About one-half of the ≈57 proteins of the photosynthetic membranes in plant chloroplasts is encoded in the chloroplast genome, translated on chloroplast ribosomes and cotranslationally inserted into the lipid bilayer of the thylakoids (flattened sacs; ref. 1). The other half is translated on cytosolic ribosomes from messenger RNA of nuclear genes and posttranslationally imported across the outer and inner chloroplast envelope membranes. The import machinery consists of smart receptor proteins, translocation channels, and chaperon proteins (2, 3). The photosynthetic proteins are assembled into five multisubunit complexes. There, the pigments for light harvesting and the electron transport molecules for the primary light-dependent charge separation and for the generation of protons and oxygen in the water-splitting reaction are positioned at the optimal distances. The two photosystems cooperate via the cytochrome b6f complex in the transfer of electrons to provide reducing power in the form of NADPH and establishment of proton gradients from the stroma side to the lumen of the thylakoid. The electrochemical gradient formed by the accumulation of protons in the thylakoid lumen provides the driving force for the phosphorylation of ADP by the ATP synthase complex. In contrast to the profound knowledge of the organization of the photosynthetic membrane and the import of the protein components into the chloroplast and their targeting to the thylakoids, progress in learning how the lipid bilayer membranes are formed is less obvious. This situation may change with the discovery of a function of the vesicle-inducing protein in plastids (VIPP) by Kroll et al. and Westphal et al. as reported in this issue of PNAS (4, 5).
In pea chloroplasts, the 37-kDa VIPP protein is located both in the vicinity of the chloroplast envelope and in the thylakoid membranes (6) and was considered by Li, Kaneko, and Keegstra (6) as a candidate for the transfer of galactolipids from their site of synthesis at the chloroplast envelope to the thylakoids. Daniela Kroll, Karin Meierhoff, Nicole Bechtold, Mikio Kinoshita, Sabine Westphal, Ute Vothknecht, Jürgen Soll, and Peter Westhoff studied a recessive Arabidosis T-DNA [portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells] insertion mutant with severe disturbances in the photosynthetic electron transport chain and the formation of the thylakoids. The insertion was identified in the gene encoding VIPP, and the mutant could be rescued by transformation with the VIPP cDNA. The cause for the disturbed development or maintenance of the thylakoids was the failure of the mutant to bud from the inner chloroplast envelope membrane vesicles, which transfer lipids from the inner envelope to the thylakoid membranes (7–10). In the transformants, the process of vesicle budding was reestablished, and the thylakoid organization normalized. The companion paper by Sabine Westphal, Lisa Heins, Jürgen Soll, and Ute Vothknecht identifies VIPP 1 genes in the genomes of Synechocystis, Anabaena, Synechococcus, and Nostoc. In these cyanobacteria, the protein is located in the plasma membrane, but its disruption in Synechocystis by insertion mutagenesis with a kanamycin cassette prevents ordered thylakoid formation and light-dependent oxygen evolution. The protein has high amino acid sequence identity with the PspA protein of Escherichia coli, but it has evolved by addition of a novel C-terminal domain of some 50 aa. In E. coli, this protein is part of an operon induced by infection with the filamentous phage f1 and by treatments that inhibit the protein translocation pathway via the Sec pathway, which is also used in chloroplasts and cyanobacteria for translocation of proteins across the thylakoid membranes. This partial identity indicates a gene duplication and recruitment for the novel function during evolution.
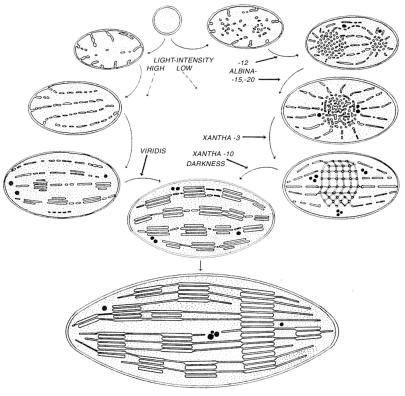
Is the vesicle transport responsible for the establishment of the lipid bilayers? In higher plants, chloroplasts develop from proplastids in the light or via the etioplast pathway after an initial dark period (Fig. 1). The primary thylakoid layers are formed by alignment of vesicles budded from the inner membrane of the plastid envelope (11, 12). Algae synthesize chlorophyll and the thylakoid system in the dark, but mutants of Chlamydomonas blocked in chlorophyll synthesis can be made to build the photosynthetic membranes starting from scratch, and it is by vesicle budding from the inner envelope membrane (13). Accumulation of envelope-budded vesicles in the plastid stroma is the hallmark of barley mutants defective in thylakoid development (14).
Figure 1.
The formation of chloroplast thylakoids. [Reproduced with permission from ref. 12 (Copyright 1959, Academic, Orlando, FL).]
The inner chloroplast envelope membrane and thylakoids are uniquely rich in monogalactosyldiacylglyceride (MGDG), a lipid prone to forming hexagonal and cubic phases and in digalactosyldiacylglyceride (DGDG), a lipid favoring planar bilayers (15). The prolamellar body formed in proplastids in flushing leaves at night in the spring (Fig. 1, right) has a very high MDGD content and contains essentially only the protochlorophyllide:NADPH-oxidoreductase complex (cf. ref. 15). In the morning, the protochlorophyllide is photoreduced to chlorophyllide a, and the membrane tubes lose their ordered arrangement and flow out into fenestrated primary lamellar layers. The perforations in the primary thylakoids are filled out, and the incipient grana are formed by attachment of flattened discs. Budding of vesicles from the inner envelope occurs throughout this process (14, 16). The possible involvement of VIPP in these processes will be of great interest. Lethal mutants in the barley locus xan-m overproduce MGDG but form only wild-type levels of protochlorophyllide oxidoreductase and reduced amounts of DGDG (14, 17). On illumination, the tubes in the large prolamellar bodies are incompletely converted into the fenestrated layer, and highly irregular membrane associations and membrane packages are formed because of the unbalanced lipid protein composition.
A crucial role in targeting precursor proteins into the chloroplast stroma and into the thylakoids is played by N-terminal transit peptides (2, 3). The cleavable transit peptide can be bipartite, its N-terminal part being cleaved off on reaching the stroma and its C-terminal part after reaching the lumen of the thylakoid. Three different mechanisms have been found for incorporation of photosynthetic proteins from the stroma into the thylakoids (18). Pinnaduwage and Bruce (19) elucidated another important role of transit peptides in thylakoid formation: Peptides containing the 50 to 60 C-terminal amino acids of the transit peptide of the small subunit of ribulose-bisphosphate carboxylase will disrupt unilamellar vesicles, but only if the vesicles contain 20 mol% MDGD, i.e., the concentration found in the chloroplast outer envelope. Addition to the vesicles of the entire transit peptide transformed them into an aggregate structure of elongated, appressed sacs similar to incipient thylakoid grana. A sensing role is played by the transit peptide for the import of the protochlorophyllide oxidoreductase precursor targeted into the prolamellar body (20). The precursor is permitted to proceed through the envelope only if protochlorophyllide synthesized in the stroma of the plastid can bind to the transit peptide and thus is also available to bind to the active site of the mature domain of the enzyme. In contrast to this protein, the precursor of the constitutive isoform of the enzyme does not require binding of protochlorophyllide to its transit peptide to gain entry permission.
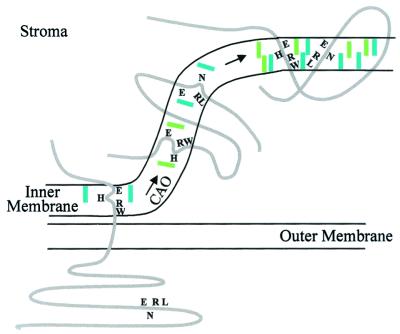
The decision whether the precursors of light-harvesting proteins of photosystem II in Chlamydomonas are permitted to continue on the import pathway through the envelope or are returned and transferred into vacuoles for degradation depends on the binding of chlorophyll a and the stabilization of this binding by conversion of some of the pigment molecules to chlorophyll b in the inner envelope membrane, as illustrated in Fig. 2 (13, 21, 22). Structure analysis (23) and site-directed mutagenesis (24) have identified the sequence motifs Glu-X-X-His/Asn-X-Arg as binding sites of chlorophyll in the first and third membrane-spanning domains of light-harvesting chlorophyll a/b and a/c proteins. Synthetic peptides of this sequence generate a loop structure by intrapeptide, electrostatic interaction between Glu and Arg. His or Asn and charge-compensated Glu-Arg pairs are ligands of the Mg atom in chlorophyll. As expected, these retention motifs bind two molecules of chlorophyll (21). Because chlorophyll a binds more readily than chlorophyll b to the motifs, it is suggested that chlorophyll a is bound first and then oxidized to chlorophyll b by chlorophyllide a oxidase (CAO; ref. 25), a stabilization of binding through increase of Lewis acid strength of the Mg atom (22). Interestingly, transcription and polyribosome formation for chloroplast-encoded reaction center proteins take place in the absence of chlorophyll, but translation is halted at the transmembrane helices that bind the reaction center chlorophyll a and electron carriers (26). Binding seems prerequisite for cotranslational incorporation into the thylakoid. Conceivably, VIPP might facilitate insertion of pigment and electron carriers at the envelope, as well as at the thylakoid. The discovery of the VIPP protein points the way to find additional proteins involved in vesicle and thylakoid membrane formation.
Figure 2.
Assembly of a light-harvesting chlorophyll a/b protein at the chloroplast envelope. Chlorophyllide a, blue-green; chlorophyllide b, olive-green; CAO, chlorophyllide a oxidase. [Reproduced with permission from ref. 22 (Copyright 2001, Elsevier Science, Oxford).]
Footnotes
References
- 1.Wollman F-A, Minai L, Nechushtai R. Biochem Biophys Acta. 1999;1411:21–85. doi: 10.1016/s0005-2728(99)00043-2. [DOI] [PubMed] [Google Scholar]
- 2.Keegstra K, Cline K. Plant Cell. 1999;11:557–570. doi: 10.1105/tpc.11.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soll J, Tien R. Plant Mol Biol. 1998;38:191–207. [PubMed] [Google Scholar]
- 4.Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht U C, Soll J, Westhoff P. Proc Natl Acad Sci USA. 2001;98:4238–4242. doi: 10.1073/pnas.061500998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Westphal S, Heins L, Soll J, Vothknecht U C. Proc Natl Acad Sci USA. 2001;98:4243–4248. doi: 10.1073/pnas.061501198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Kaneko Y, Keegstra K. Plant Mol Biol. 1994;25:619–632. doi: 10.1007/BF00029601. [DOI] [PubMed] [Google Scholar]
- 7.Douce R, Joyard J. Adv Bot Res. 1979;7:1–116. [Google Scholar]
- 8.Carde J-P, Joyard J, Douce R. Biol Cell. 1982;44:315–324. [Google Scholar]
- 9.Morré D J, Penel C, Morré D M, Sandelius A S, Morrau P, Andersson B. Protoplasma. 1991;160:49–64. [Google Scholar]
- 10.Morré D J, Selldén G, Sundqvist C, Sandelius A S. Plant Physiol. 1991;97:1558–1564. doi: 10.1104/pp.97.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mühlethaler K, Frey-Wyssling A. J Bio-phys Biochem Cytol. 1959;6:507–512. [PMC free article] [PubMed] [Google Scholar]
- 12.von Wettstein D. J Ultrastruc Res. 1959;3:234–240. [Google Scholar]
- 13.Hoober J K, Eggink L L. Photosynth Res. 1999;61:197–215. [Google Scholar]
- 14.Henningsen K W, Boynton J E, von Wettstein D. R Dan Acad Sci Lett Biol Skrifter. 1993;42:1–349. [Google Scholar]
- 15.Bruce B D. Plant Mol Biol. 1998;38:223–246. [PubMed] [Google Scholar]
- 16.von Wettstein D, Gough S, Kannangara C G. Plant Cell. 1995;7:1039–1057. doi: 10.1105/tpc.7.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Appelqist L-Å, Boynton J E, Henningsen K W, Stumpf P K, von Wettstein D. J Lipid Res. 1968;9:513–524. [PubMed] [Google Scholar]
- 18.Robinson C, Hynds P J, Robinson D, Mant A. Plant Mol Biol. 1998;38:209–221. [PubMed] [Google Scholar]
- 19.Pinnaduwage P, Bruce B D. J Biol Chem. 1996;271:32907–32915. doi: 10.1074/jbc.271.51.32907. [DOI] [PubMed] [Google Scholar]
- 20.Reinbothe C, Lebedev N, Apel K, Reinbothe S. Proc Natl Acad Sci USA. 1997;94:8890–8894. doi: 10.1073/pnas.94.16.8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eggink J K, Hoober J K. J Biol Chem. 2000;275:9087–9090. doi: 10.1074/jbc.275.13.9087. [DOI] [PubMed] [Google Scholar]
- 22.Hoober, J. K. & Eggink, L. L. (2001) FEBS Lett., in press. [DOI] [PubMed]
- 23.Kühlbrandt W, Wang D N, Fujiyoshi Y. Nature (London) 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- 24.Bassi R, Croce R, Cugini D, Sandona D. Proc Natl Acad Sci USA. 1999;96:10056–10061. doi: 10.1073/pnas.96.18.10056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oster U, Tanaka R, Tanaka A, Rüdiger W. Plant J. 2000;21:305–310. doi: 10.1046/j.1365-313x.2000.00672.x. [DOI] [PubMed] [Google Scholar]
- 26.Kim J, Gamble-Klein P, Mullet J E. J Biol Chem. 1991;266:14931–14938. [PubMed] [Google Scholar]