Dominant missense mutations in the genes encoding fibroblast growth factor receptors (FGFRs) 1–3 are the etiology of many craniosynostosis (premature fusion of the cranial sutures) and chondrodysplasia (dwarfism) syndromes (1–3). Mutations in Fgfr2 cause craniosynostosis syndromes including Crouzon syndrome, Pfeiffer syndrome, and Apert syndrome (AS). The article in this issue of PNAS by Hajihosseini et al. (4) presents the first animal model for a mutation in Fgfr2 that is associated with AS, one of the most severe of the human craniosynostosis syndromes.
FGFRs are transmembrane receptor tyrosine kinase proteins that are activated by many of the 22 members of the FGF family (5, 6). The extracellular region of the FGFR contains two or three Ig-like domains and mediates ligand binding (7). In the presence of the cofactor, heparin, or heparan sulfate, ligand binding induces receptor dimerization and subsequent activation (5, 8–10). Importantly, the affinity and specificity of FGFRs are regulated by tissue-specific alternative splicing. The paper by Hajihosseini et al. (4), and two additional studies (11, 12), show that mutations that cause AS circumvent the biochemical and developmental regulatory mechanisms that are normally imposed by tissue-specific alternative splicing of Fgfr2 and result in ectopic ligand-dependent receptor activation.
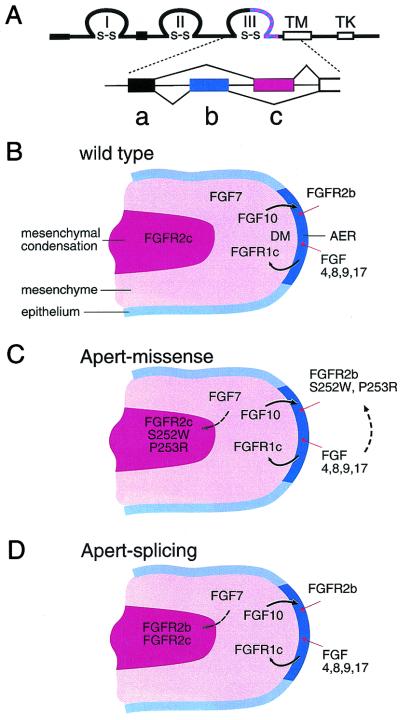
Alternative splicing in the region encoding the carboxyl-terminal half of Ig domain III creates receptor isoforms with distinct ligand binding specificity by incorporating either a b or a c exon (Fig. 1A). For FGFR2, this splicing event is very tissue-specific with b exon usage in epithelial tissue and c exon usage in mesenchymal tissue (13). Directional epithelial-mesenchymal signaling is maintained because mesenchymally expressed ligands, such as FGF7 and FGF10, can activate only epithelially spliced FGFR2b. Similarly, ligands such as FGFs 2, 4, 6, 8, 9 and 17, which tend to be expressed in epithelial tissues activate mesenchymally spliced FGFR2c (14–16). The reciprocal nature of this signaling mechanism is most eloquently illustrated in the developing limb where FGF10 is a required mesenchymal signal that induces formation of the apical ectodermal ridge and FGF8 (and possibly FGFs 4, 9, and 17) is an epithelial factor that signals to distal mesenchyme (Fig. 1B) (17–21).
Figure 1.
FGF signaling pathways in limb development. (A) Structure of the FGFR showing three disulfide linked (s-s) Ig loops (I, II, III), a transmembrane domain (TM) and an intracellular tyrosine kinase domain (TK). The striped region in the carboxyl- terminal half of Ig loop III is subject to alternative utilization of either exon b or exon c. These correspond to exons 8 and 9 of the Fgfr2 gene. (B) Schematic diagram of a developing limb showing the apical ectodermal ridge (AER), distal mesenchyme (DM), mesenchyme, and mesenchymal condensation. Sites of Fgf and Fgfr expression are shown. Limb bud initiation and outgrowth is regulated by a reciprocal signaling loop in which FGF10 signals to FGFR2b and FGFs 4, 8, 9, and 17 signal to FGFR1c (solid arrows). FGFR2c is prominently expressed in the mesenchymal condensation; however, its function and endogenous ligand in this location is not known. (C) Missense mutations causing AS could allow autocrine/juxtacrine activation of FGFR2 in the mesenchymal condensation and in the AER (dashed arrows). (D) Alu element insertions and the hemizygous deletion of exon c allow FGFR2b expression in mesenchymal tissue, effectively mimicking the missense mutations by allowing ectopic receptor activation by ligands such as FGF7 (dashed arrow).
The importance of FGFR2 signaling in organogenesis is illustrated by gene targeting studies. Mouse embryos lacking Fgfr2 die at stages before the limbs or lungs develop (22, 23). Embryos in which only the b isoform of Fgfr2 has been deleted survive until birth but also fail to develop limb buds, lung, and other organs (24). These phenotypes are remarkably similar to those seen in mice lacking Fgf10 (25–27) and demonstrate the importance of the mesenchymal to epithelial FGF signaling pathway. The effects of loss of FGFR2 signaling in mesenchymal tissues are obscured by the early embryonic lethality and the agenesis of the limbs and other structures in the null mutant.
Biochemical studies show that many of these mutations result in ligand independent FGFR activation, often involving the formation of an intermolecular disulfide bond. AS, although allelic with other craniosynostosis syndromes, is much more severe and is characterized by premature fusion of the coronal sutures, severe syndactyly in the hands and feet, brain malformations, and mental retardation. The severe syndactyly and neurological disorders are not associated with other craniosynostosis syndromes. These phenotypic differences suggest that the signals transduced by receptors harboring AS mutations differ from those in other craniosynostosis syndrome mutations. Differences could lie in the intensity of the signal or in the spatial and temporal patterns of receptor activation.
The vast majority of AS patients harbor one of two missense mutations (S252W or P253R) in the highly conserved region linking Ig domain II and III of FGFR2 (11, 28). Biochemical analysis revealed that these genetic alterations render the mesenchymally expressed mutant FGFR2c abnormally susceptible to activation by mesenchymally expressed ligands such as FGF7 and FGF10 and the epithelially expressed mutant FGFR2b abnormally susceptible to activation by epithelially expressed ligands such as FGF2, 6, and 9, thus circumventing the normal epithelial-mesenchymal signaling restrictions (Fig. 1C) (12). The mechanism by which these mutations affect receptor signaling is fundamentally different from that of other mutations that activate FGFR2, which generally cause ligand independent receptor dimerization, stabilized by intermolecular disulfide bonds (1, 3).
Recently Oldridge et al. (11) have identified two cases of AS (of 260) that do not have missense mutations in Fgfr2. Interestingly, these patients were found to have de novo Alu-insertions upstream or within the c exon (exon 9) of Fgfr2. Molecular analysis showed that these Alu insertions affect alternative splicing of Fgfr2, resulting in the ectopic expression of FGFR2b in tissues that normally would express FGFR2c (Fig. 1D). Thus mesenchymal tissue from these patients coexpresses both FGFR2c and FGFR2b. Interestingly, those authors also have identified patients with Pfeiffer syndrome with a mutation in FGFR2 that affects the 3′ splice donor site of the c exon. The phenotype of these patients is more severe than that of Pfeiffer syndrome patients with different mutations in FGFR2 and suggests that alternative splicing may be affected and could contribute to the more severe phenotype.
Hajihosseini et al. (4) have modeled these types of mutations in the mouse by specifically deleting the c exon of FGFR2 (Fgfr2Δc). The consequence of this deletion is the utilization of the alternative exon b in tissues where c normally would be used exclusively (Fig. 1D). Similar to the Alu insertions in humans, this is a dominant mutation in the mouse, and heterozygous mice (Fgfr2+/Δc) develop skeletal and visceral defects resembling those seen in patients with AS and more severe cases of Pfeiffer syndrome. Taken together, the biochemical studies and the mouse and human mutations suggest that the primary pathology in AS arises from ligand-dependent activation of FGFR2, predominantly in mesenchymal tissue.
Cartilage is formed through differentiation of condensed mesenchyme during early stages of embryonic development. Fgfr2c is highly expressed in precartilage cell condensations, and Fgf7 is highly expressed in the surrounding loose mesenchyme (29–32). Although this ligand-receptor pair is normally not functional, AS mutations or aberrations in the normal splicing pattern of FGFR2 could permit ectopic signals between different mesenchymal cell populations resulting in aberrant mesenchymal growth and differentiation. This explains why the rare Alu insertion mutations in humans and the mouse FGFR2Δc mutation develop similar phenotypes to patients with AS missense mutations.
The AS limb defects include osseous fusion of the digits and phalangeal joints and ectopic cartilage in periarticular tissues and flexor tendons (33). The skeletal defects observed by Hajihosseini et al. (4) in the Fgfr2+/Δc mice include premature fusion of the coronal sutures and precocious sternal fusion. Although limb abnormalities are not found in the mutant mice, abnormal thickening of the sternebra and ectopic ossification of intersternebral cartilage is observed. These phenotypic differences could be species specific or could result from differences in receptor signaling in epithelial tissue. For example, in the apical ectodermal ridge, missense mutations are predicted to cause increased autocrine signaling. In contrast, alternative splicing mutations would not be expected to affect signaling in epithelial tissues that already express FGFR2b.
Crouzon syndrome, unlike AS, is not associated with limb abnormalities. Most Crouzon syndrome mutations involve the gain or loss of a cysteine residue within Ig domain III of FGFR2. The consequence of this class of mutation on the function of FGFR activity is to create an unpaired-cysteine residue, which facilitates the formation of intermolecular disulphide bonds, causing ligand-independent dimerization, phosphorylation, and signaling (34). The mechanism of this mutation suggests that uniformly elevated FGFR signaling in one cell type is not sufficient to induce ectopic differentiation or that the intensity of receptor activation for this type of mutation is not sufficient to affect skeletal limb development. This mechanism supports the concept that signaling between adjacent cell layers within mesenchyme is an essential component of the pathogenesis of AS.
Hajihosseini et al. (4) also observed visceral defects in mutant mice in tissues such as lungs, kidneys, and lacrimal glands. As in the limb, these tissues also require extensive epithelial-mesenchymal interactions for normal development. In lung development, chemotactic signals (FGF10) originating from mesenchyme regulate epithelial branching. Interestingly, in mice lacking Fgf9, mesenchymal tissue is deficient and abnormal lung branching ensues (J. Colvin and D.M.O., unpublished work). Interestingly, in the Fgfr2+/Δc mice increased lung mesenchyme and defects in lung development are observed, suggesting that precise signaling within mesenchymal tissue is essential for normal development.
Most gene targeting studies result in loss of function. The neomorphic properties of the Fgfr2 allele generated by Hajihosseini et al. (4) provides a unique opportunity to explore the molecular pathogenesis of AS. This engineered mutation also has provided unique insight into the role of FGF-FGFR signaling between epithelial and mesenchymal tissues.
Acknowledgments
We thank G. Martin and I. Boime for reviewing this manuscript. This work was funded by National Institutes of Health Grant HD30652.
Footnotes
See companion article on page 3855.
References
- 1.Naski M C, Ornitz D M. Front Biosci. 1998;3:D781–D794. doi: 10.2741/a321. [DOI] [PubMed] [Google Scholar]
- 2.Muenke M, Schell U. Trends Genet. 1995;11:308–313. doi: 10.1016/s0168-9525(00)89088-5. [DOI] [PubMed] [Google Scholar]
- 3.Wilkie A O M, Morriss-Kay G M, Jones E Y, Heath J K. Curr Biol. 1995;5:500–507. doi: 10.1016/s0960-9822(95)00102-3. [DOI] [PubMed] [Google Scholar]
- 4.Hajihosseini M K, Wilson S I, DeMoerlooze L, Dickson C. Proc Natl Acad Sci USA. 2001;98:3855–3860. doi: 10.1073/pnas.071586898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Szebenyi G, Fallon J F. Int Rev Cyt. 1999;185:45–106. doi: 10.1016/s0074-7696(08)60149-7. [DOI] [PubMed] [Google Scholar]
- 6.Ornitz D M, Itoh N. Genome Biol. 2001;2:3005.1–3005.12. doi: 10.1186/gb-2001-2-3-reviews3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson D E, Williams L T. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- 8.Ornitz D M, Yayon A, Flanagan J G, Svahn C M, Levi E, Leder P. Mol Cell Biol. 1992;12:240–247. doi: 10.1128/mcb.12.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKeehan W L, Wang F, Kan M. Prog Nucleic Acid Res Mol Biol. 1998;59:135–176. doi: 10.1016/s0079-6603(08)61031-4. [DOI] [PubMed] [Google Scholar]
- 10.Ornitz D M. BioEssays. 2000;22:108–112. doi: 10.1002/(SICI)1521-1878(200002)22:2<108::AID-BIES2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 11.Oldridge M, Zackai E H, McDonald-McGinn D M, Iseki S, Morriss-Kay G M, Twigg S R, Johnson D, Wall S A, Jiang W, Theda C, et al. Am J Hum Genet. 1999;64:446–461. doi: 10.1086/302245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu K, Herr A B, Waksman G, Ornitz D M. Proc Natl Acad Sci USA. 2000;97:14536–14541. doi: 10.1073/pnas.97.26.14536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miki T, Bottaro D P, Fleming T P, Smith C L, Burgess W H, Chan A M, Aaronson S A. Proc Natl Acad Sci USA. 1992;89:246–250. doi: 10.1073/pnas.89.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ornitz D M, Xu J, Colvin J S, McEwen D G, MacArthur C A, Coulier F, Gao G, Goldfarb M. J Biol Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 15.Xu J, Lawshe A, MacArthur C A, Ornitz D M. Mech Dev. 1999;83:165–178. doi: 10.1016/s0925-4773(99)00034-9. [DOI] [PubMed] [Google Scholar]
- 16.Igarashi M, Finch P W, Aaronson S A. J Biol Chem. 1998;273:13230–13235. doi: 10.1074/jbc.273.21.13230. [DOI] [PubMed] [Google Scholar]
- 17.Martin G R. Genes Dev. 1998;12:1571–1586. doi: 10.1101/gad.12.11.1571. [DOI] [PubMed] [Google Scholar]
- 18.Lewandoski M, Sun X, Martin G R. Nat Genet. 2000;26:460–463. doi: 10.1038/82609. [DOI] [PubMed] [Google Scholar]
- 19.Sun X, Lewandoski M, Meyers E N, Liu Y H, Maxson R E, Jr, Martin G R. Nat Genet. 2000;25:83–86. doi: 10.1038/75644. [DOI] [PubMed] [Google Scholar]
- 20.Moon A M, Capecchi M R. Nat Genet. 2000;26:455–459. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moon A M, Boulet A M, Capecchi M R. Development (Cambridge, UK) 2000;127:989–996. doi: 10.1242/dev.127.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arman E, Haffnerkrausz R, Chen Y, Heath J K, Lonai P. Proc Natl Acad Sci USA. 1998;95:5082–5087. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu X, Weinstein M, Li C, Naski M, Cohen R I, Ornitz D M, Leder P, Deng C. Development (Cambridge, UK) 1998;125:753–765. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- 24.De Moerlooze L, Spencer-Dene B, Revest J, Hajihosseini M, Rosewell I, Dickson C. Development (Cambridge, UK) 2000;127:483–492. doi: 10.1242/dev.127.3.483. [DOI] [PubMed] [Google Scholar]
- 25.Ohuchi H, Hori Y, Yamasaki M, Harada H, Sekine K, Kato S, Itoh N. Biochem Biophys Res Commun. 2000;277:643–649. doi: 10.1006/bbrc.2000.3721. [DOI] [PubMed] [Google Scholar]
- 26.Min H, Danilenko D M, Scully S A, Bolon B, Ring B D, Tarpley J E, DeRose M, Simonet W S. Genes Dev. 1998;12:3156–3161. doi: 10.1101/gad.12.20.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. Nat Genet. 1999;21:138–141. doi: 10.1038/5096. [DOI] [PubMed] [Google Scholar]
- 28.Wilkie A O M, Slaney S F, Oldridge M, Poole M D, Ashworth G J, Hockley A D, Hayward R D, David D J, Pulleyn L J, Rutland P, et al. Nat Genet. 1995;9:165–172. doi: 10.1038/ng0295-165. [DOI] [PubMed] [Google Scholar]
- 29.Mason I J, Fuller-Pace F, Smith R, Dickson C. Mech Dev. 1994;45:15–30. doi: 10.1016/0925-4773(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 30.Peters K G, Werner S, Chen G, Williams L T. Development (Cambridge, UK) 1992;114:233–243. doi: 10.1242/dev.114.1.233. [DOI] [PubMed] [Google Scholar]
- 31.Orr-Urtreger A, Givol D, Yayon A, Yarden Y, Lonai P. Development (Cambridge, UK) 1991;113:1419–1434. doi: 10.1242/dev.113.4.1419. [DOI] [PubMed] [Google Scholar]
- 32.Delezoide A L, Benoistlasselin C, Legeaimallet L, Lemerrer M, Munnich A, Vekemans M, Bonaventure J. Mech Dev. 1998;77:19–30. doi: 10.1016/s0925-4773(98)00133-6. [DOI] [PubMed] [Google Scholar]
- 33.Cohen M M J. In: Craniosynostosis, Diagnosis, Evaluation, and Management. Cohen M M J, MacLean R E, editors. New York: Oxford Univ. Press; 2000. pp. 316–353. [Google Scholar]
- 34.Galvin B D, Hart K C, Meyer A N, Webster M K, Donoghue D J. Proc Natl Acad Sci USA. 1996;93:7894–7899. doi: 10.1073/pnas.93.15.7894. [DOI] [PMC free article] [PubMed] [Google Scholar]