardiovasc Med. 2000;1:88–91. doi: 10.1186/cvm-1-2-088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garson A, Jr, Dick M, 2nd, Fournier A, Gillette PC, Hamilton R, Kugler JD, van Hare GF, 3rd, Vetter V, Vick GW., 3rd The long QT syndrome in children: an international study of 287 patients. Circulation. 1993;87:1866–1872. doi: 10.1161/01.cir.87.6.1866. [DOI] [PubMed] [Google Scholar]
- 24.Trippel DL, Parsons MK, Gillette PC. Infants with long-QT syndrome and 2:1 atrioventricular block. Am Heart J. 1995;130:1130–1134. doi: 10.1016/0002-8703(95)90222-8. [DOI] [PubMed] [Google Scholar]
- 25.Gorgels AP, Al Fadley F, Zaman L, Kantoch MJ, Al Halees Z. The long QT syndrome with impaired atrioventricular conduction: a malignant variant in infants. J Cardiovasc Electrophysiol. 1998;9:1225–1232. doi: 10.1111/j.1540-8167.1998.tb00096.x. [DOI] [PubMed] [Google Scholar]
- 26.Van Hare GF, Franz MR, Roge C, Scheinman MM. Persistent functional atrioventricular block in two patients with prolonged QT intervals: elucidation of the mechanism of block. Pacing Clin Electrophysiol. 1990;13:608–618. doi: 10.1111/j.1540-8159.1990.tb02077.x. [DOI] [PubMed] [Google Scholar]
- 27.Lupoglazoff JM, Denjoy I, Villain E, Fressart V, Simon F, Bozio A, Berthet M, Benammar N, Hainque B, Guicheney P. Long QT syndrome in neonates: conduction disorders associated with HERG mutations and sinus bradycardia with KCNQ1 mutations. J Am Coll Cardiol. 2004;43:826–830. doi: 10.1016/j.jacc.2003.09.049. [DOI] [PubMed] [Google Scholar]
- 28.Lupoglazoff JM, Cheav T, Baroudi G, Berthet M, Denjoy I, Cauchemez B, Extramiana F, Chahine M, Guicheney P. Homozygous SCN5A mutation in long-QT syndrome with functional two-to-one atrioventricular block. Circ Res. 2001;89:E16–E21. doi: 10.1161/hh1401.095087. [DOI] [PubMed] [Google Scholar]
- 29.Hoorntje T, Alders M, van Tintelen P, van der Lip K, Sreeram N, van der Wal A, Mannens M, Wilde A. Homozygous premature truncation of the HERG protein: the human HERG knockout. Circulation. 1999;100:1264–1267. doi: 10.1161/01.cir.100.12.1264. [DOI] [PubMed] [Google Scholar]
- 30.Piippo K, Laitinen P, Swan H, Toivonen L, Viitasalo M, Pasternack M, Paavonen K, Chapman H, Wann KT, Hirvela E, Sajantila A, Kontula K. Homozygosity for a HERG potassium channel mutation causes a severe form of long QT syndrome: identification of an apparent founder mutation in the Finns. J Am Coll Cardiol. 2000;35:1919–1925. doi: 10.1016/s0735-1097(00)00636-7. [DOI] [PubMed] [Google Scholar]
- 31.Wehrens XH, Rossenbacker T, Jongbloed RJ, Gewillig M, Heidbuchel H, Doevendans PA, Vos MA, Wellens HJ, Kass RS. A novel mutation L619F in the cardiac Na+ channel SCN5A associated with long-QT syndrome (LQT3): a role for the I-II linker in inactivation gating. Hum Mutat. 2003;21:552. doi: 10.1002/humu.9136. [DOI] [PubMed] [Google Scholar]
- 32.Valdivia CR, Ackerman MJ, Tester DJ, Wada T, McCormack J, Ye B, Makielski JC. A novel SCN5A arrhythmia mutation, M1766L, with expression defect rescued by mexiletine. Cardiovasc Res. 2002;55:279–289. doi: 10.1016/s0008-6363(02)00445-5. [DOI] [PubMed] [Google Scholar]
- 33.Schulze-Bahr E, Fenge H, Etzrodt D, Haverkamp W, Monnig G, Wedekind H, Breithardt G, Kehl HG. Long QT syndrome and life threatening arrhythmia in a newborn: molecular diagnosis and treatment response. Heart. 2004;90:13–16. doi: 10.1136/heart.90.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Shen J, Splawski I, Atkinson D, Li Z, Robinson JL, Moss AJ, Towbin JA, Keating MT. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 35.Viswanathan PC, Benson DW, Balser JR. A common SCN5A polymorphism modulates the biophysical effects of an SCN5A mutation. J Clin Invest. 2003;111:341–346. doi: 10.1172/JCI16879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang CC, Acharfi S, Wu MH, Chiang FT, Wang JK, Sung TC, Chahine M. A novel SCN5A mutation manifests as a malignant form of long QT syndrome with perinatal onset of tachycardia/bradycardia. Cardiovasc Res. 2004;64:268–278. doi: 10.1016/j.cardiores.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 37.Johnson WH, Jr, Yang P, Yang T, Lau YR, Mostella BA, Wolff DJ, Roden DM, Benson DW. Clinical, genetic, and biophysical characterization of a homozygous HERG mutation causing severe neonatal long QT syndrome. Pediatr Res. 2003;53:744–748. doi: 10.1203/01.PDR.0000059750.17002.B6. [DOI] [PubMed] [Google Scholar]
- 38.Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, Tager-Flusberg H, Priori SG, Sanguinetti MC, Keating MT. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 39.Ackerman MJ. Genotype-phenotype relationships in congenital long QT syndrome. J Electrocardiol. 2005;38:64–68. doi: 10.1016/j.jelectrocard.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 40.Dhar Malhotra J, Chen C, Rivolta I, Abriel H, Malhotra R, Mattei LN, Brosius FC, Kass RS, Isom LL. Characterization of sodium channel alpha- and beta-subunits in rat and mouse cardiac myocytes. Circulation. 2001;103:1303–1310. doi: 10.1161/01.cir.103.9.1303. [DOI] [PubMed] [Google Scholar]
- 41.Goldin AL, Barchi RL, Caldwell JH, Hofmann F, Howe JR, Hunter JC, Kallen RG, Mandel G, Meisler MH, Netter YB, Noda M, Tamkun MM, Waxman SG, Wood JN, Catterall WA. Nomenclature of voltage-gated sodium channels. Neuron. 2000;28:365–368. doi: 10.1016/s0896-6273(00)00116-1. [DOI] [PubMed] [Google Scholar]
- 42.Wallace RH, Wang DW, Singh R, Scheffer IE, George AL, Jr, Phillips HA, Saar K, Reis A, Johnson EW, Sutherland GR, Berkovic SF, Mulley JC. Febrile seizures and generalized epilepsy associated with a mutation in the Na+-channel beta1 subunit gene SCN1B. Nat Genet. 1998;19:366–370. doi: 10.1038/1252. [DOI] [PubMed] [Google Scholar]
- 43.Isom LL, Catterall WA. Na+ channel subunits and Ig domains. Nature. 1996;383:307–308. doi: 10.1038/383307b0. [DOI] [PubMed] [Google Scholar]
- 44.Chen C, Westenbroek RE, Xu X, Edwards CA, Sorenson DR, Chen Y, McEwen DP, O’Malley HA, Bharucha V, Meadows LS, Knudsen GA, Vilaythong A, Noebels JL, Saunders TL, Scheuer T, Shrager P, Catterall WA, Isom LL. Mice lacking sodium channel beta1 subunits display defects in neuronal excitability, sodium channel expression, and nodal architecture. J Neurosci. 2004;24:4030–4042. doi: 10.1523/JNEUROSCI.4139-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen C, Bharucha V, Chen Y, Westenbroek RE, Brown A, Malhotra JD, Jones D, Avery C, Gillespie PJ, 3rd, Kazen-Gillespie KA, Kazarinova-Noyes K, Shrager P, Saunders TL, Macdonald RL, Ransom BR, Scheuer T, Catterall WA, Isom LL. Reduced sodium channel density, altered voltage dependence of inactivation, and increased susceptibility to seizures in mice lacking sodium channel beta 2-subunits. Proc Natl Acad Sci U S A. 2002;99:17072–17077. doi: 10.1073/pnas.212638099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grieco TM, Malhotra JD, Chen C, Isom LL, Raman IM. Open-channel block by the cytoplasmic tail of sodium channel beta4 as a mechanism for resurgent sodium current. Neuron. 2005;45:233–244. doi: 10.1016/j.neuron.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 47.Wang GK, Edrich T, Wang SY. Time-dependent block and resurgent tail currents induced by mouse beta4 peptide in cardiac Na+ channels. J Gen Physiol. 2006;127:277–289. doi: 10.1085/jgp.200509399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vatta M, Ackerman MJ, Ye B, Makielski JC, Ughanze EE, Taylor EW, Tester DJ, Balijepalli RC, Foell JD, Li Z, Kamp TJ, Towbin JA. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation. 2006;114:2104–2112. doi: 10.1161/CIRCULATIONAHA.106.635268. [DOI] [PubMed] [Google Scholar]